Abstract
Salmonella enterica serovar Typhimurium is a facultative intracellular pathogen that causes inflammation, necrosis, and diarrhea in pigs, as well as being an important source of food-borne diseases in humans. Probiotics and prebiotics are promising alternatives to antibiotics to control and prevent intestinal infections. The present work investigated a recently developed β-galactomannan (βGM) prebiotic compared to the proven probiotic Saccharomyces cerevisiae var. boulardii on porcine ileum intestinal epithelial cells (IECs) of the IPI-2I line and monocyte-derived dendritic cells (DCs) cocultured in vitro with Salmonella. We observed that both S. cerevisiae var. boulardii and βGM inhibited the association of Salmonella with IECs in vitro. Our data indicated that βGM has a higher ability than S. cerevisiae var. boulardii to inhibit Salmonella-induced proinflammatory mRNA (cytokines tumor necrosis factor alpha [TNF-α], interleukin-1α [IL-1α], IL-6, and granulocyte-macrophage colony-stimulating factor [GM-CSF] and chemokines CCL2, CCL20, and CXCL8) and at protein levels (IL-6 and CXCL8). Additionally, βGM and S. cerevisiae var. boulardii induced some effects on DCs that were not observed on IECs: βGM and S. cerevisiae var. boulardii showed slight upregulation of mRNA for TNF-α, GM-CSF, and CCR7 receptor on porcine monocyte-derived dendritic cells (DCs). Indeed, the addition of βGM or S. cerevisiae var. boulardii on DCs cocultured with Salmonella showed higher gene expression (mRNA) for TNF-α, GM-CSF, and CXCL8 compared to that of the control with Salmonella. In conclusion, the addition of βGM inhibits Salmonella-induced proinflammatory profiles in IECs but may promote DC activation, although associated molecular mechanisms remain to be elucidated.
INTRODUCTION
Enteropathogenic Salmonella enterica subsp. enterica serovar Typhimurium is a Gram-negative, facultative intracellular pathogen that causes inflammation and necrosis of the small and large intestines of pigs, resulting in diarrhea that may be accompanied by generalized sepsis. Although all ages are susceptible, the disease is more frequent in weaned and growing finishing pigs (22). Moreover, in European countries, Salmonella Typhimurium is the serovar most frequently isolated from slaughter pigs, and is an important source of Salmonella infections in humans due to ingestion of contaminated food (12). However, the reduction of food-borne human diseases must be coherent with the European ban on antibiotic growth promoters (AGPs) for animal feeding (regulation [EC] no. 1831/2003). AGPs have been implicated in increased on-farm prevalence of bacteria resistant to antibiotics. Such bacteria are considered to represent a health hazard owing to potential transfer of resistance to bacteria pathogenic to humans (41). Probiotic and prebiotic feed additives are promising alternatives to AGPs because they influence the intestinal microbiota, reducing colonization by pathogenic bacteria and enhancing the mucosal immune system (14).
Our work presents in vitro screening of a novel prebiotic rich in β-galactomannan (βGM) and developed from the carob bean of the Ceratonia silliqua tree, in comparison with the proven probiotic yeast Saccharomyces cerevisiae var. boulardii. For a review of the proven beneficial effects of S. cerevisiae var. boulardii in the treatment and prevention of gastrointestinal diseases, see reference 44. We studied the effects of βGM and S. cerevisiae var. boulardii on porcine intestinal epithelial cells (IECs) and dendritic cells (DCs), which are crucial to maintain gut homeostasis and to develop strong immune responses against pathogens such as Salmonella (9, 33). In order to support the development of effective prebiotics and probiotics in animal production, we characterized the ability of βGM and S. cerevisiae var. boulardii to inhibit Salmonella association with IECs, the cytokine and chemokine regulation induced by Salmonella in IECs and DCs, and the modulatory effects of βGM and S. cerevisiae var. boulardii on both cell types cocultured with the pathogen.
MATERIALS AND METHODS
IEC culture.
The porcine small intestine epithelial cell line IPI-2I (ECACC 93100622) was established from the ileum of an adult boar (SLAd/d haplotype) (17). IPI-2I cells were maintained in Dulbecco's modified Eagle's medium (DMEM)-GlutaMAX (Invitrogen, Spain) supplemented with 10% fetal calf serum (FCS) (Invitrogen) and 10 μg/ml insulin (Sigma-Aldrich, Saint-Quentin, France). In all experiments, cells were cultured in 6-well plates (Nunc, Labclinics, Spain) to confluence. For scanning electron microscopy (SEM), cells were grown onto a coverslip placed inside the well to allow removal of the monolayer. Before the addition of pre- or probiotics and/or infection, cells were washed three times, and the cell culture was replaced with DMEM supplemented with 10 μg/ml insulin (Sigma-Aldrich). Cells were used between passages 30 and 70 and periodically tested to avoid Mycoplasma contamination (MycoAlert Mycoplasma detection kit; Lonza).
Probiotic and prebiotic preparation.
Lyophilized Saccharomyces cerevisiae var. boulardii (Biocodex, Laboratoires Montrouge, France) was rehydrated with 10 ml of DMEM-GlutaMAX and incubated for 30 min at 30°C. The yeast cells were then counted with a Neubauer cell counter with methyl blue to exclude nonviable yeast cells. The yeast cells were added to the selected wells at a multiplicity of infection (MOI) of 3 and incubated overnight at 37°C and 10% CO2.
Prebiotic βGM (Salmosan; patent WO2009/144070 A2, licensed to Industrial Técnica Pecuaria, ITPSA, Barcelona, Spain) consists of a β-(1–4)-mannose backbone with branched galactose molecules (1:4 galactose/mannose ratio) (38). For these in vitro experiments, βGM was diluted in DMEM-GlutaMAX (1 mg/ml), vortexed, and incubated for 30 min at 37°C. Immediately before the Salmonella infection, βGM was added to each well at 10 μg/ml.
Host cell-pathogen assay.
