Abstract
Successful vaccination against smallpox with conventional vaccinia virus is usually determined by the development of a vesicular skin lesion at the site of vaccinia inoculation, called a “take.” Although previous vaccination is known to be associated with attenuation of the take, the immunology that underlies a no-take in vaccinia-naïve individuals is not well understood. We hypothesized that antibody profiling of individuals before and after receiving vaccinia virus would reveal differences between takes and no-takes that may help better explain the phenomenon. Using vaccinia virus proteome microarrays and recombinant protein enzyme-linked immunosorbent assays (ELISAs), we first examined the antibody response in vaccinia-naïve individuals that failed to take after receiving different doses of the replication-competent DryVax and Aventis Pasteur (APSV) smallpox vaccines. Most that received diluted vaccine failed to respond, although four no-takes receiving diluted vaccine and four receiving undiluted vaccine mounted an antibody response. Interestingly, their antibody profiles were not significantly different from those of controls that did show a take. However, we did find elevated antibody titers in no-takes prior to receiving DryVax that were significantly different from those of takes. Although the sample size studied was small, we conclude the failure to take in responders correlates with preexisting immunity of unknown etiology that may attenuate the skin reaction in a way similar to previous smallpox vaccination.
INTRODUCTION
After the last naturally occurring smallpox case in the world in Somalia in 1977, the disease was declared eradicated worldwide in 1980. Vaccinia virus (VACV), the smallpox vaccine, is an orthopoxvirus that is antigenically related to the smallpox variola virus. Vaccinia virus is conventionally inoculated into the skin of immunocompetent individuals by scarification, where it replicates, causing localized host cell lysis and the formation of a focal lesion (pock). The formation of the lesion is a useful indicator that the vaccination has been successful (called a “take”), particularly in vaccinia-naïve individuals (14, 34). However, in individuals with vaccinia immunity, revaccination is associated with a significantly lower take rate, smaller lesions, reduced incidence of fever, and reduced viral shedding (13, 14). These are all manifestations of immunological memory remaining from a previous exposure to vaccinia. Consistent with this, revaccination causes a more rapid antibody response with elevated titers compared to those of a primary infection (13), characteristic of a typical anamnestic response. The presence of a vaccination scar, the tell-tale sign of previous vaccination, is also associated with an increased chance of a no-take (39). A no-take is also more likely with increased vaccine dilution (12). However, the underlying reason is the subimmunogenic dose delivered and is distinct from a no-take caused by preexisting immunity.
DryVax (DVX; Wyeth) is a lyophilized vaccinia virus vaccine strain derived from the prototype strain deposited at the New York City Board of Health (NYCBOH) and was widely used in the Americas during the smallpox eradication campaign. In the wake of terrorist attacks on the United States in 2001, the NIAID/NIH sponsored several multicenter trials to evaluate the possibility of diluting existing stocks of smallpox vaccines to increase the national stockpile and to test the efficacy and safety of attenuated alternatives to DryVax, such as modified vaccinia virus Ankara (MVA). The Aventis Pasteur smallpox vaccine (APSV), also derived from the NYCBOH strain, is a frozen vaccine. Several million doses of APSV (or “WetVax”) that had been in storage since the eradication campaign were discovered in 2001.
Since antibodies are known to play an important role in protection engendered by vaccinia vaccination (1, 28), we have examined the vaccinia virus-specific antibody profiles in vaccinia-naïve individuals from three different dose-down clinical trials in which a take did not occur following DryVax vaccination. The aims were to determine whether there was any particular response or lack of response that correlates with a no-take. Most individuals appeared to have received a potentially subimmunogenic dose of vaccine, although eight mounted an antibody response. Interestingly, the profile in the no-take responders was indistinguishable from that in other vaccinees that did take. However, we did find an elevated incidence of preexisting vaccinia virus-reactive antibody of unknown etiology among the no-takes compared to takes.
MATERIALS AND METHODS
Human serum samples.
Sera from several completed NIAID/NIH-sponsored clinical trials were shipped on dry ice to UC Irvine for serological studies. Samples were coded as to the vaccination regimen, and no patient identifiers were provided. Once thawed, one aliquot was mixed 1:1 with glycerol and stored at −20°C and used for probing, and the remaining aliquots were maintained at −80°C. Sera from 23 individuals that did not show a take after receiving DryVax or APSV (no-takes) were drawn from three independent human clinical trials, as detailed in Table 1. All three were dose-sparing studies to evaluate whether existing DryVax or APSV stocks could be diluted to expand the supply while retaining immunogenicity. Donors with no history of smallpox vaccination were inoculated with various doses (undiluted, 1/3.2, 1/5, 1/10, 1/32, and 1/100). As controls, sera from naïves (n = 50; NCT00026611) (11) and revaccinations (n = 25; NCT00050505) from two separate protocols that had a take following vaccination (takes) were also profiled. Sera were collected before vaccination (day 0 [d0]) and at the peak response on day 28 from each donor.
Table 1.
Details of vaccine type and dilutions received by the 23 no-takes analyzed in this study
| Donor code | Triala | Vaccine type | Dilutionb | Responderc |
|---|---|---|---|---|
| 1 | A | DVX | 0.1 | |
| 2 | A | DVX | 0.1 | |
| 3 | A | DVX | 0.1 | R+ |
| 4 | A | DVX | 0.1 | |
| 5 | A | DVX | 0.1 | R+ |
| 6 | A | DVX | 1 | R+ |
| 7 | A | DVX | 1 | R+ |
| 8 | A | DVX | 1 | R+ |
| 9 | B | DVX | 0.01 | |
| 10 | B | DVX | 0.01 | |
| 11 | B | DVX | 0.01 | |
| 12 | B | DVX | 0.01 | |
| 13 | B | DVX | 0.01 | |
| 14 | B | DVX | 0.03 | |
| 15 | B | DVX | 0.1 | |
| 16 | B | DVX | 0.1 | |
| 17 | B | APSV | 0.03 | |
| 18 | B | APSV | 0.03 | |
| 19 | B | APSV | 0.1 | |
| 20 | B | APSV | 0.3 | |
| 21 | B | APSV | 1 | R+ |
| 22 | C | APSV | 0.2 | R+ |
| 23 | C | APSV | 0.2 | R+ |
Trials (NIH study ID/clinicaltrials.gov ID): A, 01-632/NCT00026611 (11); B, 02-009/NCT00038987; C, 02-054/NCT00050518.
