Abstract
Enterotoxigenic Escherichia coli (ETEC) is an important pathogenic variant (pathovar) of E. coli in developing countries from a human health perspective, causing significant morbidity and mortality. Previous studies have examined specific regulatory networks in ETEC, although little is known about the global effects of inter- and intrakingdom signaling on the expression of virulence and colonization factors in ETEC. In this study, an E. coli/Shigella pan-genome microarray, combined with quantitative reverse transcriptase PCR (qRT-PCR) and RNA sequencing (RNA-seq), was used to quantify the expression of ETEC virulence and colonization factors. Biologically relevant chemical signals were combined with ETEC isolate E24377A during growth in either Luria broth (LB) or Dulbecco's modified Eagle medium (DMEM), and transcription was examined during different phases of the growth cycle; chemical signals examined included glucose, bile salts, and preconditioned media from E. coli/Shigella isolates. The results demonstrate that the presence of bile salts, which are found in the intestine and thought to be bactericidal, upregulates the expression of many ETEC virulence factors, including heat-stable (estA) and heat-labile (eltA) enterotoxin genes. In contrast, the ETEC colonization factors CS1 and CS3 were downregulated in the presence of bile, consistent with findings in studies of other enteric pathogens. RNA-seq analysis demonstrated that one of the most differentially expressed genes in the presence of bile is a unique plasmid-encoded AraC-like transcriptional regulator (peaR); other previously unknown genetic elements were found as well. These results provide transcriptional targets and putative mechanisms that should help improve understanding of the global regulatory networks and virulence expression in this important human pathogen.
INTRODUCTION
Despite the high incidence of death resulting from enterotoxigenic Escherichia coli (ETEC) infection, especially in children in developing countries (55), the mechanisms and regulatory networks of ETEC virulence are still relatively unexplored on a genomic scale. ETEC bacteria are characterized by the production of the plasmid-encoded heat-stable (ST) and/or heat-labile (LT) enterotoxin (11). ETEC release ST and/or LT into the host epithelium, which elevates intracellular cyclic AMP (cAMP) that then activates the cystic fibrosis transmembrane regulator (CFTR) chloride channel; this activation leads to the secretion of electrolytes and, subsequently, water as diarrhea (43). In addition to ST and LT, the enteroaggregative E. coli (EAEC) heat-stable enterotoxin 1 (EAST1) protein, first identified in enteroaggregative E. coli (45), has been found in many ETEC isolates with defined colonization factor profiles (58); however, the contribution of EAST1 to ETEC diarrheal disease is currently unknown (11).
ETEC bacteria adhere to intestinal epithelium by fimbrial appendages known as colonization factors (CFs) (12). CFs are structurally and genetically diverse; to date, more than 25 CFs have been identified (12). In addition to the CFs, studies have identified other putative factors involved in ETEC pathogenesis. Recently, it was discovered that the ETEC glycoprotein EtpA can act as a bridge between the bacterial flagella and host epithelial cells (39). EtpA has also been identified as a promising target for vaccine development (37, 38). An additional virulence factor, EatA, a member of the serine protease autotransporter of the Enterobacteriaceae (SPATE) family, has been identified in a wide diversity of ETEC isolates (29). Although the function of the eatA gene product in ETEC pathogenesis is unknown, a recent study used a mouse model to demonstrate that EatA is immunogenic (36).
Many of the known virulence and colonization factors are distributed among ETEC genomes, with no universally conserved ETEC core virulence gene set (35); however, the results of gene-independent comparative genomic analyses suggest that ETEC isolates share a conserved backbone that, while not unique to ETEC, demonstrates that, irrespective of phylogeny, ETEC isolates share a common core genomic structure (44). Several putative virulence factors, primarily identified in the prototypical H10407 ETEC isolate (6), have been identified but are not widely distributed in ETEC isolates (53). There is no small-animal model of ETEC infection that recapitulates the human presentation of disease; thus, many in vivo features of virulence remain unexplored.
The ETEC global regulatory networks for virulence factors are largely unknown. To date, the majority of the ETEC transcriptional studies have focused on the ETEC rns regulon (11), which has largely been associated with the regulation of CF expression (3, 9, 23), although binding motifs have been identified adjacent to other genes (44). No regulator has been identified that is associated with the expression of the enterotoxins.
Environmental and chemical signals have been shown to regulate virulence factors in a number of enteric pathogens, including ETEC isolates. A recent study of a porcine ETEC isolate demonstrated that the addition of spent media from E. coli DH5α overexpressing LuxS resulted in reduced host cell death (60); this suggests that a quorum-sensing mechanism is present in ETEC. A quorum-sensing system for other E. coli pathovars has been described in detail (18, 25, 28, 51), and there is insufficient genomic evidence to suggest that a similar mechanism does not exist in ETEC. Additional gastrointestine-related environmental signals that have been investigated include glucose, which has been demonstrated to increase the expression of labile toxin in ETEC isolate H10407 (4). Bile salts, which are found in significant concentrations in the small intestine (15), are required for the phenotypic expression of at least 5 ETEC CFs (48). In enteropathogenic E. coli, bile stress has been shown to enhance adhesion to epithelial cells (8). Bile has been demonstrated to be a potent in vivo signal for a number of other enteric pathogens, including Salmonella, Vibrio, and Listeria species (27, 33, 42, 47, 56). In Salmonella enterica, bile was found to have a negative effect on colonization (32), but it does appear to regulate several virulence-associated pathways (31). Both Listeria (27) and Vibrio (47, 56) species respond to bile by altering surface structures to increase resistance to the detergent effects of bile, as well as by activating a number of virulence factors. Based on these observations, it is clear that bile is a relevant biological signal for multiple enteric pathogens.
Studies have also demonstrated that virulence factor expression can be affected by the presence of signals liberated by the microbiota or coinfecting pathogens. A study by Crane et al. identified that the virulence of EPEC prototype strain E2348/69 was enhanced by the release of ATP in the presence of heat-stable toxin (STa) produced by an ETEC isolate (5). Interaction of isolates from these two pathovars has also been previously noted in epidemiological studies (52). Studies of chemical signaling between bacteria (intrakingdom) and between host and pathogen (interkingdom) have been conducted on related E. coli pathovars (35), but to date no detailed genome scale transcriptional studies have been concluded on any members of the ETEC pathovar.
The goal of this study was to examine the global gene expression profiles of the reference ETEC isolate E24377A (LT positive [LT+], ST+, CS1, CS3) (35) in the presence of various relevant chemical signals. Gene expression was evaluated with a pangenome E. coli/Shigella microarray that contained the genomes of a diverse set of 32 genomes, including E24377A, as well as 46 enteric plasmids (10). In this study, the global response to signals, including glucose, previously demonstrated to enhance toxin secretion (4), and bile, previously demonstrated to increase colonization factor antigens in other isolates (48), was examined. Additionally, the exposure of ETEC to preconditioned media from a commensal E. coli isolate, a prototypical enteropathogenic E. coli (EPEC) isolate, and a Shigella boydii isolate elucidated a species-specific response. Examination of the temporal patterns of virulence gene expression demonstrated that motility was upregulated early in the growth cycle of ETEC 24377A, whereas more of the traditional virulence factors were expressed later in the growth cycle; this suggests that a virulence-related quorum-sensing mechanism may be present in this isolate. Isolate E24377A contains the gene that encodes an autoinducer 2 signal, which has been associated with quorum sensing (46). Microarray data were validated with quantitative reverse transcriptase PCR (qRT-PCR) and extended with targeted RNA sequencing (RNA-seq). The RNA-seq analysis confirmed differentially regulated genes identified using the microarray; however, the advantage of RNA-seq is the identification of novel transcripts from unannotated genomic regions. These novel transcriptional events represent genes and therapeutic targets that have yet to be evaluated. Overall, this study examined the global transcriptional mechanisms of ETEC virulence factors in response to biologically relevant signals.