Pathogenic Salmonella enterica serovar Typhimurium (Salmonella) with antigenic formulae 4,12:i:1,2 and resistant to ampicillin, chloramphenicol, streptomycin, sulfonamide, and tetracycline was provided by Ignacio Badiola, Centre Recerca en Sanitat Animal (CReSA; IRTA-UAB, Bellaterra, Spain). Aliquots of Salmonella were provided in bacterial cryopreservers (Technical Service Consultants, Ferrer International, Spain) and stored at −80°C until use. Before infection of DCs or IECs, a single Salmonella cryopreserver was added to 20 ml of Luria-Bertani (LB) medium and cultured for 3 to 4 h at 37°C with 180 rpm rotational agitation (Multitron HT; Infors). For the infection, Salmonella was used during the exponential growth phase, as determined by absorbance at 600 nm (A600). Salmonella was used at an MOI of 4, as previously determined by cytotoxic lactate dehydrogenase (LDH) assays (Roche Applied Science, Spain) (data not shown). The optimal time of coculture was previously determined by proinflammatory gene expression and protein secretion (data not shown). Therefore, in vitro challenge lasted 3 h for gene expression and bacterial adherence studies and 24 h for supernatant cytokine determination. After host cell-pathogen coculture, cells or supernatants were, respectively, sampled and stored until analysis.
Cell-associated bacterial experiment.
Inhibition of Salmonella adherence and invasion was assessed on IECs grown to confluence and incubated with Salmonella (MOI of 4), with and without S. cerevisiae var. boulardii or βGM, for 3 h. After the host-pathogen assay, supernatant was removed and cells were washed twice with sterile phosphate-buffered saline (PBS) to eliminate all nonadherent bacteria. Cells were then homogenized with 1 ml of 0.1% Triton X-100 (Sigma-Aldrich) for 15 min. This solution was serially diluted in PBS, and 100 μl (dilution, 1 × 10−4) was plated in LB agar petri dishes for 24 h at 37°C to count the CFU. The ability of Salmonella to infect IPI-2I cells was quantified as cell-associated bacteria (adhering and intracellular), calculated as follows: % cell-associated bacteria = [(adhered and intracellular salmonellae on IPI-21 cells)/(total salmonellae added/well)] × 100.
To determine differences between experimental treatments, the relative percentage of cell-associated bacteria was calculated as % relative cell-associated bacteria (%) = [(CFU/ml treatment)/(CFU/ml control infection)] × 100.
SEM.
Preventive anti-Salmonella adherent abilities of S. cerevisiae var. boulardii and βGM on IECs were visualized by SEM. The IPI-2I cell culture was prepared as previously described by Mitjans and Ferrer (24), except that cells were fixed in cacodylate buffer (0.1 M [pH 7.4]). The samples were examined in a Zeiss DSM 940A (Oberkochen, Germany) electronic microscope, operating at 15 kV. Samples were processed and examined at the Scientific and Technological Centers of the University of Barcelona.
Isolation of mRNA and cDNA synthesis.
Cells were homogenized using TRIzol reagent (Invitrogen), and total RNA was isolated using Purelink RNA minikit (Invitrogen) according to the manufacturer's instructions. The RNA concentration was determined by measuring optical density at 260 nm (OD260), and the RNA quality was assessed by calculating OD260/OD280. The samples of RNA were then treated with the DNase I amplification-grade kit (Sigma-Aldrich) (1 U/μg of RNA). A total of 1 μg of RNA was used to generate cDNA by using the Transcriptor high-fidelity cDNA synthesis kit (Roche Applied Science). Briefly, 1 μg of RNA was incubated in a final volume of 20 μl containing 2 μl deoxynucleoside triphosphate (dNTP) (final concentration of 1 mM each), 1 μl oligo(dT) (2.5 μM), 1.1 μl Transcriptor high-fidelity reverse transcriptase (10 U/μl), 4 μl 5× Transcriptor high-fidelity reverse transcriptase buffer, and 1 μl of dithiothreitol (DTT; 5 mM) and completed with ultrapure water. The reaction was maintained for 30 min at 45°C and then heat inactivated at 85°C for 10 min. The generated cDNA was stored at −80°C until analysis.
mRNA expression analysis using quantitative real-time PCR.
The mRNA and primer sequences used in this study have been published (5, 19, 22, 23, 26, 42). These primers allowed the mRNA expression analysis of various genes involved in the innate immune response (Table 1). Quantitative real-time PCR (qPCR) was performed using 2 μl of cDNA synthesized as previously described combined with primer/probe sets and IQ SYBR green supermix (Bio-Rad, Hercules, CA) according to the manufacturer's recommendations. Each qPCR included a reverse transcription negative control (RNA sample without reverse transcriptase) to check the absence of genomic DNA. The qPCR conditions were 98°C for 30 s, followed by 37 cycles with denaturation at 95°C for 15 s and annealing/elongation for 30 s (at the annealing temperatures shown in Table 1). Real-time assays were run on a Bio-Rad iQ5. The specificity of the qPCRs was assessed by analyzing the melting curves of the products and size verification of the amplicons. To minimize sample variation, we used identical numbers of cells and high-quality RNA. Samples were normalized internally using simultaneously the average cycle threshold (Cq) (7) of genes coding for hypoxanthine phosphoribosyltransferase 1 (HPRT1), ribosomal protein L19 (RPL19), and TATA box binding protein 1 (TBP-1) as reference genes in each sample to avoid any artifact of variation in the target gene. We used these three reference genes given their stability in porcine cells (5, 13) determined using calculated geNorm application (36). A standard curve was generated using diluted cDNA. The correlation coefficients of the standard curves were >0.995, and the concentrations of the test samples were calculated from the standard curves, according to the formula y = −M × Cq + B, where M is the slope of the curve, Cq is the point during the exponential phase of amplification at which the fluorescent signal is first recorded as being statistically significant above background, and B is the y-axis intercept. Cq values were used to calculate the qPCR efficiency from the given slope according to the equation qPCR efficiency = [10(−1/M) −1] × 100. All qPCRs displayed efficiency between 90% and 110% and were performed following MIQE guidelines (7). Expression data are expressed as relative values after Genex macroanalysis with three reference genes (Bio-Rad, Hercules, CA) (36).
Table 1.
