Abstract
Screening of an expression library of Leptospira interrogans with eye fluids from uveitic horses resulted in identification of a novel protein, LruC. LruC is located in the inner leaflet of the leptospiral outer membrane, and an lruC gene was detected in all tested pathogenic L. interrogans strains. LruC-specific antibody levels were significantly higher in eye fluids and sera of uveitic horses than healthy horses. These findings suggest that LruC may play a role in equine leptospiral uveitis.
TEXT
Leptospires are a common infectious cause of uveitis in horses and humans, and the immune-based sequelae are often devastating (24, 36, 15, 6, 19, 37). Pathogenic leptospires exploit the unique physiological and immunological environment of the eye to their advantage and persist in this “safe” environment during equine recurrent uveitis (ERU). However, following a lengthy incubation period during which infection is unapparent in the horse, recurring episodes of acute uveitis separated by quiescent phases of variable duration ensue. Recurrence of the disease has been explained by persistence of the inciting antigen in ocular tissue, resulting in periodic inflammation (23, 1, 16), or by delayed-type hypersensitivity. In the latter case, memory T cells in the uveal tract trigger a robust immune response on subsequent exposures, leading to acute episodes of inflammation (7, 10). Th1 bias is seen in ocular but not peripheral lymphocytes, indicating an independent ocular response (10).
In previous work, we described leptospiral proteins LruA and LruB, which are associated with very strong IgG and IgA responses in affected eyes and with reactivities to extracts of equine ocular tissue (33). Furthermore, we demonstrated that a significant proportion of human patients with leptospiral uveitis produced serum antibodies to LruA and -B (35). In this paper, we describe the identification of a novel, putative lipoprotein, LruC, and specific antibody responses directed toward this protein in the eye fluids and sera of horses with naturally acquired leptospiral uveitis.
Eye fluids and companion sera from horses of varied ages, breeds, and origins were obtained from a commercial horse slaughter plant in North America. Eyes with gross evidence of uveitis were enucleated after slaughter, and the aqueous humor, vitreous, and eye tissue were collected and frozen at −20°C (33). Eye fluids and sera were assayed for antibodies to serovars Pomona, Canicola, Icterohemorrhagiae, Hardjo, Bratislava, and Grippotyphosa with a microscopic agglutination test (MAT) and with an enzyme-linked immunosorbent assay (ELISA), and sections of eye tissues stained with hematoxylin and eosin were examined for pathological changes (33). A pool of eye fluids from five confirmed cases of leptospiral uveitis (33) was used to screen an expression library of L. interrogans to identify phage-expressing gene products reactive to antibodies in the uveitic eyes. Screening of a lambda ZAP II library of L. interrogans serovar Pomona type kennewicki JEN4 with pooled eye fluids was performed as per the manufacturer's protocol (Stratagene, La Jolla, CA) and as described previously (33, 34). Briefly, following propagation on Escherichia coli XL-1 MRF′ (Stratagene, La Jolla, CA), plaques were transferred in duplicate to IPTG (isopropyl-β-d-thiogalactopyranoside)-saturated nitrocellulose discs and immunoblotted with pooled eye fluids, diluted 1:600 (33). Bound antibody was detected with horseradish peroxidase (HRP)-labeled protein G (Zymed, San Francisco, CA) diluted 1:4,000 followed by the addition of 4-chloro-1-naphthol. Screening of the library yielded 14 reactive plaques. Positive plaques on agar plugs were allowed to elute overnight at 4°C in 500 μl of SM buffer (100 mM NaCl, 8 mM MgSO4 · 7H2O, 50 mM Tris-Cl [pH 7.5]). Reactive plaques were rescreened until clonal. Plasmids containing inserts of leptospiral DNA were rescued from selected reactive phages by using ExAssist helper phage and E. coli SOLR (Stratagene, La Jolla, CA) according to the manufacturer's protocol. Plasmids rescued from these phages were sequenced in a commercial sequencing facility (Davis Sequencing LLC, Davis, CA) using T3, T7, and custom-designed primers (Table 1) and compared with the published genomic sequences of L. interrogans serovar Lai strain 56601 (30), L. interrogans serovar Copenhageni Fiocruz L1-130 (26), and Leptospira borgpetersenii serovar Hardjo strains L550 and JB197 (5).