Dilutions of <0.1 were considered subantigenic.
Responder (R+) status determined as described in the footnote of Table 2.
Antibody profiling by protein microarrays.
Microarrays for this study were constructed essentially as described previously (6, 8, 9), with the exception that the VACV-WR plasmid collection used for protein expression in this study was generated from scratch. This new collection has been characterized and used for T cell studies (17, 18) and more recently for serological analysis (15). Briefly, vaccinia virus WR genomic DNA was used as a template for PCR to amplify individual open reading frames (ORFs), which were then cloned into a T7 expression vector by use of homologous recombination. All proteins were expressed from purified plasmids in an Escherichia coli-based coupled in vitro transcription/translation reaction (RTS 100 kits; Roche). Reactions representing 210 different vaccinia virus WR proteins were printed without further purification onto nitrocellulose-coated FAST slides (Whatman) by use of an Omni Grid 100 microarray printer (Gene Machines). Control reactions that lacked template DNA or empty expression vector were also set up. Protein expression was confirmed on the array using monoclonal antibodies to polyhistidine and hemagglutinin epitope tags engineered at the N and C termini of each protein (6). For microarray probing, human sera were used at 1/100 dilution in protein array blocking buffer (Whatman) containing E. coli lysate (Antigen Discovery Inc., Irvine, CA) to a final concentration of 10 mg/ml protein to block anti-E. coli antibodies. Sera were preincubated at room temperature (RT) for 30 min with constant mixing prior to applying to the arrays in triplicate and incubating overnight at 4°C with gentle agitation. After being washed, biotinylated anti-human IgG (Jackson) was used as secondary antibody followed by streptavidin-PBXL3 conjugate (Martek Biosciences), both at 1/200 in blocking buffer. After being washed five times, slides were air dried by brief centrifugation and then scanned and analyzed using a PerkinElmer ScanArray Express HT microarray scanner (PerkinElmer). Fluorescence intensities were quantified by using ProScanArray Express software (PerkinElmer).
Array data handling.
Data analysis was performed using the R (http://www.r-project.org) statistical software. In order to stabilize the variance observed in microarray signals (10, 16), the vsn normalization method implemented as part of the Bioconductor suite (www.bioconductor.org) is applied to the quantified array intensities. This procedure removes heteroskedasticity (2, 35, 36) and corrects for nonspecific noise effects such that control probe variance is minimized. This calibration has been shown to be effective on a number of platforms (21). In cases where d28 − d0 data were compared, the subtraction was performed with raw data prior to normalization. Intergroup comparisons were performed using a Bayes regularized t test adapted from Cyber-T for protein arrays (2, 3), which has been shown to be more effective than other differential expression techniques (25). To account for multiple comparison conditions, P values were corrected by the Benjamini and Hochberg (pBH) method to control the false discovery rate (5). After correction, P values of <0.05 were considered significant. An individual was defined as a responder if the ratio of retransformed signal intensities before and after DVX vaccination (d28/d0) was >2.0 for 2 or more of 6 signature antigens (WR101/H3L, WR113/D8L, WR148/-, WR150/A27L, WR070/I1L, and WR118/D13L [the designation to the left of the slash represents the WR name and that to the right represents the Copenhagen nomenclature; a hyphen indicates a WR gene for which there is no corresponding ortholog in the Copenhagen strain]). Seroprevalence rates on d0 and d28 were determined for each antigen based on a cutoff defined as the average + 2 standard deviations (SD) of the naïve take population (n = 50). Individuals positive for one or more of the six signature antigens were considered seropositive.
ELISAs.
Microwell enzyme-linked immunosorbent assay (ELISA) plates precoated with 6 different recombinant vaccinia proteins were constructed as described previously (15). The antigens were WR148/−, WR113/D8L, WR118/D13L, WR101/H3L, WR132/A13L, and WR070/I1L. Endpoint titers (WR113/D8L ELISA only) were determined by performing 12 2-fold serial dilutions of sera across the plate in casein blocking buffer (Blocker Thermo Scientific) and determining the nearest dilution where the signal reaches the blank. Sera were also tested at a single dilution of 1/150 against all six antigens. In both cases, bound antibodies were detected with anti-human IgG secondary antibody conjugated to horseradish peroxidase (HRP) (Bethyl Labs) followed by TMB (3,3′,5,5′-tetramethylbenzidine) developer (SureBlue reserve from KPL, Gaithersburg, MD). Optical density at 450 nm (OD450) readings were made and data sets compared by 1-tailed t tests.
Microarray data accession number.
The raw and normalized array data used in this study have been deposited in the Gene Expression Omnibus archive (http://www.dtd.nlm.nih.gov/geo/), accession number GSE34931.
RESULTS
No-takes are classified into responders and nonresponders.