Primer sequences and annealing temperatures of primer sets, expected PCR fragment sizes, and associated references
| Gene coding for protein showna | Primer sequence |
Annealing temp (°C) | Product length (bp) | Accession no. | Reference | |
|---|---|---|---|---|---|---|
| Sense | Antisense | |||||
| APRIL | TGCTCACCCGTAAACAGAAG | TAAACTCCAGCATCCCAGAC | 60 | 172 | EST BP170456 | 22 |
| BAFF | GAGAGCAGCTCCATTCAAAG | GCATGCCACTGTCTGCAATC | 60 | 103 | NM_001097498 | 22 |
| CCL2 | GTCACCAGCAGCAAGTGTC | CCAGGTGGCTTATGGAGTC | 60 | 112 | EF107669 | 23 |
| CCL17 | TGCTGCTCCTGGTTGCTCTC | ATGGCGTCCCTGGTACACTC | 67 | 169 | EST DB794536 | 5 |
| CCL20 | GCTCCTGGCTGCTTTGATGTC | CATTGGCGAGCTGCTGTGTG | 66 | 146 | NM 001024589 | 22 |
| CCR7 | AGGAGGCTCAAGACCATGAC | GATGCCGAAGATGAGTTTGC | 62 | 147 | AB090356 | |
| CXCL2 | TGCTGCTCCTGCTTCTAGTG | TGGCTATGACTTCCGTTTGG | 60 | 171 | NM_001001861 | 22 |
| GM-CSF | GAAACCGTAGACGTCGTCTG | GTGCTGCTCATAGTGCTTGG | 62 | 150 | DQ108393 | 19 |
| HPRT1 | GGACTTGAATCATGTTTGTG | CAGATGTTTCCAAACTCAAC | 60 | 91 | DQ815175 | 26 |
| IL-1α | CCCGTCAGGTCAATACCTC | GCAACACGGGTTCGTCTTC | 60 | 170 | NM 214029 | 23 |
| IL-6 | ATCAGGAGACCTGCTTGATG | TGGTGGCTTTGTCTGGATTC | 62 | 177 | NM_214399 | 23 |
| CXCL8 | TCCTGCTTTCTGCAGCTCTC | GGGTGGAAAGGTGTGGAATG | 62 | 100 | NM_213867 | 22 |
| IL-10 | GGTTGCCAAGCCTTGTCAG | AGGCACTCTTCACCTCCTC | 60 | 202 | NM_214041 | 19 |
| RPL19 | AACTCCCGTCAGCAGATCC | AGTACCCTTCCGCTTACCG | 60 | 147 | AF435591 | 22 |
| TBP-1 | AACAGTTCAGTAGTTATGAGCCAGA | AGATGTTCTCAAACGCTTCG | 60 | 153 | DQ845178 | 26 |
| TLR4 | TGTGCGTGTGAACACCAGAC | AGGTGGCGTTCCTGAAACTC | 62 | 136 | NM_001113039 | 22 |
| CXCL10 | CCCACATGTTGAGATCATTGC | CATCCTTATCAGTAGTGCCG | 60 | 168 | 42 | |
| TNF-α | CCAATGGCAGAGTGGGTATG | TGAAGAGGACCTGGGAGTAG | 62 | 116 | X54859 | 22 |
Reference gene products are underlined.
Determination of cytokine production.
Cytokine protein determination in the culture supernatant was performed by enzyme-linked immunosorbent assays (ELISAs). After 3 h of host cell-pathogen assay performed as described above, 75 μg/ml of gentamicin (Sigma-Aldrich) was added to each well to avoid bacterial overgrowth. Cell culture supernatant was collected after 24 h and stored at −80°C until analysis. Swine interleukin-6 (IL-6) and CXCL8 DuoSet ELISA kits (R&D Systems, Vitro SP, Spain) were used according to the manufacturer's recommendations.
Isolation of PBMCs and differentiation of monocyte derived dendritic cells.
Blood samples were obtained from 6- to 12-month-old large white pigs at the slaughterhouse. Blood was collected into heparinized tubes and followed the protocol described by Pilon et al. (27), with a few modifications. Briefly, peripheral blood mononuclear cells (PBMCs) were isolated by centrifugation (1,000 × g for 30 min) over Ficoll (density = 1.077; Histopaque, Sigma-Aldrich, France). Red blood lysing solution (Sigma-Aldrich) was used to remove remaining erythrocytes. Cells were then resuspended in RPMI-GlutaMAX (Gibco, Invitrogen) containing 2.5% fetal bovine serum (FBS), 1% penicillin–streptomycin, and 50 μM 2-β-mercaptoethanol (Sigma-Aldrich). Next, 150 × 106 cells/20 ml were plated in 150-cm2 cellBind flasks (Corning, Afora, Spain) and incubated for 30 min at 37°C and 5% CO2.Then, the flasks were washed with RPMI to remove all nonadherent cells (lymphocytes). To induce differentiation, monocytes were cultured with RPMI-GlutaMAX medium containing 1% penicillin–streptomycin antibiotic, 10% FBS, 50 μM β-mercaptoethanol, and swine recombinant cytokines IL-4 (100 ng/ml) and granulocyte-macrophage colony-stimulating factor (GM-CSF; 20 ng/ml) (Biosource, Invitrogen) for 6 days at 37°C and 5% CO2. On day 3, fresh medium and cytokines were added at the same concentrations used previously.
DC phenotyping.
After 6 days of culture, cells showed typical DC morphology. In addition, DCs were characterized as CD172a+ (SWC3), swine leukocyte antigen (SLA) class II-DQ+, swine leukocyte antigen (SLA) class II-DR+, CD80/86+, CD14mod, and CD11R1−. Antibodies for cell surface markers CD172a/SWC3, SLA class II-DQ, SLA class II-DR, and CD11R1 were provided by J. Domínguez (INIA, Madrid, Spain). Antibody for CD14 determination was purchased from Acris Antibodies (AntibodyBCN, Barcelona, Spain), and for CD80/CD86, we used recombinant human cytotoxic T-lymphocyte-associated molecule-4/Fc fusion protein (CTLA4-Fc IgG1; Invitrogen). The fluorescein isothiocyanate (FITC)-conjugated anti-human immunoglobulin IgG1 or Zenon tricolor mouse IgG1 and IgG2a labeling kits (Invitrogen) were used for detection by flow cytometry (FACSCanto using FACSDiva software; BD Biosciences, San José, CA).
Pathogen-induced dendritic cell activation.
After 6 days of culture, DCs were recovered and adjusted to 5 × 105 to 1 × 106 DCs/well in 24-well plates. Optimal pathogen-induced activation was previously determined by proinflammatory gene expression by qPCR (data not shown). DCs were then incubated with βGM and S. cerevisiae var. boulardii, respectively, and challenged with Salmonella (MOI of 4). After 3 h of exposure, supernatants were discarded and cells were collected in TRIzol reagent. Isolation of DC mRNA and gene expression studies were performed as described above.
Statistical analysis.
All statistical analyses were performed using the General Linear Model Procedure (PROC GLM) of SAS software version 9.1.3 (SAS Institute, Carey, NC). Means for cell-associated bacterial percentages, mRNA, and protein secretion were considered in a 2-by-3 factorial design (two infection levels × 3 experimental treatments) with Duncan's posttest for grouping analysis. The probability value P ≤ 0.05 was considered to be significant. On the figures, superscript letters are used to designate statistical significance: mean values with no common superscript letters indicate a statistically significant difference (P ≤ 0.05), and mean values with the same superscript letters indicate no statistically significant difference (P > 0.05).