Table 1.
Primers
| Primer | Sequence |
|---|---|
| a4-3F | 5′-CGC CTC GAG TGC AGT CAT AAG AAA AAA GG-3′ |
| a4-3R | 5′-GCG GAT CCT CAC TTT GAT AAA GAT GTC G-3′ |
| a4-3UF | 5′-CTT GGA TTC GTT GGA TCG CGG ATT-3′ |
| a4-3UR | 5′-TCA CTG TGG CTC CAG AGA TAG GTT-3′ |
| a4-3DF | 5′-TTC TGG ATC GTC CTC TGG TTC CAA-3′ |
| a4-3DR | 5′-AGG TTC CAT TAG ACG CCA CCC AAA-3′ |
Nucleotide and deduced amino acid sequences were analyzed with DNASIS, the Genetics Computer Group package of programs (Wisconsin Package version 10.0; Genetics Computer Group, Madison, WI), PSORT (http://psort.nibb.ac.jp/), SignalP (3), LipoP (18), SpLip (31), TMHMM (http://www.cbs.dtu.dk/), T-COFFEE (www.tcoffee.org/), and COILS (http://www.ch.embnet.org/index.html). Homologies were identified by a BLAST search using the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/BLAST/). Homologies to eight different loci were found, encoding the previously described leptospiral proteins LigA/LigB (21) (four phagemids), LigC (one phagemid), GrpE/DnaK/DnaJ (2) (four phagemids), Qlp42 (25), and LruA and LruB (three phagemids) (33), plus a novel protein. Phagemid pA4, which encodes this protein, was selected for further analysis. Sequencing of pA4 and the chromosomal locus of L. interrogans serovar Pomona type kennewicki revealed an open reading frame encoding a protein, designated LruC, of 567 amino acids with a predicted molecular mass of 57 kDa. A hexanucleotide resembling the −10 region of the σ70 bacterial promoter and a transcriptional terminator are present immediately 5′ and 3′ of lruC, respectively. The lruC structural gene of L. interrogans serovar Copenhageni strain Fiocruz L1-130 (open reading frame [ORF] LIC20172) consists of 2,097 bases encoding a protein of 699 amino acids. Multisequence alignment of this gene and that of the Lai (strain 56601; ORF LA216), and Pomona (strain JEN4 type kennewicki) versions of LruC shows that these sequences are almost identical except that the carboxy termini in the Lai and kennewicki versions are absent. In L. borgpetersenii serovar Hardjo, lruC is disrupted by an insertion sequence (5).
LruC is predicted to be a lipoprotein by the SpLip algorithm (31). The 17-residue signal peptide consists of a basic amino-terminal region (amino acids 1 through 3), a hydrophobic core (amino acids 4 through 13), and a carboxyl-terminal Ala(−4) Val(−3) Phe(−2) Gly(−1) ↓ Cys(+1) signal peptidase II cleavage site that conforms to the consensus spirochetal lipobox sequence (13). Secondary structure analysis predicts a mature protein consisting of 13% alpha helix and 21% beta sheet. The amino-terminal half of LruC is homologous to the serine-threonine rich region of the Candida agglutinin-like family of Als surface proteins (17). As is the case for the Als surface proteins, the serine-threonine rich region of LruC contains a series of tandem 31- and 32-amino-acid sequence repeats.
Recombinant His-tagged LruC (rLruC) was expressed and purified from E. coli (33, 34). Briefly, following PCR amplification of chromosomal DNA of L. interrogans serovar Pomona type kennewicki (JEN4) using the lruC-specific primers a4-3F and a4-3R (Table 1), the amplicon was digested with BamHI and XhoI and ligated into pET-15b (Novagen, Madison, WI) predigested with the same restriction endonucleases. Recombinant plasmids were transformed into E. coli Rosetta 2(DE3)(pLysS) (Novagen, Madison, WI). Expression of polyhistidine-tagged recombinant LruC was induced with 1 mM IPTG when cultures reached an optical density of 0.6 at 600 nm, and cells were harvested after 3 h. Recombinant His-tagged LruC (rLruC) was isolated using Talon metal affinity resin (Clontech Laboratories, Inc.) in buffer containing 8 M urea according to the manufacturer's recommendations. His6-LruC was dialyzed against 10 mM Tris (pH 7.5) containing 50 mM NaCl.