Sera from 23 vaccinia-naive individuals that failed to show a take after vaccination against smallpox were probed against VACV-WR proteome microarrays to define antibody profiles. Overall, this was a heterogeneous population of vaccinees. Sera were from dose-sparing studies and received different vaccine dilutions, ranging from 1/100 to undiluted DryVax (n = 15 sera) or APSV (n = 8 sera), as shown in Table 1. The heat map in Fig. 1 provides an overall impression of the antibody reactivity on d0 and d28 for each donor. Naïve takes (Fig. 1B) showed minimal reactivity before vaccination (d0) as expected, followed by a response to several antigens at the peak on d28. The previously vaccinated takes (Fig. 1C) showed elevated preexisting reactivity on d0 compared to the naïve controls, consistent with previous vaccination, and a stronger response on d28, consistent with boosting. By comparison, naïve no-takes (Fig. 1A) had variable preexisting antibody reactivity on d0 compared to the naive takes. Moreover, the no-takes also showed variable responses to the vaccine on d28. Since the sera from the no-takes were from dose-sparing studies, donors were stratified in the heat map into three groups according to vaccine dilution: <1/10 (n = 8), 1/10 to 1/3 (n = 11), and undiluted (n = 4). Visual inspection reveals that a response to vaccination is seen only in individuals receiving a vaccine dilution of 1/10 or above.
Fig 1.
Heat map overview of the antibody profiles analyzed in this study. Serum samples are listed horizontally, and arrayed antigens are listed vertically. Sera from three groups of vaccinated donors, obtained prior to vaccination (d0) and 28 days after receiving DryVax or APSV (d28), were probed against arrays. (A) Vaccinia-naïve individuals who failed to develop a take after vaccination (n = 23); (B) vaccinia-naïve individuals who developed a take after vaccination (n = 50); (C) previously vaccinated individuals who developed a take after vaccination (n = 25). Vaccine dilutions are indicated immediately above the heat map. Donors are ranked from left to right by ascending average signal intensity. The arrayed antigens comprise each individual vaccinia virus ORF in the WR genome expressed from T7 plasmids in coupled in vitro transcription/translation (IVTT) reactions. Each antigen is designated (right) by the WR number followed by the Copenhagen nomenclature. Only the top 25 most reactive antigens are shown. Control spots comprise IVTT lacking plasmid template (No DNA; used for data normalization), purified human IgG used as a control for the secondary antibody, and purified Epstein-Barr virus nuclear antigen-1 (EBNA-1) used as a control for serum quality. The heat map was generated from log-transformed and normalized data using vsn, and the data were retransformed back into approximate raw signal intensities for the heat map and converted to a color scale. *, pseudogene.
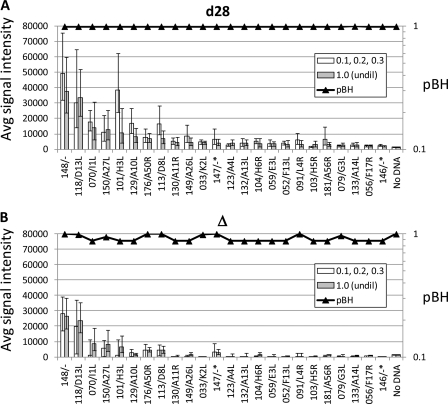
This dose-response effect is more clearly illustrated in Fig. 2A. As an indicator of the response to vaccination, the average of the signals to six “signature” vaccinia virus antigens was plotted against vaccine dilution. These antigens were selected based on their dominance in the antibody profiles of DryVax vaccinees seen in this study and as reported previously (8, 9). These comprised the intracellular mature virion (IMV) membrane proteins WR101/H3L, WR113/D8L, and WR150/A27L, core protein WR070/I1L, A-type inclusion body homolog WR148/-, and virion scaffold protein WR118/D13L. Values for d0 were subtracted from those for d28 so that only the response component (Δ) of the signal was plotted. This clearly shows a trend of increasing average response with increasing dose. A heat map of the data is also shown in Fig. 2B, with the donors ranked from left to right according to the dose of vaccine administered. It is clear that a dose below 1/10 does not engender a change (Δ) in the antibody profile, and henceforward only no-takes receiving a dilution of 1/10 or more (n = 15) are used for statistical analysis.
Fig 2.
Dose-response of antibody profiles. (A) The average signals on d28 − d0 (Δ) of six signature antigens for 23 individuals without a take (see Fig. 1A) were plotted against vaccine dilution received. Day 0 values were subtracted from d28 values to account for various backgrounds. (B) Heat map of the six signature antigens for the 23 no-takes, ranked from left to right by vaccine dilution. Maps for prevaccination (d0), postvaccination (d28), and d28 − d0 (Δ) signals are shown separately. Responders, as defined in Table 2 (n = 8), are indicated by an asterisk. Vaccinees that received APSV are indicated by a plus sign.
It is also apparent from Δ in Fig. 2B that only around half of the no-takes mounted an antibody response. To quantify this more precisely, an individual was classified as a responder if the signals increased more than 2-fold for at least two of the six signature antigens after vaccination (Table 2). Using this criterion, 8/15 no-takes were responders, which comprised all 4 receiving undiluted vaccine and 4 of the remaining 11 no-takes that received diluted vaccine. By comparison, 100% of naïves with a take (n = 50) and 88% of previously vaccinated individuals with a take (n = 25) were defined as responders. The no-take responders are indicated in Fig. 2B by the asterisks, which clearly show that responders cluster at the higher end of the dose range. We also saw no strong difference between the responses to APSV and DryVax (Fig. 2B). The data for each were thus pooled based on this observation and based on the origins and reported effectiveness of both vaccines (32, 38).
Table 2.