MATERIALS AND METHODS
Strain selection, growth conditions, and RNA extraction.
ETEC isolate E24377A was chosen because the completed genome has been sequenced (35) and the chromosomal content is printed on the microarray utilized in this study (10); the six plasmids present in the isolate, however, are not printed on the array. Initial growth conditions were compared to determine the impact of media on baseline gene expression. Cultures were grown in 5 ml of Luria broth (LB) overnight at 37°C for 16 h. Following incubation, 1:100 dilution cultures in a total volume of 50 ml were prepared from the following media: LB, LB plus glucose (4% wt/vol), Dulbecco's modified Eagle medium-high glucose (DMEM-HI; Gibco) (4.5 g/liter), or DMEM-low glucose (DMEM-LO) (1 g/liter). Cultures were grown at 37°C with rotation at 200 rpm to an optical density at 600 nm (OD600) of 0.2, 0.5, or 0.8. RNA was extracted using either a Ribo-Pure kit (catalog no. AM1925; Ambion) or a modified acid phenol-lithium chloride method. In short, 25 ml of culture was centrifuged for 10 min at 4,000 rpm at 4°C. A 5-ml volume of heated (55°C) acid phenol-chloroform (pH 5.0) and 5 ml of extraction buffer [50 mM Tris (pH 7.5), 100 mM LiCl, 5 mM EDTA (pH 8), 1% sodium dodecyl sulfate (SDS), 1% beta-mercaptoethanol] were added to the sample and mixed. Samples were then placed on a bead beater at 6 m/s for 40 s. Tubes were centrifuged (4,000 rpm for 10 min), and the aqueous phase was placed in a new tube. An equal volume of chloroform was added to the sample and mixed. The tube was centrifuged (4,000 rpm for 10 min), and the aqueous phase was removed to a new tube. A third volume of 8 M lithium chloride (catalog no. L7026; Sigma) (500 ml) was added to the solution and maintained at −20°C overnight. The next day, the sample was concentrated by centrifugation (4,000 rpm for 10 min) and the supernatant removed. The pellet was suspended in 3 ml of diethyl pyrocarbonate (DEPC)-treated H2O, and a 1/3 volume of 8 M lithium chloride was added; this solution was maintained at −20°C overnight. The following day, samples were concentrated by centrifugation and the supernatant was removed. The pellet was washed three times with 2 M LiCl and three times with 80% ethanol. The dried pellet was suspended in 200 μl of DEPC-treated H2O (catalog no. AM9906; Ambion). Following the isolation of the RNA, the samples were treated with DNase and the RNA concentrations were quantified using a NanoDrop 1000 spectrophotometer (NanoDrop). Biological replicates were processed for each set of conditions; Table 1 shows the complete experimental design. The prototype ETEC isolate H10407 (6) was also used in this study and was grown under the same conditions as described above for qRT-PCR (see below).
Table 1.
Microarray experimental design
| Condition | Organism | Mediuma | OD600 | Supplement | Supplement concn | Data type |
||
|---|---|---|---|---|---|---|---|---|
| Array | qRT-PCR | RNA-seq | ||||||
| 1 | 24377Ab | LB | 0.5 | None | N/Ac | X | X | X |
| 2 | 24377A | LB | 0.5 | Glucose | 0.4% (wt/vol) | X | X | |
| 3 | 24377A | LB | 0.5 | Bile | 3% (wt/vol) | X | X | X |
| 4 | 24377A | LB | 0.2 | Bile | 3% (wt/vol) | X | X | |
| 5 | 24377A | LB | 0.2 | None | N/A | X | X | |
| 6 | 24377A | LB | 0.8 | Bile | 3% (wt/vol) | X | X | |
| 7 | 24377A | LB | 0.8 | None | N/A | X | ||
| 8 | 24377A | DMEM-HI | 0.5 | None | N/A | X | X | |
| 9 | 24377A | DMEM-LO | 0.5 | None | N/A | X | X | |
| 10 | 24377A | DMEM-HI | 0.5 | 2348 PCM | 10% (vol/vol) | X | X | |
| 11 | 24377A | DMEM-HI | 0.5 | HS PCM | 10% (vol/vol) | X | X | |
| 12 | 24377A | DMEM-HI | 0.5 | S. boydii PCM | 10% (vol/vol) | X | X | |
| 13 | 24377A | DMEM-HI | 0.5 | HS PCM | 1% (vol/vol) | X | X | |
| 14 | H10407d | LB | 0.5 | None | N/A | X | ||
| 15 | H10407 | LB | 0.5 | Bile | 3% (wt/vol) | X | ||
| 16 | H10407 | LB | 0.5 | Glucose | 0.4% (wt/vol) | X | ||
RNA was extracted from the LB-bile mixture by the use of the standard method with RiboPure; however, the RNA extracted from the DMEM-HI–bile mixture was highly unstable with this method, and the acid phenol extraction method was therefore employed and produced RNA of sufficient quality for subsequent expression profiling.
Generation of preconditioned media.
Three isolates were used for the signaling studies that utilized preconditioned media (PCM): EPEC strain E2348/69 (16), a Shigella boydii isolate (strain BS512), and the commensal HS E. coli strain (21). The isolates were grown at 37°C with shaking at 200 rpm in LB overnight. After 16 h of growth, cells were removed by centrifugation at 8,000 × g and the supernatant was filtered with a 0.2-μm-pore-size syringe filter. PCM was aliquoted and stored at −20°C prior to use.
Medium additives.
Sodium choleate, a crude ox bile extract that contains the sodium salts of taurocholic, glycocholic, deoxycholic, and cholic acids, was obtained from Sigma (catalog no. S9875). A concentrated sodium choleate mixture (30% [wt/vol]) was made by dissolving the powder in DNA/RNA-free water and filter sterilizing (0.2 μm) it. d-Glucose was also obtained from Sigma (catalog no. G8644); a concentrated solution (10%) was subjected to sterile filterization and diluted for signaling experiments.
Microarray hybridization.
