Abstract
The microbial communities that reside within the intestinal tract in vertebrates are complex and dynamic. In this report, we establish the utility of Caenorhabditis elegans as a model system for identifying the factors that contribute to bacterial persistence and for host control of gut luminal populations. We found that for N2 worms grown on mixed lawns of bacteria, Salmonella enterica serovar Typhimurium substantially outcompeted Escherichia coli, even when E. coli was initially present at 100-fold-higher concentrations. To address whether innate immunity affects the competition, the daf-2 and daf-16 mutants were studied; their total gut bacterial levels reflect overall capacity for colonization, but Salmonella outcompeted E. coli to an extent similar to wild-type worms. To address the role of virulence properties, Salmonella Δspi-1 Δspi-2 was used to compete with E. coli. The net differential was significantly less than that for wild-type Salmonella; thus, spi-1 spi-2 encodes C. elegans colonization factors. An E. coli strain with repeated in vivo passage had an enhanced ability to compete against an in vitro-passed E. coli strain and against Salmonella. Our data provide evidence of active competition for colonization niches in the C. elegans gut, as determined by bacterial factors and subject to in vivo selection.
INTRODUCTION
The microbial communities that reside within the intestinal tract in vertebrates are complex, and their populations are dynamic (11, 25, 43–45, 49). However, little is known about how microorganisms establish and maintain stable colonization of their hosts. How do different species found and maintain a presence in the intestinal ecosystem? How do they interact with their host and with other microbial community members? In which regions of the intestine is colonization preferred? Why are some strains favored over others? Attempts to establish suitable animal models to address these questions using pigs, chicks, mice and other hosts are limited by difficulties in animal handling, including needs for xenobiosis, and establishing colonization with limited numbers of bacterial species is expensive (51, 59). However, reliable animal models are needed for assessing the contribution of both bacterial and host factors to the process of intestinal colonization.
Simplified invertebrate model systems facilitate the understanding of the mechanisms underlying the interactions between gut microorganisms and their host. Caenorhabditis elegans, a free-living soil nematode that feeds primarily on bacteria (10), possesses important attributes, permitting it to serve as a model host to address such questions. Its rapid generation time, ease of propagation, well-defined cell lineage, fully sequenced genome containing a large number of vertebrate orthologues (50), and genetic tractability has aided the study of many biological processes, including microbial pathogenesis (1, 2, 8, 14, 15, 21, 22, 24, 29, 31, 39, 57) and immunity (26, 30, 32, 36, 40, 41, 52, 55, 56). C. elegans possesses evolutionarily conserved signaling pathways for innate immunity, especially those involving the DAF-2 insulin/IGF-I-like receptor (12, 19, 20, 48), p38 MAP kinase (37, 38, 58), and transforming growth factor β (TGF-β) (16, 46), which regulate an array of antibacterial effector molecules, including lysozymes, lipases, and C-type lectins (18, 26, 47).
The C. elegans gut, composed of 20 epithelial cells arranged to form a tube with a central lumen (53), can harbor large bacterial populations during adult life and thus can be used as a model for commensalism. When C. elegans is grown on bacterial lawns, most of the ingested bacterial cells are killed by the pharyngeal grinder (6, 7); however, some escape and can retain viability within the C. elegans gut, phenomena observed for both Escherichia coli and Salmonella enterica serovar Typhimurium (3, 42). We sought to harness this property as a way to understand both bacterial persistence and competition within the C. elegans lumen. We hypothesized that selection occurs within the C. elegans gut, leading to dominance, coexistence, or replacement of competing bacteria. After hypothesizing that both host selection and particular strain characteristics affect the outcome of the competition, we used competition between E. coli and Salmonella in hosts with specific immune phenotypes to address questions that pertain to the establishment of commensalism.
MATERIALS AND METHODS
C. elegans strains and growth conditions.
All strains were provided by the Caenorhabditis Genetic Center and maintained on modified (0.35% peptone) nematode growth medium (mNGM), using standard procedures (54).
Bacterial strains, plasmids, and growth conditions.