The predicted molecular masses of LruC in strains JEN4 and Fiocruz L1-130 are approximately 57 and 72 kDa, respectively (data not shown). LruC in both strains migrates in sodium dodecyl sulfate (SDS)-polyacrylamide gels more slowly than expected from the predicted masses. The discrepancy in predicted and observed mass may be due to either posttranslational modifications, possibly by glycosylation at the serine residues in the serine-rich region, as is observed in some streptococcal adhesins (38). Another possibility is that LruC is an acidic protein (pI ∼ 5.1). This may result in LruC binding proportionally less SDS, resulting in slowed migration in SDS-polyacrylamide gels (4).
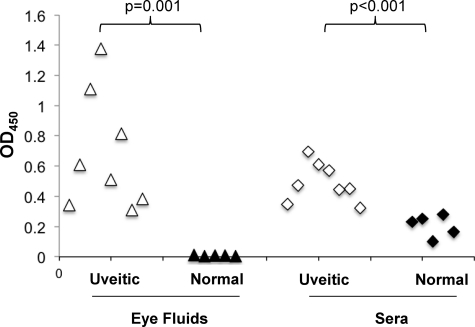
Next, recombinant LruC was used in ELISA to measure antibody levels in eye fluids and sera of leptospiral uveitic and healthy horses. A checkerboard titration was performed in Maxisorp 96-well plates (Nalge-Nunc, Rochester, NY) to determine the optimum concentration of recombinant LruC. Wells were coated with 100 ng protein, followed by blocking with 4% nonfat dry milk. Diluted eye fluids or sera (1:400) were added and incubated for 1 h at 37°C. Bound immunoglobulin G (IgG) was detected using HRP-protein G (Zymed, San Francisco, CA). Plates were developed with a ready-to-use 3,3′,5,5′-tetramethyl benzidine substrate solution (1-Step Turbo TMB-ELISA; Thermo Scientific, Rockford, IL). Absorbance was read at 450 nm with a Spectramax plate reader using SoftMax Pro software (Molecular Devices, Sunnyvale, CA). Statistical analyses were performed using Student's t test assuming unequal variances. Uveitic eye fluids (n = 8) contained significantly (P = 0.001) higher levels of LruC-specific IgG than fluids from healthy animals (n = 5) (Fig. 1). Specific IgG levels in uveitic sera (n = 8) were also significantly (P < 0.001) higher than those in sera from normal horses (n = 5) (Fig. 1).
Fig 1.
Measurement of LruC-specific antibody levels by ELISA. LruC-specific IgG levels in eye fluids and sera of leptospiral uveitic horses (n = 8) were significantly higher than the levels in eye fluids and sera of healthy control animals (n = 5). Wells were coated with 100 ng protein, and fluids and sera were diluted 1:400. OD450, optical density at 450 nm.
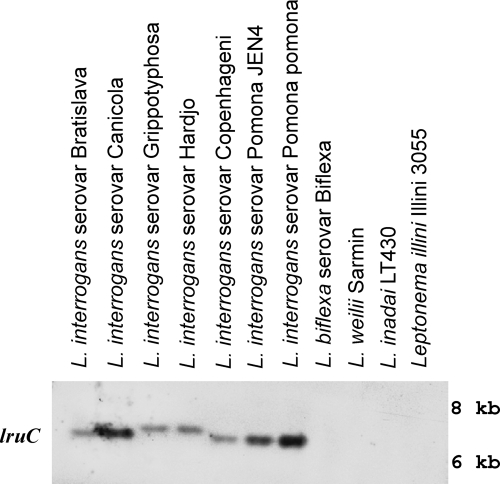
Next, Southern blot analyses were performed to examine the distribution of lruC in pathogenic and nonpathogenic leptospires. DNAs of L. interrogans serovars Pomona (strains Pomona and JEN4), Canicola (Hond Utrecht IV), Grippotyphosa (Andaman), Hardjo (Hardjoprajitno), and Bratislava (Jez Bratislava), L. biflexa serovar Biflexa, Leptospira weilii (Sarmin), Leptospira inadai (LT430), and Leptonema illini (Illini 3055) were isolated from 5-ml mid-log-phase cultures (33). DNAs were digested overnight with HindIII at 37°C and separated on a 0.8% agarose gel for 4 h at 50 V, transferred to a Hybond-N nylon membrane (Amersham, Piscataway, NJ), and fixed by UV cross-linking according to the manufacturer's protocol. Primers a4-3F and a4-3R (Table 1), specific for lruC were used in