For whole-genome transcriptional analyses, cDNA was synthesized from 16 μg of total RNA from biological rep-licates as listed in Table 1, using a Superscript III cDNA synthesis kit (Life Technologies) with random hexamers. cDNA was partially digested with a DNA-free DNase kit (catalog no. AM1906; ABI) for various time periods until a desired size range (20 to 200 bp) was obtained. Approximately 1.5 μg of digested, labeled cDNA was hybridized per array. DNA fragment size distributions were visualized using Tris-borate-EDTA (TBE) gradient polyacrylamide gels (catalog no. 161-1235; Bio-Rad) (4% to 20%). The optimal digest for each sample was end labeled with biotin-11-ddATP (catalog no. NEL548001EA; Perkin Elmer) using terminal deoxynucleotidyl transferase and verified by a gel shift assay according to Affymetrix standard protocols. A hybridization mixture was prepared that contained labeled cDNA, 7.8% dimethyl sulfoxide (DMSO), and an OligoB2 mixture (catalog no. 900301; Affymetrix) according to the manufacturer's protocols. GeneChips were prepared by prehybridization for 10 min at 45°C with the kit prehybridization solution; samples were denatured at 95°C for 1 min, equilibrated to 45°C, centrifuged, and transferred to prepared arrays (10). Hybridization was continued in a rotating incubator for 16 h at 60 rpm and 45°C. Posthybridization, the hybridization cocktail was recovered and the arrays were washed and stained using the standard Affymetrix ProkGE-WS2v3_450 fluidics protocol and GeneChip hybridization, wash, and stain kit reagents (catalog no. 900720; Affymetrix).
Microarray data analysis.
Following a scan of each array with the Affymetrix GCOS software, expression was normalized using the Simpleaffy Bioconductor R package (57). Comparisons were double filtered according to the fold change value (FC ≥ 2) and the Student t test P value (P ≤ 0.005). Biological replicates were processed for each set of conditions, and microarrays were analyzed for the number of present calls and the intensity of signal compared to all other arrays. Arrays that were determined to be anomalous, compared to the corresponding biological replicate, were not used for downstream analysis; additional replicates were then processed so that biological replicates were processed at least in duplicate for each set of conditions. Further comparisons between sets of conditions for transcriptionally altered genes were performed with custom Perl and Python scripts.
Quantitative reverse transcriptase PCR (qRT-PCR) assays.
Differentially regulated chromosomal genes were identified through the microarray analysis. To validate these findings, primers were designed with Primer 3 software (40) to correspond to genes from the E24377A genome (Table 2). E24377A plasmids are not contained on the microarray; therefore, plasmid-encoded virulence genes were selected for interrogation via only qRT-PCR. The constitutively expressed ribosomal subunit gene (rpoA) was used as an internal control for relative quantification. The constitutive expression of rpoA under the examined conditions was verified using the microarray analysis.
Table 2.
Oligonucleotides designed from E24377A and used in qRT-PCR assays
| Gene | Forward (5′–3′) sequence | Reverse (5′–3′) sequence | No. of nucleotides |
|---|---|---|---|
| estA | GTGGTCCTGAAAGCATGAATAG | AATAGCACCCGGTACAAGCA | 74 |
| eltA | TATAGCTCCGGCAGAGGATG | TCCAGGGTTCTTCTCTCCAA | 76 |
| eatA | GCCAACGACTCCCAAATAGA | CAGTTTGCCGGTAACATTCA | 140 |
| etpA | ATGCCGGTGGCTATATTGTG | GTACGGATGGTGCCACTGTT | 60 |
| csoA | GTTGACCCGACTGTTGACCT | ACAACGGTGTTGATGGTGTG | 96 |
| cstA | GGCCCACTCTAACCAAAGAAC | AGCCCAAGTTGCATCCAG | 67 |
| astA | TGCATCGTGCATATGGTG | GCGAGTGACGGCTTTGTAGT | 51 |
| peaR | AATGCAATCAAGAATGAGCAAA | GATGCACTAGAAATCCCAATCA | 95 |
| fliC | CAGCTCCATCGACAAATTCC | TGGTGTTGTTCAGGTTGGTG | 74 |
| rpoA | TGTAGGCAATACGCTCCACA | GGTTATGTGCCGGCTTCTAC | 105 |
Reactions for qRT-PCR were completed using Power SYBR green PCR Master Mix (Applied Biosystems) with a two-step reaction protocol consisting of 40 cycles of 94°C for 30 s and 60°C for 1 min, followed by a dissociation phase for quality control. The 10-μl qPCR mixtures contained 0.2 μM specific primers, Power SYBR green (catalog no. 4367659; Applied Biosystems) (2×), and template (2.5 μl of 1:100-diluted cDNA). Two biological replicates, with 4 technical replicates, were processed for each set of conditions. LinRegPCR (41) was used to calculate efficiency (E) and threshold cycle (CT) values based on background corrected fluorescence data (30, 54).
Pairwise comparisons were conducted with the Student t test to calculate a P value. Delta CT values were calculated by subtracting the rpoA CT value from the target CT value. Eight delta CT values from the reference conditions were then compared to eight delta CT values from the experimental conditions in the t test.
RNA-seq.
To examine the transcriptome in an unbiased manner, RNA-seq analysis was performed. RNA was extracted from E. coli E24377A grown in LB and LB-bile. Three biological replicates were combined into a single sample for each set of conditions. The rRNA was depleted using a Ribo-Zero kit according to the manufacturer's specifications (catalog no. RZC1046; Epicentre). Fifty base-pair reads were obtained from each sample using standard operating procedures for the Illumina GA-II platform (Illumina) at the Institute for Genome Sciences Genomics Resource Center (http://www.igs.umaryland.edu/research/grc/intro.php). Reads were then mapped to the E24377A chromosome and six plasmids (20) with the BWA aligner (22). Counts for each annotated genomic feature were determined by htseq-count (http://www-huber.embl.de/users/anders/HTSeq/doc/count.html). Differential expression between counts for each feature was then calculated with DESeq (1) using the false-detection rate-adjusted Benjamini Hochberg P value (2). The relative expression of differentially expressed genes was plotted with Circos (19).
In an effort to identify novel transcribed features, reads that mapped to the E24377A genome, but were not assigned to an annotated genomic feature, were extracted from the analysis. These transcripts were pooled and assembled with Oases (http://www.ebi.ac.uk/∼zerbino/oases/), which utilizes Velvet assemblies (59), but was developed to assemble transcriptomics data. These assembled transcripts were then used to search the Kegg database (http://www.genome.jp/kegg/) for homologous genes. Differential expression for these novel transcripts was calculated using the same protocols as reads that mapped to a feature.
Microarray data accession number.
RNA-seq and microarray data were submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under GEO accession no. GSE31806.
RESULTS
Global transcriptional response in baseline medium selection.
Due to the paucity of whole-genome transcriptional data for any ETEC isolate, initial microarray assays were performed with ETEC isolate E24377A (35) to determine the transcriptional response to commonly utilized medium conditions that would induce virulence factor expression. The medium conditions selected were mid-log-phase growth (OD600 = 0.5) in DMEM-high glucose (DMEM-HI; 4.5 g/liter), DMEM-low glucose (DMEM-LO; 1.0 g/liter), and Luria broth (LB), with and without glucose added at 4 g/liter; DMEM was chosen as a growth medium as it is required for virulence factor expression in other E. coli pathovars (17). Comparisons of the growth in DMEM-HI to that in DMEM-LO identified 61 features with differential expression profiles (≥2-fold change; P value < 0.005) and 52 features upregulated in DMEM-HI (see Table S1 in the supplemental material). Transcriptional analysis identified 288 features in E24377A that were differentially regulated between LB and DMEM-HI (see Table S2 in the supplemental material). Features upregulated in DMEM-HI include the outer membrane receptor gene fepA and the siderophore components entE and entF; features upregulated in LB include genes encoding flagellar components as well as the nitrate reductase and transporter protein genes narH and narK.