E. coli OP50 (10) and S. enterica serovar Typhimurium SL1344 (60) have been described. S. Typhimurium SL1344 containing plasmid pSMC21 (9) and the Δspi-1 Δspi-2 mutant were kindly provided by Fred Ausubel (3) and Heran Darwin, respectively. Cultures were grown in Luria-Bertani (LB) broth at 37°C supplemented or not with ampicillin (100 μg/ml), kanamycin (50 μg/ml), streptomycin (60 μg/ml), and tetracycline (20 μg/ml). Bacterial lawns used for C. elegans life span assays were prepared by spreading 25 μl of an overnight culture of the bacterial strains on 3.5-cm-diameter mNGM agar plates. Plates were incubated overnight at 37°C and cooled to room temperature before use. Bacterial strains and plasmids used in this study are listed in Table 1.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype | Source and reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| OP50 | Uracil auxotroph | Caenorhabditis Genetics Center (10), University of Minnesota |
| OP50mgh | Uracil auxotroph, Strr | F. M. Ausubel, Harvard Medical School |
| S. Typhimurium | ||
| SL1344 | Wild type, Strr | F. M. Ausubel (3) |
| spi-1 spi-2 strain | ΔSPI-I and ΔSPI-II, Kmr Cmr | K. H. Darwin, NYU School of Medicine |
| Plasmids | ||
| pSMC21 | Ampr Kmr, GFP | G. V. Bloemberg (9), Harvard Medical School |
| pGB5 | Ampr Tetr, GFP | G. V. Bloemberg (9) |
| pRZT3 | Tetr, DsRed | W. Bitter (49), VU Medical Centre, Amsterdam, Netherlands |
In vitro competition assays.
To determine in vitro competition between bacterial strains at 25°C, the temperature used for the in vivo studies, single colonies of E. coli and S. Typhimurium SL1344 or the Salmonella Δspi-1 Δspi-2 mutant were used to inoculate LB broth. Cultures were incubated overnight at 37°C and, the next morning, adjusted to the same optical density (0.250). One milliliter of each culture was then used to inoculate 10 ml of fresh LB broth supplemented with streptomycin (60 μg/ml) to control contamination, since both strains are streptomycin resistant. Tubes then were incubated at 25°C, and samples were obtained every 3 to 6 h to determine the number of bacterial CFU. Dilutions were plated on MacConkey agar supplemented with streptomycin (60 μg/ml) to identify the two competing bacterial populations.
Life span assays.
C. elegans life span determinations essentially followed established methods (3, 23). However, to avoid competition between introduced bacterial strains, nematodes were age synchronized by a bleaching procedure as described previously (54), and then embryos were incubated at 25°C on mNGM agar plates containing E. coli OP50, S. Typhimurium SL1344, or its Δspi-1 Δspi-2 mutant. The fourth larval stage (L4) was designated day 0 for our studies, and worms were transferred daily to fresh plates to eliminate overcrowding by progeny and until they laid no further eggs. Worm mortality was scored over time, with death defined when a worm no longer responded to touch (35). Worms that died of protruding/bursting vulva, bagging, or crawling off the agar were excluded from the analysis (5). Kaplan-Meier survival analysis was performed using GraphPad Prism. For each bacterial lawn, the time required for 50% of the worms to die (TD50) in each mutant population was compared to that in the wild-type population, using a paired t test. A P value of <0.05 was considered to reflect significant differences from the control. A total of 100 worms were used in each life span experiment, and all experiments were performed at least in duplicate.
Bacterial colonization assay.
Nematodes were age synchronized by bleaching (54), and embryos were incubated at 25°C on mNGM agar plates containing E. coli OP50, S. Typhimurium SL1344, or SL1344 Δspi-1 Δspi-2, as described above, to prepare for the bacterial colonization assays. Bacterial colonization of C. elegans was determined using a method adapted from Garsin et al. (24) and others (4; R. A. Alegado, personal communication). At each time point tested, 10 worms were picked and placed on an agar plate containing 100 μg/ml gentamicin to remove surface bacteria. The worms then were washed in 5-μl drops of 25 mM levamisole in M9 buffer (LM buffer) for paralysis and inhibition of pharyngeal pumping and expulsion, were washed twice more with LM buffer containing 100 μg/ml gentamicin, and washed twice more with M9 buffer alone. The washed nematodes then were placed in a 1.5-ml Eppendorf tube containing 50 μl of phosphate-buffered saline (PBS) buffer with 1% Triton X-100 and mechanically disrupted using a motor pestle. Worm lysates were diluted in PBS buffer and incubated overnight at 37°C on MacConkey agar. Lactose-fermenting (E. coli) and nonfermenting (Salmonella) colonies were quantified and used to calculate the number of bacteria per nematode. We have previously reported the bacterial intestinal colonization of wild-type, daf-2, and daf-16 worms (C. Portal-Celhay, E. R. Bradley, and M. J. Blaser, submitted for publication).