Numbers of responders to smallpox vaccine in no-takes and takes
| Vaccine dilution | No. (%) of responders to smallpox vaccine/totala |
||
|---|---|---|---|
| Naïve, no take | Naïve, take | Previous vaccination, take | |
| All | 8/15 (53.3) | 50/50 (100.0) | 22/25 (88.0) |
| 1/10, 1/5, 1/3 | 4/11 (36.4) | NA | 18/19 (94.7) |
| Undiluted | 4/4 (100.0) | 50/50 (100.0) | 4/6 (66.6) |
An individual was defined as a responder if the ratio of retransformed signal intensities before and after vaccination (d28/d0) was >2.0 for 2 or more of 6 signature antigens (WR101/H3L, WR113/D8L, WR148/-, WR150/A27L, WR070/I1L, and WR118/D13L). NA, not applicable.
Antibody profiles after vaccination in no-take responders are not significantly different from those of takes.
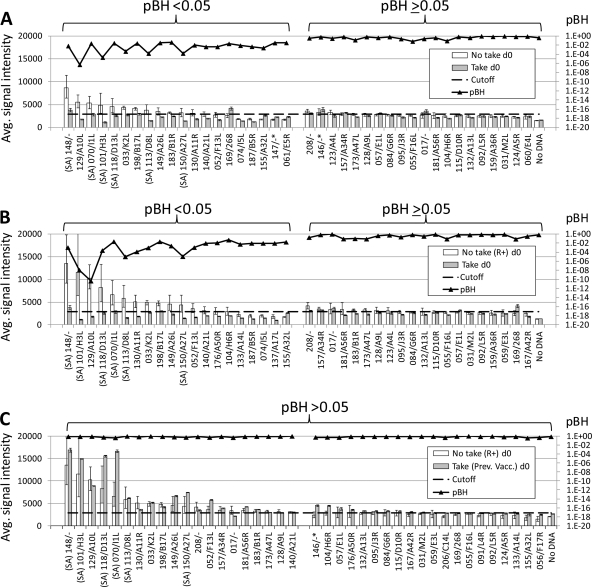
The next aim was to determine whether there was any particular antibody, or lack of antibody, characteristic of the response in a no-take. We first used t tests to compare profiles of responders receiving undiluted vaccine (n = 4) with responders receiving dilutions of 1/10, 1/5, and 1/3 (n = 4). No significant differences were revealed, using either the d28 signals (Fig. 3A) or using d28 − d0 (Δ) signals (Fig. 3B). The no-take responders were then pooled (n = 8) and compared to takes (n = 50) using t tests (Fig. 4A). Several antigens were significant, although their signals were low and close to the baseline. When the t test was repeated using only those receiving undiluted vaccine (n = 4), the number of significant antigens was reduced to only two, both of which had low signals (Fig. 4B). We concluded that the peak antibody responses in the takes and no-takes sampled are essentially the same.
Fig 3.
t test comparison of antibody profiles in no-take responders receiving undiluted versus diluted smallpox vaccine. t tests were performed comparing undiluted vaccine profiles (n = 4) to profiles obtained with vaccine dilutions of 1/10, 1/5, and 1/3 (n = 4) using log-transformed and normalized data. Benjamini and Hochberg-corrected P values (pBH) are overlaid onto histograms generated from retransformed average signal intensities ± standard errors of the means (SEM). Significant differences are indicated by pBH values of <0.05, nonsignificant differences are ≥0.05. (A) d28 profiles; (B) d28 − d0 (Δ) profiles.
Fig 4.
t test comparisons of d28 − d0 (Δ) signals of no-take responders versus takes. t tests were performed as described in the legend to Fig. 3. (A) All no-take responders (n = 8) versus naïve takes (n = 50); (B) no-take responders receiving undiluted DryVax only (n = 4) versus takes (n = 50). R+, responders only.
Antibody profiles in naïve individuals with a take compared to those with no-takes are significantly different before vaccination.
It is known that previous vaccination against smallpox is associated with an attenuated take upon a revaccination (13, 14). Although the no-take individuals studied here were all vaccinia naive (i.e., they had no exclusion criteria for the clinical trial, such as a vaccination scar or birth date during the eradication campaign), we hypothesized that they had preexisting vaccinia reactivity (or cross-reactivity) of unknown etiology that may be accounting for the attenuated take. This hypothesis was supported by higher seroprevalence on d0 in the no-take population (Table 3). Here, 8 of 15 (53.3%) no-takes were seropositive for at least one of the six signature antigens on d0. This compares to 3 of 50 (6%) takes and 24 of 25 (96%) previously vaccinated individuals. Importantly, the seroprevalence in no-takes on d0 is even higher when the nonresponders are excluded (increases from 62.5% to 75.0%; not shown). The latter figure is likely to be a more accurate figure for seroprevalence in individuals that fail to take if the nonresponders in this population simply received a subantigenic dose. After vaccination, the proportion of seropositives increases in all three groups. The smallest number of seropositives is the no-take group (62.5%), compared to 84% for the naïve takes and 100% for previously vaccinated takes. If only the responders are considered (n = 8), the seropositive rate for no-takes on d28 rises to 100%.
Table 3.
Numbers of seropositive donors before and after DryVax vaccinationa
| Day | Naïve |
Previous vaccination, take |
||||||
|---|---|---|---|---|---|---|---|---|
| No-take |
Take |
|||||||
| Sum (n = 15) | % | Sum R+ (n = 8) | % | Sum (n = 50) | % | Sum (n = 25) | % | |
| 0 | 8 | 53.3 | 6 | 75.0 | 3 | 6.0 | 24 | 96.0 |
| 28 | 10 | 62.5 | 8 | 100.0 | 42 | 84.0 | 25 | 100.0 |
An individual was defined as seropositive if log-normalized signals for one or more of six signature antigens (see Table 2) was above a cutoff, defined for each antigen as the average + 3 SD of the naïve/take population on d0. R+, responders only.