RESULTS
Determination of cell-associated bacteria.
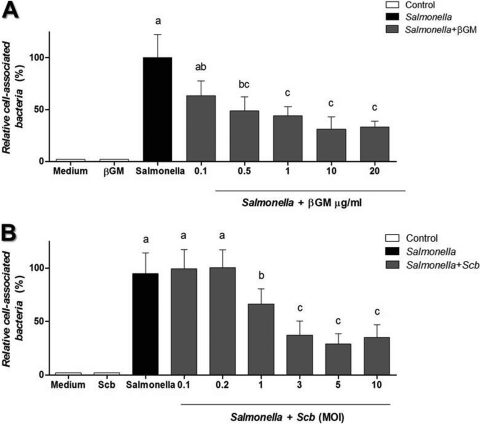
Cell-associated bacteria on IECs were measured to assess the ability of S. cerevisiae var. boulardii and βGM to bind to Salmonella, as an indicator of the potential of S. cerevisiae var. boulardii and βGM to inhibit Salmonella colonization of the intestinal tract. The coculture of Salmonella (∼4 × 106 CFU) and IECs (∼1 × 106 cells/well) showed that approximately 45% of added salmonellae became cell associated (data not shown). The presence of βGM (10 μg/ml) (Fig. 1A) or S. cerevisiae var. boulardii (MOI of 3) (Fig. 1B) significantly inhibited Salmonella association to around 50% of control values (P < 0.001) (Fig. 1). These optimal doses of S. cerevisiae var. boulardii or βGM were chosen for the following assays.
Fig 1.
Cell-associated Salmonella on IECs in the presence of βGM or S. cerevisiae var. boulardii (Scb). Adherence and/or invasion of Salmonella on IECs cocultured with βGM (A) or S. cerevisiae var. boulardii (B) is inhibited in a dose-dependent manner. Data (n = 5) are expressed as mean percentages ± standard deviations (SDs). Columns within each histogram with no common superscripts are significantly different (P < 0.05).
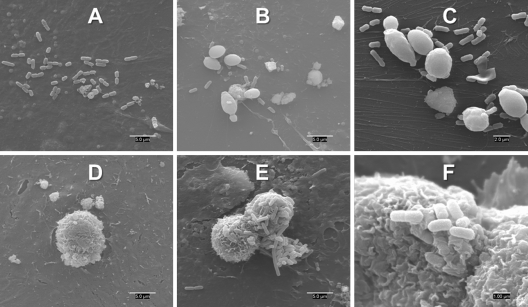
Scanning electronic microscopy.
SEM images of IECs infected with Salmonella confirm bacterial attachment to the cell surface (Fig. 2A). The images also reveal that βGM mainly shows a spherical structure (Fig. 2D) with salmonellae attached to the surface and thus reducing the density in surrounding bacteria adhered to the epithelium (Fig. 2E to F). Regarding the incubation with S. cerevisiae var. boulardii, the yeast appears with its characteristic ovoid structure (Fig. 2B and C). In this case, there is also a reduction in the surrounding attached bacteria due to the concentration of microorganisms near the yeast.
Fig 2.
Interaction of Salmonella with βGM or S. cerevisiae var. boulardii on the surface of IPI-2I cells assessed by scanning electron microscopy. Images show Salmonella attachment on control IPI-2I cells (A), Salmonella with S. cerevisiae var. boulardii (B and C), control βGM (D), and Salmonella with βGM (E and F).
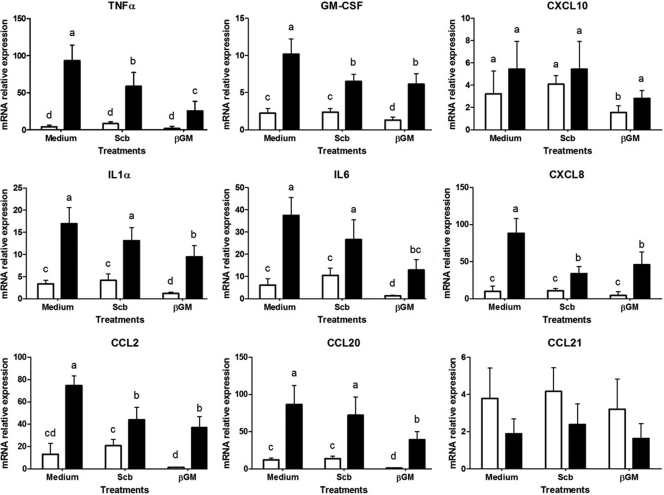
Cytokine and chemokine mRNA expression on IECs.
To assess the preventive effect of S. cerevisiae var. boulardii and βGM on the early immune response to Salmonella, we studied the mRNA expression of several proinflammatory cytokines and chemokines. The Salmonella coculture induced a large upregulation in mRNA levels of proinflammatory cytokines compared to controls without Salmonella (Fig. 3) (P < 0.001) for tumor necrosis factor-α (TNF-α; 22.6-fold), granulocyte-macrophage colony-stimulating factor (GM-CSF; 4.5-fold), interleukin-1α (IL-1α; 5-fold), and IL-6 (6-fold), as well as for chemokine (C-X-C motif) ligand 8 (CXCL8; 7-fold), chemokine (C-C motif) ligand 2 (CCL2; 5.7-fold), and CCL20 (7.2-fold). Despite there being no statistically significant differences, there was a slight increase in expression of chemokine CXCL10 (1.48-fold), whereas CCL21 tended to decrease (P < 0.07, 2-fold decrease) after Salmonella coculture.
Fig 3.
Effects of S. cerevisiae var. boulardii (Scb) and βGM on cytokine and chemokine mRNA expression in IECs cultured with Salmonella. IECs (1 × 106 cells/well) were cocultured with S. cerevisiae var. boulardii (3 yeast cells/cell) or βGM (10 μg/ml) with Salmonella (MOI of 4) for 3 h. Data (n = 6) are presented as means of mRNA relative expression ± SDs. Columns within each histogram with no common superscripts are significantly different (P < 0.05). Results are representative of 3 independent experiments. ☐, control; ■, Salmonella.