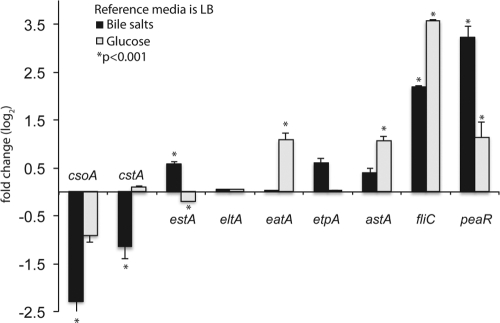
The lack of the plasmid sequences from E24377A on the microarray prevented a complete global analysis. Therefore, we used targeted qRT-PCR to examine the plasmid-borne virulence-associated genes; a complete list of all qRT-PCR expression values for each set of conditions is shown in Table S3 in the supplemental material. Comparisons of the medium conditions determined that the genes associated with the CS1 (csoA) and CS3 (cstA) colonization factors involved in ETEC adhesion displayed enhanced gene expression in DMEM compared to LB irrespective of the glucose concentration, whereas flagellar genes (e.g., fliC) were upregulated in LB (Table 3). A previous study performed with ETEC isolate H10407 demonstrated that the expression of labile-toxin and adhesive mechanisms was upregulated in the presence of glucose (3, 4). Comparisons of the transcriptome of E24377A grown in LB and in LB plus glucose identified 21 differentially regulated features (see Table S4 in the supplemental material). However, the ETEC E24377A expression profiles, as determined by the qRT-PCR assay, did not identify significant glucose-induced expression of the enterotoxin genes estA and eltA (Fig. 1). Glucose did upregulate the expression of astA, which encodes the enterotoxin EAST1 (45). Glucose also upregulated the expression of the autotransporter eatA, a plasmid-encoded araC family transcriptional regulator (peaR), and the flagellar gene fliC. The presence of glucose had no apparent effect on the expression of the other virulence factors encoded by csoA (CS1), cstA (CS3), or etpA (EtpA) (Fig. 1).
Table 3.
Microarray results from a comparison of E24377A grown in DMEM-HI versus LB
| Gene | Annotation | Fold change (log2)a | P value |
|---|---|---|---|
| fepA | Outer membrane receptor | 6.72 | 0.0007 |
| entF | Enterobactin synthase subunit F | 3.55 | <0.0001 |
| cstA | Carbon starvation protein | 2.13 | 0.0004 |
| cooA | CS1 major subunit | 1.34 | 0.0007 |
| rstA | Transcriptional regulator | −1.62 | <0.0001 |
| fliN | Flagellar motor switch protein | −3.57 | 0.0005 |
| narH | Nitrate reductase 1, beta subunit | −4.58 | 0.0020 |
| fliC | Flagellin | −5.24 | 0.0003 |
LB was used as the reference medium and DMEM-HI as the query medium.
Fig 1.
Effect of the addition of glucose or bile to LB media. A column chart of fold changes of expression values (qRT-PCR) between ETEC E24377A grown in bile salts and grown in glucose to an OD600 of 0.5 is shown. Values are based on fold change calculated from ΔΔCT values. All values are log2 transformed. Columns with an asterisk indicate a significant P value of <0.001 as calculated by Student's t test. Error bars represent standard deviations of the results determined with 2 biological replicates. The reference is E24377A grown in LB alone to an OD600 of 0.5. Variable patterns of gene expression were observed with the selected virulence factors.
ETEC transcriptional response to preconditioned media.
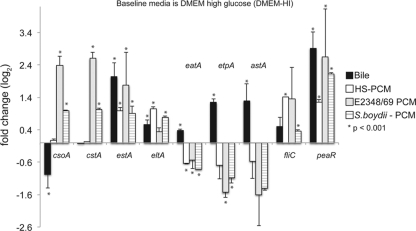
The secreted components from related bacterial community members have the potential to increase virulence in other known enteric pathogens (5). In the present study, preconditioned media (PCM) from a commensal isolate of E. coli, E. coli HS (21, 35), were added to media that contained E24377A to represent the normal E. coli component of the human gastrointestinal tract microbiota. PCM from two pathogens were also added: the typical EPEC isolate, E. coli E2348/69, was added to determine whether ETEC virulence expression was affected by signals from a different pathovar, and Shigella boydii PCM was added to identify changes in gene expression due to the presence of an intracellular pathogen. The microarray analysis demonstrated that only one feature, an uncharacterized GntR family transcriptional regulator (locus ABV18196), was consistently transcriptionally regulated in E24377A in response to each PCM compared to the baseline condition (DMEM-HI). While there was only one differentially regulated chromosomal gene common to all PCM transcriptomes, there was a specific response to each of the PCM (see Tables S5 to S7 in the supplemental material). Additional plasmid-focused studies with qRT-PCR demonstrated that expression of estA and eltA was significantly greater under all PCM conditions compared to the baseline condition (Fig. 2). The plasmid-encoded AraC-like regulator (peaR) gene was also upregulated in all preconditioned media. In contrast, the presence of PCM from the two pathogenic strains upregulated the expression of genes that encode colonization factors CS1 and CS3, whereas PCM from the commensal strain HS had no effect on colonization factor expression.
Fig 2.
E. coli E24377A gene expression profiles in response to biologically relevant environmental signals A column chart showing fold change expression values (qRT-PCR) between ETEC E24377A grown in DMEM media and biologically relevant additives: bile (3% [wt/vol]; black bars), preconditioned media from commensal E. coli isolate HS (white bars), enteropathogenic E. coli isolate E2348/69 (gray bars), and Shigella boydii BS512 (horizontally striped bars). Values are based on fold change calculated from ΔΔCT values. All values are log2 transformed. Columns with an asterisk indicate a significant P value of <0.001 as calculated by Student's t test. Error bars represent standard deviations of the results determined with at least 2 biological replicates. The reference is E24377A grown in DMEM alone to an OD600 of 0.5. PCM, preconditioned media. The responses differed among the various preconditioned media, indicating a species-specific response, whereas the bile response was similar to that observed with LB media as described for Fig. 1.
To determine whether the concentration of PCM was inhibitory toward gene expression, an additional set of experimental conditions used only a 1% (vol/vol) concentration of PCM extracted from the commensal E. coli HS rather than the 10% used in all other conditions (Table 1). Microarray analysis demonstrated that the relative expression results were essentially unchanged in comparisons of these conditions (1% versus 10%), as only 11 features exhibited differential expression results (see Table S8 in the supplemental material).
ETEC transcriptional response to bile salts.
Bile salts have been demonstrated previously as a regulator of virulence phenotypes in ETEC (14, 49). To determine the global transcriptional response to bile salts, rather than a phenotype response or single gene expression analysis, we utilized microarray, qRT-PCR, and RNA-seq technologies.
Based on the microarray analysis, the largest effect of a chemical addition on differential gene expression was seen with the addition of bile salts to LB and DMEM-HI. Fifteen array features were consistently upregulated in both LB-bile and DMEM-HI–bile compared to the results seen with a pairwise comparison of all other conditions combined (see Table S9 in the supplemental material). Further studies of the plasmid-encoded virulence factors with qRT-PCR demonstrated that the addition of bile salts to LB increased the expression of the toxin gene, estA (Fig. 1), whereas in the DMEM experiments, the toxin and adhesion genes estA, eltA, etpA, and astA were all significantly upregulated (Fig. 2) (P < 0.001). Expression of plasmid-borne genes that encode the colonization factors CS1 and CS3 was downregulated or not significantly changed, depending on the media, in the presence of bile salts (Fig. 1 and 2); in DMEM, cstA (CS3) was not significantly downregulated in bile salts.