We performed t test comparisons of preexisting (d0) profiles of naïve no-takes and takes to identify significantly different antigens (Fig. 5A). The aim was to identify markers that could be used to predict a no-take prior to vaccination. A number of antigens were significantly different, including all five of the signature antigens (SA). A cutoff based on d0 of the takes showed that ∼12 significant antigens were seropositive. When the test was repeated with the no-take responders only (n = 8), the average signal intensities of the no-takes generally increased, although the number of significant antigens above the cutoff remained unchanged (Fig. 5B). These antigens are listed in Table 4. The table shows the amino acid homology between the WR prototype and other related orthopoxviruses. Although the etiology of the preexisting antibodies is not known, all of the viruses shown are possible candidates, and all of them show high homology with WR. Interestingly, if these no-take responders are compared with the previously vaccinated individuals by t tests, the number of significant differences decreases to zero (Fig. 5C), which further supports the notion of a prior exposure to an orthopoxvirus in the no-take responders.
Fig 5.
t test comparisons of d0 signals in no-takes versus takes. t tests were performed as described in the legend to Fig. 3. (A) All no-takes (n = 15) versus takes (n = 50); (B) no-take responders only (n = 8) versus takes (n = 50); (C) no-take responders only (n = 8) versus previously vaccinated takes. Horizontal dashed line, cutoff defined as the mean + 2 SD of d0 signals in the naïve/take population. R+, responders only; SA, signature antigen.
Table 4.
Functions of VACV-WR proteins recognized by preexisting antibodies in no-takes who responded to vaccinationa
| VACV name (WR/COP) | Type | Class | Function | Amino acid sequence homology (% identity) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| HSPV | CMLV | CPXV | MPXV | VACV-Lister | VACV-ACAM | ||||
| WR148 | L | Virion assembly | Cowpox A-type inclusion protein homolog | 98.9 | 95.5 | 92.9 | 95.3 | 99.5 | 98.8 |
| WR101/H3L | L | IMV membrane | Cell attachment, herparin binding | 99.4 | 99.4 | 96.0 | 93.8 | 98.8 | 99.1 |
| WR129/A10L | L | Core | Precursor of core protein 4a | 98.9 | 97.3 | 96.8 | 97.0 | 99.6 | 98.7 |
| WR118/D13L | L | Virion assembly | Rifampin target, inner surface of IMV membrane | 99.5 | 99.3 | 99.1 | 98.9 | 99.8 | 99.6 |
| WR070/I1L | L | Core | DNA binding protein | 98.4 | 98.4 | 100.0 | 99.4 | 100.0 | 99.7 |
| WR113/D8L | L | IMV membrane | Cell attachment, chondroitin binding | 98.4 | 96.1 | 94.4 | 94.1 | 100.0 | 98.7 |
| WR130/A11R | L | Unknown | Unknown | 99.7 | 98.1 | 99.4 | 99.1 | 99.7 | 99.7 |
| WR033/K2L | E | Host range/defense | Serine protease inhibitor-like | 97.9 | 93.8 | 94.9 | 92.3 | 98.9 | 99.7 |
| WR198/B17L | E/L | Unknown | Unknown | 98.5 | 98.0 | 97.4 | 97.3 | 98.8 | 98.2 |
| WR149/A26L | L | IMV membrane associated | Cell attachment | 98.8 | 95.0 | 93.3 | 91.4 | 99.4 | 99.4 |
| WR150/A27L | L | IMV membrane associated | Cell attachment | 100.0 | 97.3 | 98.2 | 94.6 | 99.1 | 100.0 |
| WR052/F13L | L | EEV membrane | Phospholipase | 99.3 | 98.7 | 98.4 | 97.7 | 100.0 | 99.3 |
| WR140/A21L | L | IMV membrane associated | Component of entry/fusion complex | 98.3 | 99.2 | 98.3 | 97.4 | 100.0 | 98.3 |
| WR176/A50R | E | Replication | DNA ligase | 98.0 | 97.1 | 98.2 | 98.2 | 98.9 | 98.2 |
The ranking of antigens is as defined in the legend to Fig. 4B. All protein descriptions and sequence homologies were obtained from the Poxvirus Bioinformatics Resource Center (www.poxvirus.org). WR, Western Reserve; COP, Copenhagen; E, early promoter; L, late promoter; HSPV, horsepox virus (MNR76 strain); CMLV, camelpox virus (CMS strain); CPXV, cowpox virus (Brighton Red strain); MPXV, monkeypox virus (Zaire strain); VACV-Lister, vaccinia virus (Lister vaccine strain); VACV-ACAM, vaccinia virus (ACAM2000 vaccine strain).
Elevated levels of preexisting antibodies in no-takes is confirmed by recombinant protein ELISAs.
We aimed to validate the above-described findings on an alternative immunoassay platform by purifying six different vaccinia virus antigens expressed in E. coli and coating them onto ELISA plates: WR101/H3L, WR118/D13L, WR113/D8L, WR148/-, WR070/I1L, or WR132/A13L. The performance of these ELISAs has been reported previously (15). Here, the most sensitive of the six antigens for detection of DryVax vaccination was WR113/D8L, which was able to detect reactivity 28 days after DryVax in 96% of naïve and 100% of previously vaccinated individuals. Thus, WR113/D8L was used to measure antibody endpoint titers of pre- and postvaccination sera from the naïve no-takes and takes for comparison (Fig. 6A). Endpoint titers were determined by performing 12 2-fold dilutions of each serum sample starting at 1/20. The average titer on d0 of the whole no-take population was ∼5-fold higher than that of the takes, although this failed to reach significance in a t test (P = 0.055; Fig. 6B). However, if the nonresponders were excluded, the titers in the remaining responder no-takes (n = 8) reached significance compared to those of takes (P = 0.006). This finding is consistent with the array data that suggested no-take responders had higher preexisting antibody levels than naïve takes (Fig. 5B), at least for the D8 antigen. After vaccination, titers to WR113/D8L were significantly higher in takes than in all no-takes (P = 0.048), although nonsignificant if nonresponders were excluded. Again, this was consistent with array data showing that no-takes that responded to DryVax had profiles that were not significantly different from those of controls with a take (Fig. 5C).