The addition of S. cerevisiae var. boulardii or βGM did not induce proinflammatory effects per se compared to control cells (Fig. 3). Indeed, βGM showed a slight anti-inflammatory effect (2-fold decreases) for GM-CSF, IL-1α, IL-6, and CCL20 compared to control cells (P < 0.05) (Fig. 3). We observed up to 70% inhibition of Salmonella-induced mRNA expression of TNF-α, GM-CSF, and CCL20 in IECs treated with 10 μg/ml βGM (P < 0.001) (Fig. 3). Furthermore, gene expression of IL-1α, IL-6, CCL2, and CXCL8 was between 1.5- and 3-fold-decreased in βGM-treated cells compared to the Salmonella control. Likewise, the addition of S. cerevisiae var. boulardii induced between 2.6- and 1.4-fold inhibition of Salmonella-induced mRNA for the TNF-α, GM-CSF, CXCL8, and CCL2 genes (P < 0.05) (Fig. 3). However, no significant differences were observed for IL-1α, IL-6, and CCL20 in S. cerevisiae var. boulardii-treated cells compared to the Salmonella control (Fig. 3).
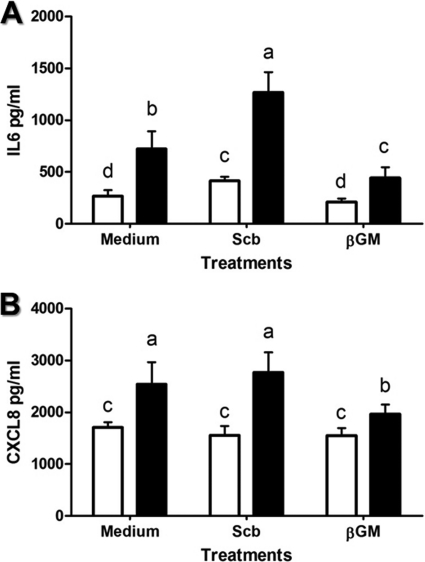
Cytokine production.
The preventive effect of βGM and S. cerevisiae var. boulardii on the Salmonella-induced proinflammatory IEC response was evaluated by the determination of secreted cytokine IL-6 and chemokine CXCL8, determined by ELISA 24 h after infection. The addition of βGM did not induce proinflammatory effects (Fig. 4). However, secretion of IL-6 was upregulated (1.5-fold; P < 0.05) in S. cerevisiae var. boulardii-treated cells, but no changes were observed for CXCL8. Coculture with Salmonella triggered up to 3-fold upregulation for IL-6 concentration (P < 0.01) (Fig. 4A) and 1.4-fold increase for CXCL8 (Fig. 4B) compared to control wells. Salmonella-induced secretion of IL-6 and CXCL8 was 38% (Fig. 4A) and 20% inhibited, respectively, in βGM-treated cells compared to the Salmonella group (P < 0.05) (Fig. 4B). On the other hand, coculture of S. cerevisiae var. boulardii before Salmonella infection did not prevent reduced Salmonella-induced secretion of IL-6 and CXCL8 (Fig. 4A and B).
Fig 4.
Effect of S. cerevisiae var. boulardii (Scb) and βGM on IL-6 and CXCL8 secretion induced by Salmonella. The cytokine IL-6 (A) and chemokine CXCL8 (B) concentrations in supernatants from IECs (1 × 106 cells/well) cocultured for 24 h with Salmonella (MOI of 4) are decreased by βGM (10 μg/ml). Data (n = 6) are presented as means ± SDs. Columns within each histogram with different superscripts are significantly different (P < 0.05). Data are representative of 3 independent experiments. ☐, control; ■, Salmonella.
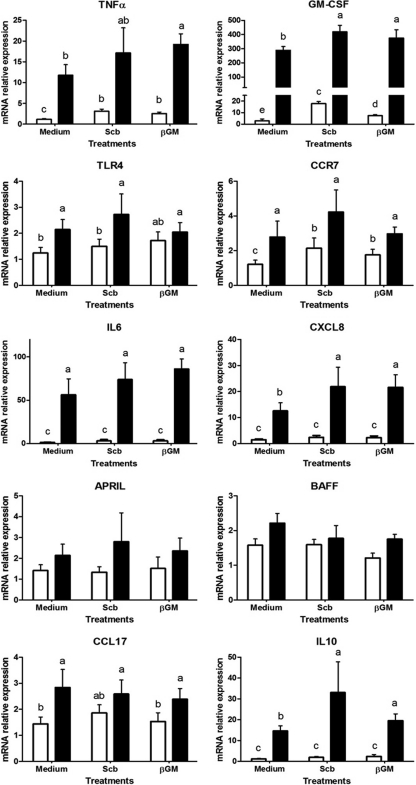
Modulation of mRNA expression of porcine monocyte-derived DCs.
Modulation of DCs by S. cerevisiae var. boulardii and βGM was studied by mRNA gene expression in DCs after Salmonella coculture. We observed Salmonella-induced mRNA upregulation for TNF-α (10-fold), GM-CSF (100-fold), Toll-like receptor 4 (TLR4; 1.7-fold), CCR7 (2.3-fold), IL-6 (38.5-fold), CXCL8 (8.4-fold), CCL17 (2-fold), and IL-10 (12-fold) genes compared to the control DCs (P < 0.001) (Fig. 5). The highest upregulation for these proinflammatory cytokines, chemokines, and immune-related receptors was obtained with Salmonella at an MOI of 5 (data not shown). A near-significant trend (P < 0.06) was observed for other genes, such as A proliferation-inducing ligand (APRIL) and B-cell activating factor (BAFF) (Fig. 5).
Fig 5.
Salmonella-induced gene expression in porcine DCs cocultured with S. cerevisiae var. boulardii (Scb) or βGM. Relative mRNA expression of proinflammatory cytokines (TNF-α, GM-CSF, IL-6, and IL-10), chemokines (CXCL8 and CCL17), receptors (CCR7 and TLR4), and regulatory factors (APRIL and BAFF) in DCs is enhanced by Salmonella. Data (n = 6) are presented as means of mRNA relative expression ± SDs. Columns with no common superscripts are significantly different (P < 0.05). ☐, control; ■, Salmonella.
Both S. cerevisiae var. boulardii and βGM induced upregulation of the TNF-α (2.5-fold), GM-CSF (3-fold and 6-fold, respectively) and CCR7 (1.7-fold) genes compared to untreated DCs (P < 0.05) (Fig. 5). The coculture with βGM or S. cerevisiae var. boulardii increased between 1.5- and 2.3-fold Salmonella-induced mRNA for TNF-α, GM-CSF, CXCL8, and IL-10 compared to the Salmonella-challenged DCs (Fig. 5) (P < 0.05). However, this effect was not observed for CCR7, TLR4, and CCL17 (Fig. 5).