Comparison of the glucose and bile responses of ETEC isolates E24377A and H10407.
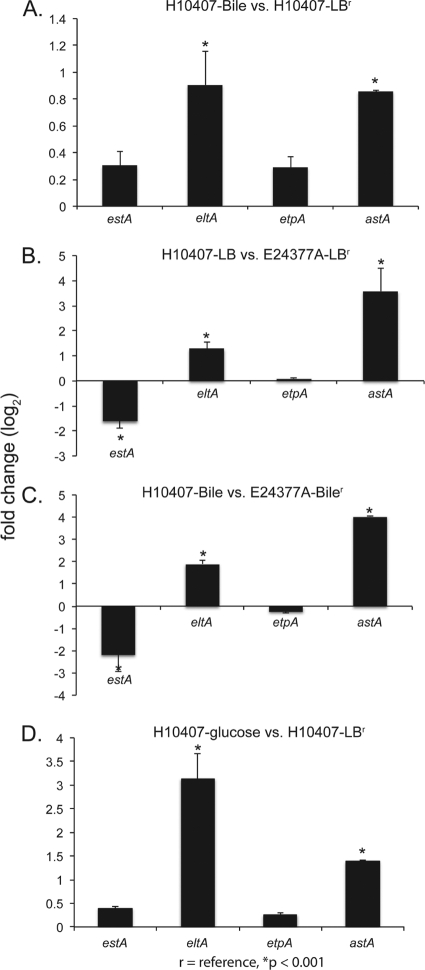
The apparent discrepancy between the published H10407 response to glucose (3, 4) and the E24377A response observed in this study was examined using qRT-PCR assays. This observation was expanded to examine the bile-related response of these two isolates. The similarity between these two isolates with respect to plasmids allowed direct comparison of the virulence-associated genes, including etpA, estA, elta, and astA. The results demonstrate that in the presence of bile salts, H10407 does not increase the expression of estA or etpA at mid-log-phase growth (OD600 = 0.5) but does increase the expression of eltA and astA (Fig. 3A). This is in contrast to the transcriptional response in E24377A, where expression of astA, eltA, and etpA were unchanged (Fig. 1) with the addition of bile; only estA shows similar patterns for the two strains examined. The baseline expression characteristics in LB of homologous genes were generally not similar between strains (Fig. 3B); for example, differential expression of eltA was observed in ETEC H10407 compared to that of E24377A in the presence of bile salts (Fig. 3C). The estA primers used in this study amplified only the STh gene on the p948 plasmid (GenBank accession no. FN649418) in H10407; the STp gene on the p666 plasmid (accession no. FN649417) was not tested for expression in this study, as no identical gene is present in E24377A.
Fig 3.
Identification of transcriptional differences between prototype ETEC strains. To determine the impact of strain on the transcriptional response in ETEC isolates E24377A and H10407, we used qRT-PCR to examine 4 genes. Columns with an asterisk indicate a significant P value of <0.001 as calculated by Student's t test. Error bars represent standard deviations of the results determined with at least 2 biological replicates. (A) Differential expression between H10407 grown in LB and in LB-bile, demonstrating significantly increased expression of eltA and astA. (B) H10407 expression in LB compared to E24377A expression in LB, with significantly increased expression of eltA and astA. (C) Differential expression between H10407 and E24377A grown in LB-bile, demonstrating a significantly greater bile-related response of the eltA and astA genes in H10407. (D) Differential expression of H10407 grown in either LB or LB-glucose. This panel highlights the previously published response of H10407 to glucose, which differs significantly from that in E24377A (Fig. 1).
In both isolates, glucose increased the expression of astA. An upregulation of eltA was observed in the presence of glucose in H10407 (Fig. 3D), while no increase was observed for either enterotoxin in E24377A (Fig. 1).
Temporal aspects of transcription.
A growth time course was conducted to determine whether known and suspected virulence factors were expressed at a particular growth stage. ETEC E24377A was grown in LB and LB-bile and harvested at OD600s of 0.2, 0.5, and 0.8 to represent early, middle, and late log-phase growth. The qRT-PCR assays were then performed, and the relative levels of gene expression of known and suspected virulence factors were examined. The results demonstrated that expression of the virulence-associated genes is generally upregulated at later stages of growth in the presence of bile salts (Fig. 4). A comparison of fold changes seen in the various growth stages in bile, compared to the corresponding growth density results seen in LB alone, demonstrated that the greatest levels of gene expression occurred at OD600 = 0.8 (Fig. 4). qRT-PCR analysis also confirmed the RNA-seq result demonstrating that the plasmid-encoded AraC-like regulator (peaR) is upregulated in the presence of bile (see below). The time course analysis also demonstrated that expression of this regulator begins early in the growth curve and is greatest at OD600 = 0.8. Expression of csoA (CS1) stayed low during the entire growth curve, while that of cstA (CS3) expression was upregulated only at OD600 = 0.8 (Fig. 4). The qRT-PCR analysis showed that expression of the flagellar gene fliC was temporally increased in the presence of bile salts at all growth densities. These results were confirmed at OD600 = 0.5 in LB with both RNA-seq and microarray analyses.
Fig 4.
Temporal expression of ETEC virulence factors in bile. A time course column chart of E24377A grown in LB-bile to different growth densities (OD600 = 0.2, 0.5, and 0.8) is shown. Values are based on fold change calculated from ΔΔCT values. Columns with an asterisk indicate a significant P value of <0.001 as calculated by Student's t test. Error bars indicate standard deviations of the results determined with biological replicates. Overall, we observed generally increased gene expression as the cultures reached the stationary phase of growth, suggesting that a quorum-sensing mechanism is involved in global ETEC regulation.
Overall, the temporal aspects of gene expression in response to bile indicate that there is a complex regulatory cascade involved that includes a bile response as well as a quorum-sensing response.
RNA-seq analysis to identify novel ETEC transcriptional events.