the PCR to amplify the lruC gene, which was then labeled with digoxigenin by using a DIG High Prime DNA labeling and detection kit (Roche Applied Science, Indianapolis, IN). Prior to digoxigenin labeling, the lruC PCR amplicon was digested with HindIII, and the larger fragment was extracted from the gel. The UV-cross-linked nylon membrane was subjected to prehybridization at 42°C for 30 min in DIG Easy hybridization solution. After denaturation, approximately 25 ng/ml of probe was mixed with DIG Easy hybridization solution and incubated with the membrane at 42°C with gentle agitation. The next day, the membrane was washed for 15 min at room temperature with three changes of the buffer containing 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% SDS and then washed three times for 15 min at 65°C in 0.5× SSC with 0.1% SDS, prewarmed to 65°C. After stringency washes, the membrane was treated with antidigoxigenin-alkaline phosphatase, followed by a chemiluminescent substrate (DIG High Prime DNA labeling and detection kit). Hybridization was detected by exposing the membrane to an X-ray film (Pierce, Rockford, IL). lruC genes were found by Southern blotting in L. interrogans serogroup Icterohemorrhagiae serovar Copenhageni and serovars Pomona (strains Pomona and JEN4), Canicola, Hardjo, Bratislava, and Grippotyphosa but not in the saprophytic species L. biflexa, the intermediate pathogen L. inadai, or the non-L. interrogans pathogenic species L. weilii (Fig. 2).
Fig 2.
Distribution of lruC in pathogenic and nonpathogenic Leptospira spp. Detection of lruC sequences by Southern blotting in pathogenic serovars of L. interrogans or other Leptospira spp.; Leptonema illini and L. interrogans serovar Pomona JEN4 were included as negative and positive controls, respectively.
Polyclonal antiserum was raised in New Zealand White rabbits by subcutaneous administration of 100 μg of recombinant protein and 1 μl of N-acetylmuramyl-l-alanyl-d-isoglutamine (Sigma, St. Louis, MO) adsorbed to aluminum hydroxide (Alhydrogel; Accurate Chemical & Scientific Corp., Westbury, NY). Booster injections contained 100 μg (subcutaneous) and 5 μg (intravenous) of the antigen and were administered 14 and 28 days after the primary immunization. Serum was obtained 35 days after the primary immunization. Rabbits used in this study were handled in accordance with relevant national and international guidelines, and all animal work was approved by the University of Kentucky Institutional Animal Care and Use Committee (970025A).
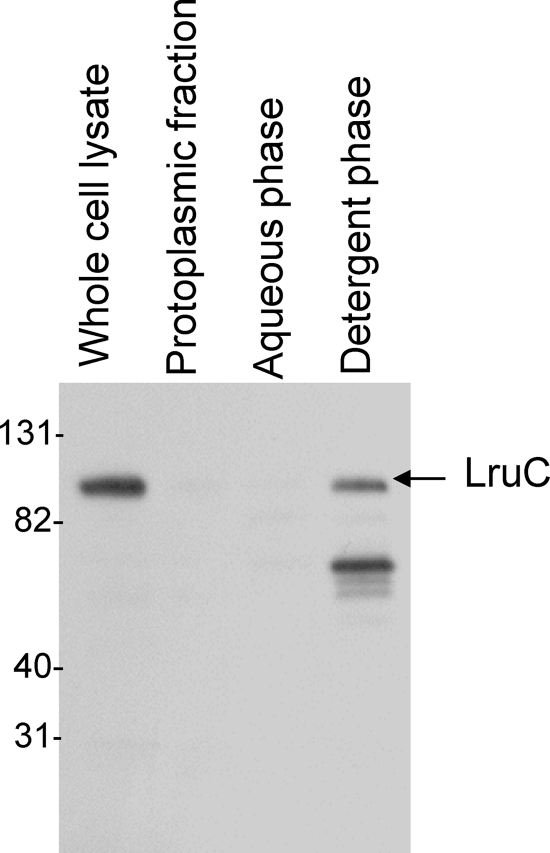
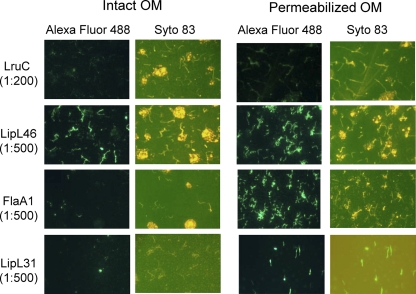
The outer membrane fraction of low (fourth)-passage L. interrogans serovar Copenhageni Fiocruz L1-130 was extracted by Triton X-114 solubilization and phase partitioning as described previously (11). Briefly, leptospires washed in phosphate-buffered saline containing 5 mM MgCl2 were extracted in 0.5% protein-grade Triton X-114 (Calbiochem), 150 mM NaCl, 10 mM Tris (pH 8.0), and 1 mM EDTA at 4°C. Insoluble material (protoplasmic cylinder) was pelleted by centrifugation at 16,000 × g for 10 