DISCUSSION
Developing probiotic and prebiotic alternatives to AGPs is especially challenging in the area of prevention of intestinal infections, particularly Salmonella and Escherichia coli (6). Prebiotics and probiotics are believed to combat pathogens using less costly resources, reducing the drain on energy due to innate immune responses (3), and modulating IEC and DC functionality (14), thus helping to preserve gut homeostasis.
The spread of Salmonella from the intestinal lumen to other host tissues mainly occurs in Peyer's patches (PP) of the distal ileum, through compromised M cells, via enterocytes, or through the DC dendrites of the follicle-associated epithelium (FAE) (34), causing a huge proinflammatory profile in IECs and DCs. These cells orchestrate a rapid innate immune response to confine invading bacteria and to prevent Salmonella dissemination to other tissues (see references 10 and 11 for review). Besides constituting a physical barrier, IECs sense pathogens through pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), and secrete several signaling cytokines and chemokines (11). Arce et al. (2) showed that recognition of Salmonella lipopolysaccharide (LPS) by TLR4 of intestinal IPI-2I cells increases proinflammatory mRNA for TNF-α and CXCL8 (neutrophil recruitment) and slightly upregulates CCL2 (monocyte chemotaxis). The present data update our knowledge about IEC response after Salmonella exposure. We detected that Salmonella also induces an important mRNA proinflammatory response in IECs, involving IL-1α, IL-6 (acute-phase reactions, proliferation, and differentiation of macrophages and B cells), GM-CSF (proliferation and activation of neutrophils and macrophages), and a trend for CXCL10, which is a potent chemoattractant of Th1-type CD4+ and NK cells. These results confirm mucosal immune orientation toward a Th1 response related to STAT-4 transcription factor after Salmonella infection assessed in the porcine in vivo intestinal gut loop model (22). Furthermore, challenged IECs enhance immature DC chemotaxis (upregulation of CCL20) versus the mature DC response (downregulation of CCL21) to promote bacterial uptake across the epithelial barrier. Additionally, our data show that Salmonella induces an activated phenotype of porcine monocyte-derived DCs, as shown for the expression of CCR7 receptor and several proinflammatory cytokines (TNF-α, IL-6, and GM-CSF) and chemokines (CXCL8 and CCL17) (Fig. 5). However, no changes were observed for A proliferation-inducing ligand (APRIL) and B-cell activating factor (BAFF), which in humans are related to direct modulation of the local immunoglobulin A switch and enhanced B-cell survival and proliferation, respectively (20).
In our study, βGM and S. cerevisiae var. boulardii showed different immune modulatory abilities when IECs and DCs were infected with Salmonella. We observed that βGM and S. cerevisiae var. boulardii behave differently with respect to inhibition of Salmonella-induced mRNA and secretion of proteins containing genes involved in inflammation (TNF-α, GM-CSF, IL-1α, and IL-6) and recruitment/activation of immune cells (CXCL8, CCL2, CXCL10, and CCL20). The anti-inflammatory properties of the probiotic S. cerevisiae var. boulardii have been related to the secretion of small molecules (<10 kDa) that interfere with phosphorylation of mitogen-activated protein kinase (MAPK), extracellular signal-regulated kinase 1/2 (ERK1/2), and p38, inhibiting nuclear factor κB (NF-κB) transcription factor, which regulates the expression of proinflammatory genes in human colonic T-84 cells cocultured with Salmonella (21) and porcine intestinal epithelial cells of the IPEC-1 line upon enterotoxigenic E. coli F4 (K88) in vitro infection (42). Our data are partially in agreement with those of Martins et al. (21) describing inhibitory effects of S. cerevisiae var. boulardii on Salmonella-induced mRNA for CXCL8 in T-84 cells. However, we observed a reduced anti-inflammatory effect of S. cerevisiae var. boulardii on IECs cocultured with Salmonella. Indeed, βGM has higher inhibitory effects on Salmonella-induced mRNA (TNF-α, IL-1α, and IL-6) and protein IL-6 and CXCL8 secretion compared to S. cerevisiae var. boulardii. To our knowledge, few data suggest direct modulation of proinflammatory gene expression by mannan polysaccharides (25). Nevertheless, their anti-inflammatory effects are mainly related to the prevention of pathogen adhesion and invasion, as discussed below.
Present results for cell-associated bacteria and SEM images for IECs describe how βGM and S. cerevisiae var. boulardii interact with Salmonella and reduce pathogen attachment and invasion of porcine intestinal epithelial cells in a dose-dependent manner. Previous studies in our laboratory showed that βGM and S. cerevisiae var. boulardii also block pathogenic E. coli attachment on IPI-2I cells (2a) in a similar way to other commercially available galactoligosaccharides, using Caco-2 and Hep-2 cells (32). These previous data also confirmed the preventive properties of S. cerevisiae var. boulardii against E. coli infections (43), related to species- or strain-specific effects of Saccharomyces yeasts (35). Enteropathogenic species of Salmonella and E. coli share a type I fimbria or pilus structure that contains multiple subunits of bacterial lectins that bind to mannan units of the glycoproteins on the surface of host cells (1, 8, 30). As a source of mannans, βGM and S. cerevisiae var. boulardii may mimic the host cell receptor to which the pathogen adheres (32, 39). In that sense, Searle et al. (28, 29) showed that nondigestible oligosaccharides (NDOs) such as galactoligosaccharides reduce Salmonella in vitro adhesion and invasion in the human colonic HT-29-19A cell line and also in the ileum gut loop model in mice. Furthermore, S. cerevisiae var. boulardii has already been described to bind Salmonella on its surface (15), thus preserving intestinal barrier function through inhibition of pathogen adhesion and invasion of T-84 cells (21). Together, these data suggest that products rich in mannanoligosaccharides, such as βGM and S. cerevisiae var. boulardii, may have prophylactic roles against porcine pathogens bearing type I fimbriae (4).