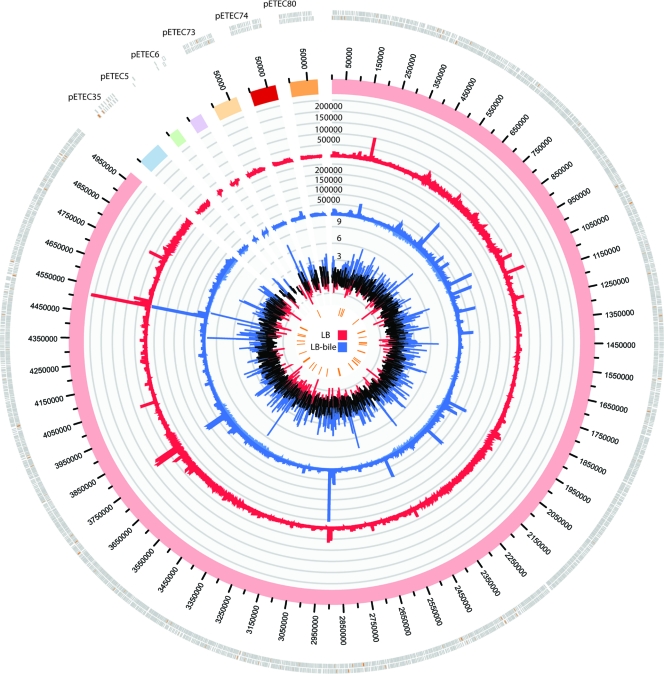
RNA-seq is a novel and unbiased technology for examining the complete transcriptome of any biological system (recently reviewed by Sorek and Cossart [50]). This technology was utilized to examine the transcriptional response to bile in ETEC isolate E24377A at mid-log-phase growth (OD600 = 0.5). More than 30 million Illumina sequences (50 base-paired end reads) were obtained for each set of conditions; of these sequences, approximately 74% were of sufficient quality and could be mapped to an annotated genomic feature. Sequence counts for each annotated feature were compared between samples using the negative binomial method implemented in DESeq (1). In this study, the DESeq analysis identified multiple features that demonstrate significant, differential expression between growth conditions (Table 4 and Fig. 5). One of the most significantly differentially expressed genes (corrected P value = 0.002) observed was a plasmid-borne AraC family transcriptional regulator (peaR) encoded on the pETEC73 plasmid (EcE24377A_D0031). A bioinformatic analysis of this peptide against the GenBank database identified a homologous regulator (csvR) (96% amino acid similarity) associated with regulation of the colonization factors CS1 and CS5 in an unrelated ETEC isolate (7).
Table 4.
Most differentially expressed genes in the RNA-seq analysis of LB (reference) and LB-bile
| Gene annotation | Accession no. | Counta |
Fold change (log2) | P valueb | |
|---|---|---|---|---|---|
| LB | LB-bile | ||||
| Hypothetical protein | YP_001464430 | 1 | 463 | 9.56 | 0.001 |
| Hypothetical protein | YP_001463383.1 | 13 | 2,241 | 7.44 | 0.002 |
| Hypothetical protein | YP_001462569 | 1 | 201 | 7.35 | 0.014 |
| Plasmid-encoded araC (peaR) | YP_001451499.1 | 26 | 3,769 | 7.19 | 0.002 |
| Hypothetical protein | YP_001462412 | 3 | 309 | 6.65 | 0.016 |
| Hypothetical protein | YP_001462237 | 10 | 951 | 6.51 | 0.013 |
| Hypothetical protein | YP_001463716 | 3 | 280 | 6.51 | 0.019 |
| Hypothetical protein | YP_001463386 | 31 | 2,448 | 6.31 | 0.007 |
| Multiple antibiotic resistance (marB) | YP_001462817 | 2 | 179 | 6.18 | 0.031 |
| Hypothetical protein | ABV16359 | 7 | 306 | 5.37 | 0.065 |
| Hypothetical protein | YP_001463094 | 78 | 2,906 | 5.22 | 0.030 |
| Hypothetical protein | YP_001462826 | 94 | 2,885 | 4.96 | 0.049 |
Results represent normalization with DESeq.
Results represent Benjamini-Hochberg correction.
Fig 5.
Global transcriptional analysis as determined by RNA-seq and circular visualization. A circular plot showing the RNA-seq mapping data for E24377A chromosome and its 6 plasmids is shown. The outer histogram, colored red, represents the normalized mapped reads from E24377A grown in LB. The blue histogram represents the normalized mapped reads from E24377A grown in LB-bile. The inner histogram shows all fold changes for each annotated genomic feature. Peaks tipped in blue represent upregulation in LB-bile (log2 fold change > 2), while those tipped in red represent downregulation in LB-bile (log2 fold change < −2). The inner data track ticks (orange) include those features that showed significant differential regulation (P < 0.08) according to DESeq.
An increase in normalized count numbers was also observed for the enterotoxin gene estA with the addition of bile. While the expression of these genes was relatively low and a significant P value was not observed, the fold change results are congruent with the results obtained with qRT-PCR. These analyses demonstrate the ability to use RNA-seq and qRT-PCR data hand in hand for expression analysis.
Approximately 2 million sequence reads in each set of conditions mapped to the E24377A genome but were not assigned to any annotated genomic feature. These regions represent transcriptionally active but uncharacterized regions of the genome. The reads corresponding to these areas were extracted from the alignment file, and the transcripts were assembled using Oases software. This analysis identified more than 300 regions that were greater than 150 nucleotides in length; eight of these potential transcripts were significantly upregulated (false-discovery rate corrected P value < 0.08) in the presence of bile; a list of these previously unannotated transcripts is shown in Table 5. Most of these transcripts are associated with intergenic regions that may include gene promoters, whereas other novel transcripts were annotated as hypothetical proteins, as they have not been predicted or annotated in other genome sequences to date; these regions represent transcriptionally active units and possible novel functional peptides in E24377A.
Table 5.
Differential expression of nonannotated transcripts by RNA-seq analysis
| pETEC_74 location (chromosome nucleotide no.)a |
Countb |
Fold change (log2) | P valuec | Annotationd | ||
|---|---|---|---|---|---|---|
| Start | End | LB | Bile | |||
| 11572 | 11674 | 1 | 12,150 | 13.2 | 0.0000 | Hypothetical protein |
| 898175 | 898323 | 1 | 3,379 | 12.4 | 0.0000 | intergenic region |
| 1708879 | 1709036 | 1 | 683 | 9.1 | 0.0025 | Intergenic region |
| 1867008 | 1867176 | 8 | 1,376 | 7.5 | 0.0050 | Intergenic region |
| 2979643 | 2979873 | 154 | 17,582 | 6.8 | 0.0071 | Intergenic region |
| 4008052 | 4008248 | 11 | 1,239 | 6.8 | 0.0094 | Intergenic region |
| 1925715 | 1925840 | 49 | 4,268 | 6.4 | 0.0094 | Hypothetical protein |
| 4265429 | 4265561 | 1,375 | 95,084 | 6.1 | 0.0759 | Hypothetical protein |
E24377A isolate.
Results represent normalization with DESeq.
Results represent Benjamini Hochberg correction.
Based on best BLAST hit against the KEGG Genes database.
DISCUSSION
The paucity of global transcriptional data from the ETEC pathovar provided the impetus to undertake the current study, which utilized a combination of pan-genome microarray, qRT-PCR, and RNA-seq technologies to investigate the in vitro gene expression of ETEC isolates in the presence of biologically relevant chemical signals. It is unclear whether in vitro results are representative of in vivo expression; however, a representative small-animal model for ETEC diarrheal disease is not currently available, and comprehensive in vitro studies are therefore a necessary first approximation of in vivo expression.
An E. coli/Shigella pan-genome microarray, containing the chromosomal content of E24377A, was used to examine the transcriptional profiles seen under different sets of experimental conditions. An initial analysis of microarray results in baseline growth media showed an upregulation of flagellar genes in LB compared to DMEM (Table 3). Previous studies of E. coli gene expression have demonstrated that in nutrient-limited conditions, E. coli downregulates expression of flagellar genes and often upregulates virulence factors (13). The same result observed in this study with ETEC suggests that the observed response is part of a conserved E. coli transcriptional program in response to nutrient depletion but also suggests that the observed gene expression profile is valid and that a common E. coli transcriptional network exists for these processes.