min. Phase separation of the supernatant was performed by warming it to 37°C after the addition of 20 mM CaCl2, followed by centrifugation for 10 min at 1,000 × g. Proteins in aqueous and detergent phases were precipitated with acetone. Purity of separation of hydrophobic outer membrane proteins, hydrophilic periplasmic proteins, and the protoplasmic cylinder was confirmed by using antisera to known outer and inner membrane proteins LipL32 and LipL31, respectively (data not shown) (12, 14). The protoplasmic cylinder comprises the cytoplasm, the inner membrane, and the anchored periplasmic flagella (9). The detergent-soluble phase contains outer membrane constituents, and periplasmic proteins separate into the aqueous phase (20). These Triton X-114 fractions were separated on an SDS gel, transferred to a nitrocellulose membrane, and blotted with antiserum to LruC. LruC was detected exclusively in the outer membrane-rich detergent phase, indicating that it is an outer membrane protein (Fig. 3). The lower-molecular-weight band in the detergent phase lane is most likely a breakdown product of LruC. An outer membrane protein can be transported either to the outer surface or to the inner leaflet of the outer membrane of leptospires. To address this question, surface immunofluorescence studies were performed as described before (28, 29) with a minor modification. 4′6-Diamidino-2-phenyl-indole dihydrochloride (DAPI) was replaced with Syto 83 (Molecular Probes, Eugene, OR) DNA stain. Syto 83 was added to the leptospiral culture at a 1:1,000 dilution, and cells were incubated in the dark at 30°C for 1 h before the organisms were harvested as described previously (28, 29). LruC was undetectable in intact leptospires by this technique (Fig. 4), suggesting that either it was restricted to the inner leaflet of the leptospiral outer membrane, similar to what has been proposed for LipL36 (32), or LruC may be expressed on the surfaces of leptospires at a level that is insufficient for detection by this method.
Fig 3.
Localization of LruC by cellular fractionation with Triton X-114. Fractions of the detergent phase (outer membrane fraction), aqueous phase (periplasmic fraction), and protoplasmic cylinder (inner membrane plus cytoplasm fraction) were separated by SDS-polyacrylamide gel electrophoresis and analyzed by immunoblotting using rabbit antiserum specific for LruC. As a control for fractionation purity, fractions were also analyzed using rabbit antiserum to LipL32 and LipL31 (data not shown).
Fig 4.
Surface immunofluorescence assay (IFA). Intact or membrane-permeabilized L. interrogans was probed with rabbit immune sera specific for either LruC, outer membrane protein LipL46 (22), periplasmic flagellar protein FlaA1 (8), or intermembrane protein LipL31 (14). Binding of rabbit sera to leptospires was detected with Alexa Fluor 488-conjugated goat anti-rabbit IgG fragments. A Syto 83 counterstain was used to monitor the presence of spirochetes. The identities of individual proteins recognized by the particular antiserum and their dilutions are indicated on the left.