The ability of βGM and S. cerevisiae var. boulardii to modulate DC maturation directly correlates with previous published data (40) indicating that their role in immune regulation is related to the structure and size of mannans or β-glucans (40). The present study shows that Salmonella-induced maturation is slightly enhanced in βGM-treated DCs compared to the control group for some of the studied genes (coding for TNF-α, IL-6, CXCL8, and IL-10). We hypothesize that βGM may modify Salmonella structures or antigens, and this might increase their recognition by PRRs of DCs. In contrast to our data, mannan-coated structures have been described (31) to activate TLR4 signaling pathways in a dose-dependent manner in murine DCs, triggering the expression of costimulatory molecules CD40, CD80, and CD86 and leading to an mRNA upregulation for IL-1-β and TNF-α cytokines among other Th1/Th2 cytokines. Since no differences were observed in TLR4 gene expression of DCs treated with S. cerevisiae var. boulardii or βGM, we alternatively propose C-type lectin receptors (CLRs) expressed on DCs, such as mannose receptor (MR) or dectin-2 (16), which have high affinity for mannose residues and are associated with the Th17 response (37) against intracellular pathogens (18) for the recognition of mannan-coated structures that lead to DC activation and maturation. Further studies may determine the contribution of C-type lectin receptors, especially MR, to sense mannan-coated structures upon Salmonella infection. To elucidate further possible modes of actions of βGM and S. cerevisiae var. boulardii in animal production, the biological relevance of these in vitro results may be characterized by future research approaches—for example, three-dimensional coculture (45) or gut loop intestinal models (22)—to establish the cross talk of IECs, DCs, and other cell types involved in mucosal immune responses, such as monocytes/macrophages, neutrophils, and intraepithelial lymphocytes.
In conclusion, this study demonstrates that prebiotic βGM and probiotic S. cerevisiae var. boulardii interact with Salmonella. The prebiotic βGM has higher anti-inflammatory properties in IECs cocultured with Salmonella compared to the proven S. cerevisiae var. boulardii probiotic. The present work also provides some evidence about how βGM and S. cerevisiae var. boulardii may modulate DC maturation directly and also when faced with Salmonella infection. These studies provide some interesting data about the in vitro effect of prebiotic βGM and probiotic S. cerevisiae var. boulardii, both considered as natural alternatives to AGPs in combatting Salmonella infection.
ACKNOWLEDGMENTS
This work was supported by grant AGL 2009-11936 from the Ministerio de Ciencia e Innovación (MICIIN; Spain). We gratefully acknowledge I. Badiola (CReSA) for providing the Salmonella Typhimurium strain and J. Domínguez-Juncal (Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria, INIA, Madrid, Spain) for providing pig cell surface marker antibodies.
J.B. is one of the inventors of the patent Salmosan WO2009/144070 A2 commercially licensed to Industrial Técnica Pecuaria (ITPSA, Barcelona, Spain). This does not alter the author's adherence to all the Clinical and Vaccine Immunology policies on data collection and analysis, preparation of the manuscript, or sharing data and materials. The other authors declare that they have no competing interests.
R.B. conceived the study and carried out IEC and DC host cell-pathogen cocultures, molecular studies, and immunoassays. I.D. participated in IEC host cell-pathogen assays and molecular analysis. R.M.-V., M.T.B., and A.M.G.-Z. performed scanning microscopy studies. R.L. participated in experimental design and statistical analysis. R.M.-V., R.F., P.M., H.S., and J.B. equally conceived and coordinated the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Footnotes
Published ahead of print 1 February 2012
REFERENCES
- 1. Althouse C, Patterson S, Fedorka-Cray P, Isaacson RE. 2003. Type 1 fimbriae of Salmonella enterica serovar Typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect. Immun. 71: 6446–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arce C, Ramírez-Boo M, Lucena C, Garrido JJ. 2010. Innate immune activation of swine intestinal epithelial cell lines (IPEC-J2 and IPI-2I) in response to LPS from Salmonella typhimurium. Comp. Immunol. Microbiol. Infect. Dis. 33: 161–174 [DOI] [PubMed] [Google Scholar]
- 2a. Badia R, et al. 25 January 2012. Effect of Saccharomyces cerevisiae var. Boulardii and beta-galactomannan oligosaccharide on porcine intestinal epithelial and dendritic cells challenged in vitro with Escherichia coli F4 (K88). Vet. Res. [Epub ahead of print.]doi: 10.1186/1297-9716-43-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey M. 2009. The mucosal immune system: recent developments and future directions in the pig. Dev. Comp. Immunol. 33: 375–383 [DOI] [PubMed] [Google Scholar]
- 4. Becker PM, Galletti S. 2008. Food and feed components for gut health-promoting adhesion of E. coli and Salmonella enterica. J. Sci. Food Agric. 88: 2026–2035 [Google Scholar]
- 5. Bruel T, et al. 2010. Epithelial induction of porcine suppressor of cytokine signaling 2 (SOCS2) gene expression in response to Entamoeba histolytica. Dev. Comp. Immunol. 34: 562–571 [DOI] [PubMed] [Google Scholar]
- 6. Brufau J. 2003. Animal feeding in Europe: challenges and opportunities, p 15–4 In Lyons TP, Jacques KA. (ed), Nutritional biotechnology in the feed and food industries. Notthingham University Press, Nottingham, United Kingdom [Google Scholar]
- 7. Bustin SA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55: 611–622 [DOI] [PubMed] [Google Scholar]
- 8. Capitani G, Eidam O, Glockshuber R, Grütter MG. 2006. Structural and functional insights into the assembly of type 1 pili from Escherichia coli. Microbes Infect. 8: 2284–2290 [DOI] [PubMed] [Google Scholar]
- 9. Devriendt B, Stuyven E, Verdonck F, Goddeeris BM, Cox E. 2010. Enterotoxigenic Escherichia coli (K88) induce proinflammatory responses in porcine intestinal epithelial cells. Dev. Comp. Immunol. 34: 1175–1182 [DOI] [PubMed] [Google Scholar]
- 10. Eckmann L, Kagnoff MF. 2001. Cytokines in host defense against Salmonella. Microbes Infect. 3: 1191–1200 [DOI] [PubMed] [Google Scholar]
- 11. Eckmann L, Kagnoff MF. 2005. Intestinal mucosal responses to microbial infection. Springer Semin. Immunopathol. 27: 181–196 [DOI] [PubMed] [Google Scholar]
- 12. European Food Safety Authority 2006. The Community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2005. EFSA J. 94: 1–228 [Google Scholar]