Following the establishment of a baseline set of conditions, exogenous environmental signals were introduced into the growth medium and expression of virulence factors was characterized with qRT-PCR; the first chemical signal tested was glucose. The presence of glucose in the growth medium did not affect expression of either enterotoxin gene, estA or eltA, in E24377A but did upregulate the expression of astA and eatA (Fig. 1); glucose significantly upregulated eltA expression in H10407 (Fig. 3D), confirming previous observations (4) and highlighting the difficulty of working with diverse isolates of ETEC. These two ETEC strains are not closely related phylogenetically (44) and each may have evolved a unique response to glucose (see below). This result also suggests that a comparative analysis of expression profiles from additional ETEC isolates is necessary to determine the effect of chemical signals on a set of phylogenetically diverse ETEC isolates to identify common responses, if they exist. This concept is being examined in other E. coli pathovars in studies where one or two type strains have been used for extended periods of time but may not represent the entire group of pathogens.
Since bacterial infections occur in a community setting rather than a monoculture, the next set of signals examined consisted of preconditioned media (PCM) from related E. coli and Shigella isolates. The preconditioned media were obtained from E. coli HS, a commensal isolate that represents the normal microbiota; E. coli E2348/69, a pathogenic E. coli isolate and the prototype of EPEC isolates; and a Shigella boydii isolate, which represents an intracellular pathogen. Microarray analysis demonstrated that more genes were downregulated by the presence of PCM than were upregulated (see Tables S5 to S7 in the supplemental material) and suggested that the presence of PCM may actually be slightly inhibitory on a global scale. However, qRT-PCR analysis showed that the presence of each of the three PCM added at 10% significantly increased the expression of the virulence genes estA and eltA encoding the enterotoxins LT and ST relative to the baseline set of conditions (Fig. 2). There were a number of PCM-specific responses, such as the significant increase in the expression of colonization factors in response to the PCM from E. coli E2348/69 and the S. boydii isolate; this suggests that coinfection with these pathogens may increase colonization. Interestingly, PCM from the human commensal strain HS had a negligible effect on colonization factor expression. While this suggests that intrakingdom signaling from pathogenic strains enhances colonization factor expression whereas those from commensal strains do not, additional studies are required to extend this observation.
Bile is a significant host signal that enteric bacteria encounter as they transit the gastrointestinal tract. It is therefore not surprising that a number of enteric pathogens respond to bile by increasing the expression of virulence factors (24, 27, 31–33, 56). This includes the enhancement of ETEC colonization factors in some isolates by the addition of bile to the media (14, 21, 49). Temporal analysis demonstrated that expression of the enterotoxins, as well as that of other known or predicted virulence factors, is highly dependent on the growth density. At an OD600 of 0.8, many of the known or suspected virulence factors are upregulated in the presence of bile compared to LB alone (Fig. 4). These data suggest that a quorum-sensing mechanism in ETEC may control virulence gene expression. While quorum sensing and virulence have been linked in other enteric pathogens (25, 28, 51), it has not been previously identified in ETEC. Additional transcriptional analyses are needed to determine how all other genes are regulated at this growth density in the presence of bile.
The interrogation of transcriptional patterns with RNA-seq technology provides the opportunity for the identification of novel transcriptional events; in some cases, novel transcripts represent promoter structures, and in other cases, they represent expressed genes that have not been previously identified. In the quest for vaccines and therapeutics, these newly identified transcripts are promising targets for future study. We have identified more than 300 such regions in the genome that are longer than 150 bp; a list of the most differentially regulated novel transcripts is shown in Table 5. Additional information that can be derived from the RNA-seq data includes promoter and operon structures. Continued examination of these data set will yield further insight into the transcriptional networks of ETEC.
The isolate used in this study was E. coli E24377A, the first ETEC whose genome was completely sequenced (34). The “prototypical” ETEC isolate most often studied is ETEC H10407 (6), despite the fact that it represents a less representative phylogeny of sequenced ETEC genomes (44) and contains a gene repertoire that is significantly different from those of other known ETEC isolates (53). The comparison of H10407 and E24377A identified a number of transcriptional differences. Previous work by Bodero et al. identified a number of features that are altered by addition of glucose (3, 4); however, we did not observe the same transcriptional patterns with E24377A (Fig. 1 and 3). The results demonstrate that estA, encoding the heat-stable toxin, is upregulated by bile salts in E24377A whereas eltA, encoding the heat-labile toxin, is upregulated by bile salts in H10407 (Fig. 1 and 3); the STp gene in H10407 was not investigated, because a comparable gene does not exist in E24377A. These results highlight that the use of one single isolate as a “prototype” for a pathovar, especially for a diverse pathovar such as ETEC, should not be extrapolated to the entire pathovar without further investigation.
Despite the bactericidal effects of bile in the intestine (24), growth curves for ETEC E24377A grown in LB and in LB-bile showed similar slopes during the log growth phase (data not shown). Additionally, the presence of bile salts in both LB and DMEM media significantly increased the expression of the enterotoxin gene estA in E24377A; this result was observed using qRT-PCR and verified with RNA-seq. These observations suggest that ETEC not only has adapted to bile but that bile may be a signal that the bacterium has entered the small intestine and triggers virulence. An upregulation of the plasmid-encoded AraC regulator (peaR) was observed in the presence of bile, glucose, and PCM; homologous regulators in other ETEC strains, such as the Rns regulon, have been associated with expression of colonization factors (3, 26). However, in E24377A, the upregulation of peaR was generally accompanied by a downregulation of csoA (CS1) and no significant change in cstA (CS3) expression (Fig. 1 and 2). Therefore, it is likely that peaR in E24377A is not involved in colonization factor regulation but could potentially regulate other novel virulence factors. Additional work, including the deletion of the peaR gene and subsequent expression profiling, is required to determine the exact role of this regulator.
This report provides preliminary insights into the expression of known and predicted ETEC virulence and colonization factors in the presence of chemical and environmental signals. The results suggest that gene expression is indeed affected by chemical signals in vitro and indicate that future in vivo studies from human diarrheal samples are required to verify the expression networks observed in this study.
Supplementary Material
ACKNOWLEDGMENTS
These studies were funded in whole or in part by the National Institute of Allergy and Infectious Diseases (NIAID, NIH; RO1 AI089894) and startup funds from the state of Maryland.