In our previous work, LruA and LruB were shown to cross-react with equine ocular components (33, 35a). In another study, ELISA using recombinant LruA and LruB as antigens also gave positive reactions with the sera from patients with the autoimmune conditions Behçet's disease and Fuchs uveitis, which may be due to the demonstrated cross-reactivity between antibodies to ocular autoantigens and LruA/B. Since these uveitis-associated leptospiral proteins cross-react with ocular components, we asked if LruC similarly cross-reacts with equine ocular tissues. Aqueous extracts were prepared from the ciliary body, cornea, lens, and retina of a normal eye from a young horse serologically negative for Leptospira spp. (27) and separated on a 12% polyacrylamide gel. Proteins were electrotransferred to nitrocellulose membrane and blocked with 4% nonfat dry milk in Tris-buffered saline–Tween (TBS-T). Membranes were incubated with either preimmunization serum or LruC antiserum followed by HRP-conjugated protein G (Invitrogen) and developed as described elsewhere (33). Monospecific antiserum to LruC did not react with tissue extracts of the ciliary body, cornea, lens, or retina (data not shown). Thus, LruC is probably not directly associated with autoimmune aspects of leptospiral uveitis.
In conclusion, we describe a novel leptospiral protein, LruC, expressed in the eyes of horses with lesions of chronic leptospiral uveitis. The gene encoding the protein is present in most of the pathogenic Leptospira spp. tested. LruC-specific antibodies are produced at significantly higher levels in eye fluids and sera of leptospiral uveitic horses. Together, these data suggest that LruC may have a role in the pathogenesis of leptospiral uveitis. Exploring this possibility in equine and human uveitis should be the focus of future studies.
Nucleotide sequence accession number.
The L. interrogans JEN4 lruC nucleotide sequence has been deposited in the GenBank database under accession number AY803757.
ACKNOWLEDGMENTS
This work was supported by the Keeneland Endowment. A. Verma was funded by a Paul Mellon Fellowship in Equine Studies. The support of Public Health Service grant AI-34431 (to D.A.H.) from the National Institute of Allergy and Infectious Diseases and VA Medical Research Funds (to D.A.H. and J.M.) is also acknowledged.
We thank Michael Donahue for Leptospira strains and Claire Adams, Amy Bowman, Catherine Brissette, Logan Burns, Alicia Chenail, Brandon Jutras, and Michael Woodman for helpful comments.
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Abrams K, Brooks DE. 1990. Equine recurrent uveitis: current concepts in diagnosis and treatment. Equine Pract. 12:27–35 [Google Scholar]
- 2. Ballard SA, Go M, Segers RP, Adler B. 1998. Molecular analysis of the dnaK locus of Leptospira interrogans serovar Copenhageni. Gene 216:21–29 [DOI] [PubMed] [Google Scholar]
- 3. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 4. Bryan J. 1989. Caldesmon, acidic amino acids and molecular weight determinations. J. Muscle Res. Cell Motil. 10:95–96 [DOI] [PubMed] [Google Scholar]
- 5. Bulach DM, et al. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. U. S. A. 103:14560–14565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chu KM, Rathinam R, Namperumalsamy P, Dean D. 1998. Identification of Leptospira species in the pathogenesis of uveitis and determination of clinical ocular characteristics in south India. J. Infect. Dis. 177:1314–1321 [DOI] [PubMed] [Google Scholar]
- 7. Cook CS, Harling DE. 1983. Equine recurrent uveitis. Equine Vet. J. 2:2–15 [Google Scholar]
- 8. Cullen PA, et al. 2005. Surfaceome of Leptospira spp. Infect. Immun. 73:4853–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Faine S, Adler B, Bolin C, Perolat P. 1999. Leptospira and leptospirosis, 2nd ed MediSci, Melbourne, Australia [Google Scholar]
- 10. Gilger BC, et al. 1999. Characterization of T-lymphocytes in the anterior uvea of eyes with chronic equine recurrent uveitis. Vet. Immunol. Immunopathol. 77:17–28 [DOI] [PubMed] [Google Scholar]
- 11. Haake DA, et al. 1991. Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect. Immun. 59:1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haake DA, et al. 2000. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 68:2276–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haake DA. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haake DA, Matsunaga J. 2002. Characterization of the leptospiral outer membrane and description of three novel leptospiral membrane proteins. Infect. Immun. 70:4936–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Halliwell RE, Brim TA, Hines MT, Wolf D, White FH. 1985. Studies on equine recurrent uveitis. II. The role of infection with Leptospira interrogans serovar Pomona. Curr. Eye Res. 4:1033–1040 [DOI] [PubMed] [Google Scholar]