- 13. Facci MR, et al. 2011. Stability of expression of reference genes in porcine peripheral blood mononuclear and dendritic cells. Vet. Immunol. Immunopathol. 141: 11–15 [DOI] [PubMed] [Google Scholar]
- 14. Gaggìa F, Mattarelli P, Biavati B. 2010. Probiotics and prebiotics in animal feeding for safe food production. Int. J. Food Microbiol. 141(Suppl 1): S15–S28 [DOI] [PubMed] [Google Scholar]
- 15. Gedek BR. 1999. Adherence of Escherichia coli serogroup O 157 and the Salmonella typhimurium mutant DT 104 to the surface of Saccharomyces boulardii. Mycoses 42: 261–264 [DOI] [PubMed] [Google Scholar]
- 16. Hollmig ST, Ariizumi K, Cruz PD. 2009. Recognition of non-self-polysaccharides by C-type lectin receptors dectin-1 and dectin-2. Glycobiology 19: 568–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaeffer B, Bottreau E, Velge P, Pardon P. 1993. Epithelioid and fibroblastic cell lines derived from the ileum of an adult histocompatible miniature boar (d/d haplotype) and immortalized by SV40 plasmid. Eur. J. Cell Biol. 62: 152–162 [PubMed] [Google Scholar]
- 18. Khader SA, Gopal R. 2010. IL-17 in protective immunity to intracellular pathogens. Virulence 1: 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levast B, et al. 2010. Ultra-early weaning in piglets results in low serum IgA concentration and IL17 mRNA expression. Vet. Immunol. Immunopathol. 137: 261–268 [DOI] [PubMed] [Google Scholar]
- 20. Mackay F, Schneider P, Rennert P, Browning J. 2003. BAFF and APRIL: a tutorial on B cell survival. Annu. Rev. Immunol. 21: 231–264 [DOI] [PubMed] [Google Scholar]
- 21. Martins FS, et al. 2010. Interaction of Saccharomyces boulardii with Salmonella enterica serovar Typhimurium protects mice and modifies T84 cell response to the infection. PLoS One 5: e8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meurens F, et al. 2009. Early immune response following Salmonella enterica subspecies enterica serovar Typhimurium infection in porcine jejunal gut loops. Vet. Res. 40: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meurens F, et al. 2007. Commensal bacteria and expression of two major intestinal chemokines, TECK/CCL25 and MEC/CCL28, and their receptors. PLoS One 2: e677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mitjans M, Ferrer R. 2004. Morphometric study of the guinea pig small intestine during development. Microsc. Res. Tech. 63: 206–214 [DOI] [PubMed] [Google Scholar]
- 25. Naito Y, et al. 2006. Partially hydrolyzed guar gum down-regulates colonic inflammatory response in dextran sulfate sodium-induced colitis in mice. J. Nutr. Biochem. 17: 402–409 [DOI] [PubMed] [Google Scholar]
- 26. Nygard AB, Jørgensen CB, Cirera S, Fredholm M. 2007. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol. Biol. 8: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pilon C, et al. 2009. Induction of porcine regulatory cells by mycophenolic acid-treated dendritic cells. Transplant Proc. 41: 700–702 [DOI] [PubMed] [Google Scholar]
- 28. Searle LE, et al. 2009. A mixture containing galactooligosaccharide, produced by the enzymic activity of Bifidobacterium bifidum, reduces Salmonella enterica serovar Typhimurium infection in mice. J. Med. Microbiol. 58: 37–48 [DOI] [PubMed] [Google Scholar]
- 29. Searle LE, et al. 2010. Purified galactooligosaccharide, derived from a mixture produced by the enzymic activity of Bifidobacterium bifidum, reduces Salmonella enterica serovar Typhimurium adhesion and invasion in vitro and in vivo. J. Med. Microbiol. 59: 1428–1439 [DOI] [PubMed] [Google Scholar]
- 30. Sharon N. 2006. Carbohydrates as future anti-adhesion drugs for infectious diseases. Biochim. Biophys. Acta 1760: 527–537 [DOI] [PubMed] [Google Scholar]
- 31. Sheng KC, et al. 2006. Mannan derivatives induce phenotypic and functional maturation of mouse dendritic cells. Immunology 118: 372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shoaf K, Mulvey GL, Armstrong GD, Hutkins RW. 2006. Prebiotic galactooligosaccharides reduce adherence of enteropathogenic Escherichia coli to tissue culture cells. Infect. Immun. 74: 6920–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Summerfield A, McCullough KC. 2009. The porcine dendritic cell family. Dev. Comp. Immunol. 33: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tam MA, Rydström A, Sundquist M, Wick MJ. 2008. Early cellular responses to Salmonella infection: dendritic cells, monocytes, and more. Immunol. Rev. 225: 140–162 [DOI] [PubMed] [Google Scholar]
- 35. van der Aa Kühle A, Skovgaard K, Jespersen L. 2005. In vitro screening of probiotic properties of Saccharomyces cerevisiae var. boulardii and food-borne Saccharomyces cerevisiae strains. Int. J. Food Microbiol. 101: 29–39 [DOI] [PubMed] [Google Scholar]
- 36. Vandesompele J, et al. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vautier S, Ma Sousa G, Brown GD. 2010. C-type lectins, fungi and Th17 responses. Cytokine Growth Factor Rev. 21: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Warrand J. 2006. Healthy polysaccharides. Food Technol. Biotechnol. 44: 355–370 [Google Scholar]
- 39. White LA, Newman MC, Cromwell GL, Lindemann MD. 2002. Brewers dried yeast as a source of mannan oligosaccharides for weanling pigs. J. Anim. Sci. 80: 2619–2628 [DOI] [PubMed] [Google Scholar]
- 40. Wismar R, Brix S, Laerke HN, Frøkiaer H. 2011. Comparative analysis of a large panel of non-starch polysaccharides reveals structures with selective regulatory properties in dendritic cells. Mol. Nutr. Food Res. 55: 443–454. [DOI] [PubMed] [Google Scholar]
- 41. World Health Organization 1997. The medical impact of the use of antimicrobials in food animals: Report of a WHO meeting, Berlin, Germany. Document no. WHO/EMC/ZOO/97.4 World Health Organization, Geneva, Switzerland [Google Scholar]
- 42. Zanello G, et al. 2011. Saccharomyces cerevisiae modulates immune gene expressions and inhibits ETEC-mediated ERK1/2 and p38 signaling pathways in intestinal epithelial cells. PLoS One 6: e18573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zanello G, et al. 2011. Saccharomyces cerevisiae decreases inflammatory responses induced by F4+ enterotoxigenic Escherichia coli in porcine intestinal epithelial cells. Vet. Immunol. Immunopathol. 141: 133–138 [DOI] [PubMed] [Google Scholar]
- 44. Zanello G, Meurens F, Berri M, Salmon H. 2009. Saccharomyces boulardii effects on gastrointestinal diseases. Curr. Issues Mol. Biol. 11: 47–58 [PubMed] [Google Scholar]
- 45. Zoumpopoulou G, Tsakalidou E, Dewulf J, Pot B, Grangette C. 2009. Differential crosstalk between epithelial cells, dendritic cells and bacteria in a co-culture model. Int. J. Food Microbiol. 131: 40–51 [DOI] [PubMed] [Google Scholar]