Fig 6.
Comparisons of antibody titers of naïve no-takes versus naïve takes by WR113/D8L ELISA. (A) Individual donor endpoint titers at d0 and d28 postvaccination are plotted for no-takes (n = 15) and takes (n = 17). Vaccine dilution received is indicated below each sample, and responders (as defined by arrays in Table 2) are indicated by asterisks. Horizontal dashed line, average d0 titer; horizontal solid line, average d28 titer. (B) P values for different t test comparisons using ELISA data. Each test was performed with naïve takes (n = 50) versus all no-takes (n = 15) or no-take responders only (R+; n = 8).
We then extended the ELISA analysis to all six purified antigens using a single dilution of sera (Fig. 7) and compared OD450 readings by t tests (Table 5). Again, signals to the WR113/D8L were significantly higher on d0 in no-take responders than in takes (P = 0.002). Of the other five antigens tested, signals to WR118/D13L and WR101/H3L were also significantly higher on d0 in the no-take responders (P = 0.012 and 0.006, respectively). In contrast, none of these were significantly different on d28. These data are consistent with the earlier observations that differences between no-take responders and takes lie mainly in the preexisting antibody levels rather than after vaccination. Of particular interest here was WR132/A13L, which was the only antigen of the six tested that was significantly different between no-take responders and takes on d28. This is consistent with data shown in Fig. 1 and 4. Similarly, we reported previously (15) that while WR132/A13L is strongly recognized by vaccinia-naïve individuals undergoing a primary response to DryVax, it is not recognized by previously vaccinated individuals after boosting with DryVax. Thus, the lack of response to this antigen by no-takes here, although not diagnostic, is consistent with preexisting immunity.
Fig 7.
Recombinant vaccinia virus protein ELISAs. OD450 values for pre- and postvaccination sera (diluted to 1/150) from the three donor groups and assayed against six different recombinant vaccinia virus antigens by ELISA. The individual donors within each group are ranked from left to right by ascending vaccine dose. ND, not done.
Table 5.
Summary of recombinant vaccinia virus antigen ELISAsa
| t test |
P value |
|||||
|---|---|---|---|---|---|---|
| WR148/− | WR113/D8L | WR118/D13L | WR101/H3L | WR132/A13L | WR070/I1L | |
| Pre (no-take, R+) vs pre (take) | 0.0760 | 0.0020 | 0.0119 | 0.0057 | 0.2497 | 0.2196 |
| Post (no-take, R+) vs post (take) | 0.4557 | 0.0551 | 0.1245 | 0.4852 | 0.0061 | 0.3545 |
| Pre vs post (no-take, R+) | 0.0003 | 0.0572 | 0.0032 | 0.1341 | 0.4295 | 0.1801 |
| Pre vs post (take) | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0007 |
Shown are P values when ELISA data (OD450) from different groups of serum samples were compared by t tests, as shown in the first column. ELISA data were obtained for samples before (d0) and after (d28) DryVax vaccination. For t tests, data from no-take responders only (n = 8) were compared with data from takes (n = 45). Significant P values (<0.05) are underlined. R+, responders only.
DISCUSSION
Successful vaccination against smallpox is conventionally determined by the presence of a take at the site of inoculation. In primary vaccinations, the take involves a 2- to 3-week progression from papule to vesicle, to pustule, to scab (27) but is more rapid, around 8 days, in revaccinations (23). The lesion is thought to be a result of viral damage and inflammation at the site of inoculation (26). In this study, we were able to compare the vaccinia virus-specific antibody profiles of 23 no-takes with naïve and previously vaccinated individuals with a take, both before and after vaccination. We used objective criteria to define preexisting immunity and response to vaccine based on six signature vaccinia virus antigens on microarrays (three membrane proteins and three other structural proteins), shown previously to dominate the response to DryVax in humans (6, 8, 15). The membrane proteins comprised WR150/A27L, WR101/H3L, and WR113/D8L. These are the membrane targets of antibodies that neutralize intracellular mature virions (IMVs) and contribute to protection (7, 22, 31, 33). The remaining three signature antigens comprise the WR070/I1L core protein (20), WR118/D13L, which is a membrane scaffold protein involved in intracellular morphogenesis (37), and WR148/-, an A-type inclusion protein homolog that is strongly recognized by most vaccinated subjects (9, 19, 24, 29).
The no-takes studied here were a heterogeneous population, receiving lyophilized (DryVax) or frozen (APSV) vaccine at doses ranging from undiluted to 1/100, and with differing antibody titers, both before and after vaccination. We pooled data from DryVax and APSV vaccinees to increase the sample size. This was done on the basis that we did not see obvious differences between the DryVax and APSV responder profiles (see Fig. 2B). Moreover, both are derived from the NYCBOH parent strain and retain high take rates after dilution to 1/10 (11, 38). However, we are aware that while diluted APSV (1/10) induces comparable antibody and T cell responses to undiluted vaccine (32, 38), dilution of DryVax has dose-dependent effects on both (4, 12). The studies described here are presented with this caveat. No-takes that failed to respond are true cases of vaccination failure, for which the skin reaction is a good indicator. This could be due to insufficient dose, problems with vaccine delivery, etc. In contrast, no-take responders are likely to have other underlying reasons for the lack of a take. When these were segregated and compared with the takes, the analyses consistently showed that the profiles after vaccination are essentially identical. This suggests that in no-take responders, the presence or absence of particular antibody responses is not a major determinant in whether a skin take develops.