Footnotes
Published ahead of print 3 January 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300 [Google Scholar]
- 3. Bodero MD, Harden EA, Munson GP. 2008. Transcriptional regulation of subclass 5b fimbriae. BMC Microbiol. 8:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bodero MD, Munson GP. 2009. Cyclic AMP receptor protein-dependent repression of heat-labile enterotoxin. Infect. Immun. 77:791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crane JK, Choudhari SS, Naeher TM, Duffey ME. 2006. Mutual enhancement of virulence by enterotoxigenic and enteropathogenic Escherichia coli. Infect. Immun. 74:1505–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crossman LC, et al. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J. Bacteriol. 192:5822–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Haan LA, Willshaw GA, van der Zeijst BA, Gaastra W. 1991. The nucleotide sequence of a regulatory gene present on a plasmid in an enterotoxigenic Escherichia coli strain of serotype O167:H5. FEMS Microbiol. Lett. 67:341–346 [DOI] [PubMed] [Google Scholar]
- 8. de Jesus MC, Urban AA, Marasigan ME, Barnett Foster DE. 2005. Acid and bile-salt stress of enteropathogenic Escherichia coli enhances adhesion to epithelial cells and alters glycolipid receptor binding specificity. J. Infect. Dis. 192:1430–1440 [DOI] [PubMed] [Google Scholar]
- 9. Espert SM, Elsinghorst EA, Munson GP. 2011. The tib adherence locus of enterotoxigenic Escherichia coli is regulated by cyclic AMP receptor protein. J. Bacteriol. 193:1369–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fang H, et al. 2010. An FDA bioinformatics tool for microbial genomics research on molecular characterization of bacterial foodborne pathogens using microarrays. BMC Bioinformatics 11(Suppl. 6):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fleckenstein JM, et al. 2010. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 12:89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaastra W, Svennerholm AM. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444–452 [DOI] [PubMed] [Google Scholar]
- 13. Girón JA, Torres AG, Freer E, Kaper JB. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361–379 [DOI] [PubMed] [Google Scholar]
- 14. Grewal HM, et al. 1997. A new putative fimbrial colonization factor, CS19, of human enterotoxigenic Escherichia coli. Infect. Immun. 65:507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harries JT, Sladen GE. 1972. The effects of different bile salts on the absorption of fluid, electrolytes, and monosaccharides in the small intestine of the rat in vivo. Gut 13:596–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iguchi A, et al. 2009. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191:347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jarvis KG, et al. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. U. S. A. 92:7996–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kendall MM, Rasko DA, Sperandio V. 2007. Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 75:4875–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levine MM, et al. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119–1122 [DOI] [PubMed] [Google Scholar]
- 22. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mahon V, Smyth CJ, Smith SG. 2010. Mutagenesis of the Rns regulator of enterotoxigenic Escherichia coli reveals roles for a linker sequence and two helix-turn-helix motifs. Microbiology 156:2796–2806 [DOI] [PubMed] [Google Scholar]
- 24. Merritt ME, Donaldson JR. 2009. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J. Med. Microbiol. 58:1533–1541 [DOI] [PubMed] [Google Scholar]
- 25. Moreira CG, Weinshenker D, Sperandio V. 2010. QseC mediates Salmonella enterica serovar Typhimurium virulence in vitro and in vivo. Infect. Immun. 78:914–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Munson GP, Holcomb LG, Scott JR. 2001. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infect. Immun. 69:186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Olier M, et al. 2004. Screening of glutamate decarboxylase activity and bile salt resistance of human asymptomatic carriage, clinical, food, and environmental isolates of Listeria monocytogenes. Int. J. Food Microbiol. 93:87–99 [DOI] [PubMed] [Google Scholar]
- 28. Parker CT, Sperandio V. 2009. Cell-to-cell signalling during pathogenesis. Cell. Microbiol. 11:363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel SK, Dotson J, Allen KP, Fleckenstein JM. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 72:1786–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prouty AM, et al. 2004. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol. Med. Microbiol. 41:177–185 [DOI] [PubMed] [Google Scholar]
- 32. Prouty AM, Gunn JS. 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 68:6763–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pumbwe L, et al. 2007. Bile salts enhance bacterial co-aggregation, bacterial-intestinal epithelial cell adhesion, biofilm formation and antimicrobial resistance of Bacteroides fragilis. Microb. Pathog. 43:78–87 [DOI] [PubMed] [Google Scholar]
- 34. Rasko DA, et al. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rasko DA, et al. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881–6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roy K, Bartels S, Qadri F, Fleckenstein JM. 2010. Enterotoxigenic Escherichia coli elicits immune responses to multiple surface proteins. Infect. Immun. 78:3027–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roy K, Hamilton D, Allen KP, Randolph MP, Fleckenstein JM. 2008. The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect. Immun. 76:2106–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roy K, Hamilton D, Ostmann MM, Fleckenstein JM. 2009. Vaccination with EtpA glycoprotein or flagellin protects against colonization with enterotoxigenic Escherichia coli in a murine model. Vaccine 27:4601–4608 [DOI] [PubMed] [Google Scholar]
- 39. Roy K, et al. 2009. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457:594–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rozen S, Skaletsky HJ. 2000. Primer3 on the WWW for general users and for biologist programmers, p 365–386 In Krawetz S, Misener S. (ed), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 41. Ruijter JM, et al. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rychlik I, Barrow PA. 2005. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol. Rev. 29:1021–1040 [DOI] [PubMed] [Google Scholar]
- 43. Sack RB, et al. 1971. Enterotoxigenic Escherichia coli isolated from patients with severe cholera-like disease. J. Infect. Dis. 123:378–385 [DOI] [PubMed] [Google Scholar]
- 44. Sahl JW, et al. 2011. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect. Immun. 79:950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Savarino SJ, Fasano A, Robertson DC, Levine MM. 1991. Enteroaggregative Escherichia coli elaborate a heat-stable enterotoxin demonstrable in an in vitro rabbit intestinal model. J. Clin. Invest. 87:1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schauder S, Shokat K, Surette MG, Bassler BL. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463–476 [DOI] [PubMed] [Google Scholar]
- 47. Schuhmacher DA, Klose KE. 1999. Environmental signals modulate ToxT-dependent virulence factor expression in Vibrio cholerae. J. Bacteriol. 181:1508–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sjöling A, Wiklund G, Savarino SJ, Cohen DI, Svennerholm AM. 2007. Comparative analyses of phenotypic and genotypic methods for detection of enterotoxigenic Escherichia coli toxins and colonization factors. J. Clin. Microbiol. 45:3295–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sommerfelt H, et al. 1992. Genetic relationship of putative colonization factor O166 to colonization factor antigen I and coli surface antigen 4 of enterotoxigenic Escherichia coli. Infect. Immun. 60:3799–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sorek R, Cossart P. 2010. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 11:9–16 [DOI] [PubMed] [Google Scholar]
- 51. Sperandio V, Mellies JL, Nguyen W, Shin S, Kaper JB. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Torres ME, et al. 2001. Etiology of children's diarrhea in Montevideo, Uruguay: associated pathogens and unusual isolates. J. Clin. Microbiol. 39:2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Turner SM, et al. 2006. Phylogenetic comparisons reveal multiple acquisitions of the toxin genes by enterotoxigenic Escherichia coli strains of different evolutionary lineages. J. Clin. Microbiol. 44:4528–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vandesompele J, et al. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. WHO 2006. Future directions for research on enterotoxigenic Escherichia coli vaccines for developing countries. Wkly. Epidemiol. Rec. 81:97–104 [PubMed] [Google Scholar]
- 56. Wibbenmeyer JA, Provenzano D, Landry CF, Klose KE, Delcour AH. 2002. Vibrio cholerae OmpU and OmpT porins are differentially affected by bile. Infect. Immun. 70:121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wilson CL, Miller CJ. 2005. Simpleaffy: a BioConductor package for Affymetrix Quality Control and data analysis. Bioinformatics 21:3683–3685 [DOI] [PubMed] [Google Scholar]
- 58. Yamamoto T, Echeverria P. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64:1441–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhu J, et al. 2011. Involvement of quorum sensing and heat-stable enterotoxin A in cell damage caused by a porcine enterotoxigenic Escherichia coli strain. Infect. Immun. 79:1688–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.