- 16. Hartskeerl RA, et al. 2004. Classification of leptospira from the eyes of horses suffering from recurrent uveitis. J. Vet. Med. B 51:110–115 [DOI] [PubMed] [Google Scholar]
- 17. Hoyer LL. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176–180 [DOI] [PubMed] [Google Scholar]
- 18. Juncker AS, et al. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kalsow CM, Dwyer AE. 1998. Retinal immunopathology in horses with uveitis. Ocul. Immunol. Inflamm. 6:239–251 [DOI] [PubMed] [Google Scholar]
- 20. Maher PA, Singer SJ. 1985. Anomalous interaction of the acetylcholine receptor protein with the nonionic detergent Triton X-114. Proc. Natl. Acad. Sci. U. S. A. 82:958–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsunaga J, et al. 2003. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 49:929–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsunaga J, Werneid K, Zuerner RL, Frank A, Haake DA. 2006. LipL46 is a novel surface-exposed lipoprotein expressed during leptospiral dissemination in the mammalian host. Microbiology 152:3777–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller TR, Whitley RD. 1987. Uveitis in horses. Mod. Vet. Pract. 351-357. [Google Scholar]
- 24. Morter RL, Williams RD, Bolte H, Freeman MJ. 1969. Equine leptospirosis. J. Am. Vet. Med. Assoc. 155:436–442 [PubMed] [Google Scholar]
- 25. Nally JE, Artiushin S, Timoney JF. 2001. Molecular characterization of thermoinduced immunogenic proteins Qlp42 and Hsp15 of Leptospira interrogans. Infect. Immun. 69:7616–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nascimento AL, et al. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parma AE, Santisteban CG, Villalba JS, Bowden RA. 1985. Experimental detection of an antigenic relationship between Leptospira and equine cornea. Vet. Immunol. Immunopathol. 10:215–224 [DOI] [PubMed] [Google Scholar]
- 28. Pinne M, Haake DA. 2009. A comprehensive approach to identification of surface-exposed, outer membrane-spanning proteins of Leptospira interrogans. PLoS One 4:e6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pinne M, Haake DA. 2011. Immuno-fluorescence assay of leptospiral surface-exposed proteins. J. Vis. Exp. 2011:2805 doi:10.3791/2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ren SX, et al. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888–893 [DOI] [PubMed] [Google Scholar]
- 31. Setubal JC, Reis M, Matsunaga J, Haake DA. 2006. Lipoprotein computational prediction in spirochaetal genomes. Microbiology 152:113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shang ES, Summers TA, Haake DA. 1996. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 64:2322–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Verma A, Artiushin S, Matsunaga J, Haake DA, Timoney JF. 2005. LruA and LruB, novel lipoproteins of pathogenic Leptospira interrogans associated with equine recurrent uveitis. Infect. Immun. 73:7259–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Verma A, et al. 2006. LfhA, a novel factor H-binding protein of Leptospira interrogans. Infect. Immun. 74:2659–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Verma A, et al. 2008. LruA and LruB antibodies in sera of humans with leptospiral uveitis. Clin. Vaccine Immunol. 15:1019–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a. Verma A, Kumar P, Babb K, Timoney JF, Stevenson B. 2010. Cross reactivity of antibodies against leptospiral recurrent uveitis-associated proteins A and B (LruA and LurB) with eye proteins. PLoS Negl. 4(8):e778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Williams RD, Morter RL, Freeman MJ, Lavignette AM. 1971. Experimental chronic uveitis: ophthalmic signs following equine leptospirosis. Invest. Ophthalmol. 10:948–954 [PubMed] [Google Scholar]
- 37. Wollanke B, Rohrbach BW, Gerhards H. 2001. Serum and vitreous humor antibody titers in and isolation of Leptospira interrogans from horses with recurrent uveitis. J. Am. Vet. Med. Assoc. 219:795–800 [DOI] [PubMed] [Google Scholar]
- 38. Zhou M, Wu H. 2009. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology 155:317–327 [DOI] [PubMed] [Google Scholar]