Unlike the response component, significant differences in preexisting antibodies were found to exist, with the no-take responders having the highest level of preexisting antibody. Thus, 53% of no-takes and 75% of no-take responders had preexisting antibody by arrays, compared to 6% of takes (Table 3). The elevated preexisting antibody in the no-takes was also supported by ELISAs using purified proteins. Currently, the origin of these preexisting antibodies is not known. Several lines of evidence argue in favor of exposure to a related orthopoxvirus as the origin, rather than an unrelated but antigenically cross-reactive source. First, the preexisting antibodies are known hallmarks of a vaccinia infection, particularly of previously vaccinated individuals, for which there is no significant difference (Fig. 5C). The preexisting antibody profile is dominated by late structural proteins and includes antibodies to IMV membrane proteins WR101/H3L, WR113/D8L, WR149/A26L, and WR150/A27L and core proteins WR129/A10L and WR070/I1L. Second, these preexisting antibodies are not against novel targets, ruling out an unrelated but cross-reactive etiological agent. Third, the lack of response to WR132/A13L in ELISA is also consistent with preexisting immunity to orthopox (15). Finally, none of the vaccinees had exclusion criteria indicating previous smallpox vaccination. Overall, the data are consistent with previous exposure to a related orthopoxvirus. We have no information about exposure to other environmental orthopoxviruses by the individuals studied here. However, natural exposure to such viruses may have the same effect as previous smallpox vaccination and contribute to the attenuation of the take.
Attenuation of the take may be antibody or T cell mediated. Although we have not measured T cell responses in the this study, we favor the notion that preexisting T cell memory leads to rapid clearance of infection at the site of inoculation and the attenuation of the take without interfering with the boosting effect on the antibody arm of the response. In another study, numbers of residual vaccinia virus-specific CD4+ T cells in individuals vaccinated against smallpox 30 years previously are inversely associated with the size of the skin take upon revaccination (30). Thus, memory T cells remaining after the primary vaccination are likely to contribute to attenuation of the take after revaccination. A similar process may be operating in the vaccinia-naïve no-takes studied here if they were already exposed to a vaccinia virus-like orthopox virus. The same study (30) also found no relationship between neutralizing antibody titers after revaccination and lesion size. This is consistent with the findings presented here in which we saw no difference in antibody profiles between takes and no-takes after vaccination. Interestingly, the authors found no association between residual (day 0) vaccinia virus-neutralizing antibody titers and lesion size after revaccination, whereas our data are consistent with a role of d0 titers and lack of a take. The discrepancy does highlight that there are likely to be subtle differences between bona fide revaccinations and the no-take population studied here. For example, the etiological agent postulated here, or when our no-take population may have been exposed, may differ from that of the other study. Moreover, the different serological assays used and the different sample sizes may also be influential.
In summary, we have profiled the antibody responses of vaccinia takes and no-takes. The main findings are that the no-takes can be classified into responder and nonresponder populations. In this study, nonresponsiveness was a vaccine dose-related phenomenon, and the skin test provides an accurate indicator of vaccination failure in these individuals. In no-takes that responded to the vaccine, the skin reaction is not an accurate indicator of vaccination failure. No-take responders do not differ significantly from takes after vaccination, suggesting that the failure to take is not related to the antibody response to vaccination. In contrast, no-takes tend to have higher preexisting antibody titers before vaccination than takes, with profiles not dissimilar to those of previously vaccinated individuals. The origin of these preexisting antibodies is not thought to be vaccinia, but they nevertheless appear to be having an attenuating effect on the take, similar to previous vaccination.
ACKNOWLEDGMENTS
We thank Robert Johnson and Mark Challberg, Division of Microbiology and Infectious Diseases, NIAID/NIH, for initiating the project and for insightful comments on the work. We also thank Sharon Frey, Department of Internal Medicine at Saint Louis University, for providing patient metadata.
This study was supported by a contract from the NIAID/NIH to Antigen Discovery Inc. and in part by grants R44-AI-58365, N01-AI-05413, N01-AI-40097, U54AI065359, and U01AI078213.
D.H.D., P.F., and X.L. declare a financial interest in Antigen Discovery, Inc. D.H.D., P.F., X.L., and the University of California, Irvine, may financially benefit from this interest if the company is successful in marketing its products that are related to this research. The terms of this arrangement have been reviewed and approved by the University of California, Irvine, in accordance with its conflict of interest policies.
Footnotes
Published ahead of print 18 January 2012
REFERENCES
- 1. Amanna IJ, Slifka MK, Crotty S. 2006. Immunity and immunological memory following smallpox vaccination. Immunol. Rev. 211: 320–337 [DOI] [PubMed] [Google Scholar]
- 2. Baldi P, Hatfield GW. 2002. DNA microarrays and gene expression: from experiments to data analysis and modeling. Cambridge University Press,Cambridge, United Kingdom [Google Scholar]
- 3. Baldi P, Long AD. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519 [DOI] [PubMed] [Google Scholar]
- 4. Belshe RB, et al. 2004. Dose-dependent neutralizing-antibody responses to vaccinia. J. Infect. Dis. 189: 493–497 [DOI] [PubMed] [Google Scholar]
- 5. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57: 289–300 [Google Scholar]
- 6. Davies DH, et al. 2005. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. U. S. A. 102: 547–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davies DH, et al. 2005. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J. Virol. 79: 11724–11733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davies DH, et al. 2007. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics 7: 1678–1686 [DOI] [PubMed] [Google Scholar]
- 9. Davies DH, et al. 2008. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus Ankara is comparable to that of Dryvax. J. Virol. 82: 652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Durbin BP, Hardin JS, Hawkins DM, Rocke DM. 2002. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics 18(Suppl 1): S105–S110 [DOI] [PubMed] [Google Scholar]
- 11. Frey SE, et al. 2002. Clinical responses to undiluted and diluted smallpox vaccine. N. Engl. J. Med. 346: 1265–1274 [DOI] [PubMed] [Google Scholar]
- 12. Frey SE, et al. 2002. Dose-related effects of smallpox vaccine. N. Engl. J. Med. 346: 1275–1280 [DOI] [PubMed] [Google Scholar]
- 13. Frey SE, Newman FK, Yan L, Lottenbach KR, Belshe RB. 2003. Response to smallpox vaccine in persons immunized in the distant past. JAMA 289: 3295–3299 [DOI] [PubMed] [Google Scholar]
- 14. Gassmann R, et al. 2008. Clinical and immune response to undiluted and diluted smallpox vaccine. Swiss Med. Wkly. 138: 392–397 [DOI] [PubMed] [Google Scholar]
- 15. Hermanson G, et al. 2012. Measurement of antibody responses to modified vaccinia virus Ankara (MVA) and Dryvax® using proteome microarrays and development of recombinant protein ELISAs. Vaccine 30: 614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl 1): S96–S104 [DOI] [PubMed] [Google Scholar]
- 17. Jing L, et al. 2008. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J. Virol. 82: 7120–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jing L, et al. 2009. ORFeome approach to the clonal, HLA allele-specific CD4 T-cell response to a complex pathogen in humans. J. Immunol. Methods 347: 36–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones-Trower A, et al. 2005. Identification and preliminary characterization of vaccinia virus (Dryvax) antigens recognized by vaccinia immune globulin. Virology 343: 128–140 [DOI] [PubMed] [Google Scholar]
- 20. Klemperer N, Ward J, Evans E, Traktman P. 1997. The vaccinia virus I1 protein is essential for the assembly of mature virions. J. Virol. 71: 9285–9294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kreil DP, Karp NA, Lilley KS. 2004. DNA microarray normalization methods can remove bias from differential protein expression analysis of 2D difference gel electrophoresis results. Bioinformatics 20: 2026–2034 [DOI] [PubMed] [Google Scholar]
- 22. Lai CF, Gong SC, Esteban M. 1991. The purified 14-kilodalton envelope protein of vaccinia virus produced in Escherichia coli induces virus immunity in animals. J. Virol. 65: 5631–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lane JM, Goldstein J. 2003. Evaluation of 21st-century risks of smallpox vaccination and policy options. Ann. Intern. Med. 138: 488–493 [DOI] [PubMed] [Google Scholar]
- 24. Liu X, Kremer M, Broyles SS. 2004. A natural vaccinia virus promoter with exceptional capacity to direct protein synthesis. J. Virol. Methods 122: 141–145 [DOI] [PubMed] [Google Scholar]
- 25. Long AD, et al. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K-12. J. Biol. Chem. 276: 19937–19944 [DOI] [PubMed] [Google Scholar]
- 26. Martin DB. 2002. The cause of death in smallpox: an examination of the pathology record. Mil. Med. 167: 546–551 [PubMed] [Google Scholar]
- 27. Maurer DM, Harrington B, Lane JM. 2003. Smallpox vaccine: contraindications, administration, and adverse reactions. Am. Fam. Physician 68: 889–896 [PubMed] [Google Scholar]
- 28. Moss B. 2011. Smallpox vaccines: targets of protective immunity. Immunol. Rev. 239: 8–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel DD, Pickup DJ, Joklik WK. 1986. Isolation of cowpox virus A-type inclusions and characterization of their major protein component. Virology 149: 174–189 [DOI] [PubMed] [Google Scholar]
- 30. Puissant-Lubrano B, et al. 2010. Control of vaccinia virus skin lesions by long-term-maintained IFN-gamma+TNF-alpha+ effector/memory CD4+ lymphocytes in humans. J. Clin. Invest. 120: 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Putz MM, Midgley CM, Law M, Smith GL. 2006. Quantification of antibody responses against multiple antigens of the two infectious forms of vaccinia virus provides a benchmark for smallpox vaccination. Nat. Med. 12: 1310–1315 [DOI] [PubMed] [Google Scholar]
- 32. Rock MT, Yoder SM, Talbot TR, Edwards KM, Crowe JE., Jr 2006. Cellular immune responses to diluted and undiluted Aventis Pasteur smallpox vaccine. J. Infect. Dis. 194: 435–443 [DOI] [PubMed] [Google Scholar]
- 33. Sakhatskyy P, Wang S, Chou TH, Lu S. 2006. Immunogenicity and protection efficacy of monovalent and polyvalent poxvirus vaccines that include the D8 antigen. Virology 355: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seaman MS, et al. 2010. Effect of vaccination with modified vaccinia Ankara (ACAM3000) on subsequent challenge with Dryvax. J. Infect. Dis. 201: 1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sundaresh S, et al. 2006. Identification of humoral immune responses in protein microarrays using DNA microarray data analysis techniques. Bioinformatics 22: 1760–1766 [DOI] [PubMed] [Google Scholar]
- 36. Sundaresh S, et al. 2007. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics 23: i508–i518 [DOI] [PubMed] [Google Scholar]
- 37. Szajner P, Weisberg AS, Lebowitz J, Heuser J, Moss B. 2005. External scaffold of spherical immature poxvirus particles is made of protein trimers, forming a honeycomb lattice. J. Cell Biol. 170: 971–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Talbot TR, et al. 2004. Vaccination success rate and reaction profile with diluted and undiluted smallpox vaccine: a randomized controlled trial. JAMA 292: 1205–1212 [DOI] [PubMed] [Google Scholar]
- 39. Thomas G, Frenzel E, Hanna H. 2004. Smallpox vaccine: “nontake” responses in previously vaccinated adults. Infect. Control Hosp. Epidemiol. 25: 613–615 [DOI] [PubMed] [Google Scholar]