Shift and competition assays.
To examine the persistence of colonization after a change (“shift”) in bacterial lawns, C. elegans embryos were grown on NGM agar plates seeded with a founder bacterial strain (either E. coli OP50-pSMC21 or S. Typhimurium SL1344-pSMC21). At either day 2 (48 h) or day 4 (96 h) of adulthood (L4 plus 2 or L4 plus 4, respectively), worms were washed as described above and transferred to plates seeded with a second (introduced) bacterial strain (either E. coli OP50-pGB5 or S. Typhimurium SL1344-pGB5). After 24 h, 10 worms were washed, homogenized by grinding, and plated on selective agars to quantify the two distinct (founder and introduced residual) bacterial populations.
To evaluate the competition to colonize C. elegans, lawns were composed of two bacterial species. Embryos were initially grown on NGM agar plates seeded with E. coli OP50 (Strs). At day 3 of adulthood, worms were washed and transferred to new plates containing streptomycin and mixed lawns of E. coli OP50mgh (Strr) and S. Typhimurium SL1344 (Strr), with different ratios (0.1:10, 10:10, and 10:0.1) of the two bacterial strains. Worms were allowed to feed on the mixed lawns at 25°C for 6 h and then transferred back to plates seeded with E. coli OP50 (Strs). After an additional 24 h, 10 worms for each condition were washed, homogenized by grinding, and plated on MacConkey agar supplemented with streptomycin (to eliminate the Strs E. coli cells) to quantitate the two founding and competing Strr bacterial populations.
Adaptation assay.
C. elegans strain N2 was fed on mixed lawns of E. coli OP50mgh (Strr) and S. Typhimurium SL1344 (Strr) at a 10:10 ratio, allowed to feed for 6 h, and transferred back to plates seeded with E. coli OP50 (Strs). After an additional 24 h, 10 worms were washed, homogenized by grinding, and plated on MacConkey agar supplemented with streptomycin. A single colony of E. coli was reisolated and expanded in vitro by subculturing. This in vivo passage was performed 10 times, and the sequentially in vivo-passaged E. coli cells were subsequently used for the creation of mixed (Salmonella and E. coli) bacterial lawns and for competition assays. In this way, in vitro- and in vivo-passaged E. coli cells could be compared in terms of in vivo fitness vis a vis Salmonella cells.
Fluorescence microscopy.
Worms were washed and placed on a pad of 2% agarose in 5 μl of M9 buffer, with 30 mM sodium azide as an anesthetic. When the worms stopped moving, a coverslip was placed over the pad, and worms were examined by fluorescence microscopy using a Leica DMI 6000B inverted microscope.
Statistical analysis.
All assays were repeated multiple times. Linear regression analysis was done using Sigma Plot V.10. Data were analyzed using two-sample t tests, assuming equal variances. A P value of <0.05 was considered significantly different from the control.
RESULTS
Host age and genotype determine the nature of persistent bacterial colonization of the C. elegans gut.
In prior work, we characterized bacterial proliferation inside the C. elegans gut by quantifying viable bacteria and assessing variation in mutants with specific immune phenotypes (Portal-Celhay et al., submitted). As C. elegans ages, its intestinal bacterial load increases, and we found an inverse relationship with life span. We now confirm that the E. coli OP50 intestinal colonization of the long-lived daf-2 mutants is an order of magnitude lower than that of N2 worms (Fig. 1). There were significantly higher E. coli luminal densities in daf-16 worms, and those in the pharynx-defective phm-2 mutants were higher than in N2 worms, differences consistent with variation in worm longevity (Fig. 1A and B). Studies of the phm-2 worms confirm the critical role (6) of an intact grinder in limiting introduction of viable bacteria into the C. elegans gut (Fig. 1B). Using the daf-16(m26) daf-2(el370) double mutant, we previously showed that the daf-16 mutation suppresses the low levels of bacterial colonization in daf-2 strains (Portal-Celhay et al., submitted). Notably, in each worm background we observed, we could enumerate viable bacterial colonization of the gut lumen persisting beyond a week at the host-specific level, permitting studies of the determinants of colonization.
Fig 1.
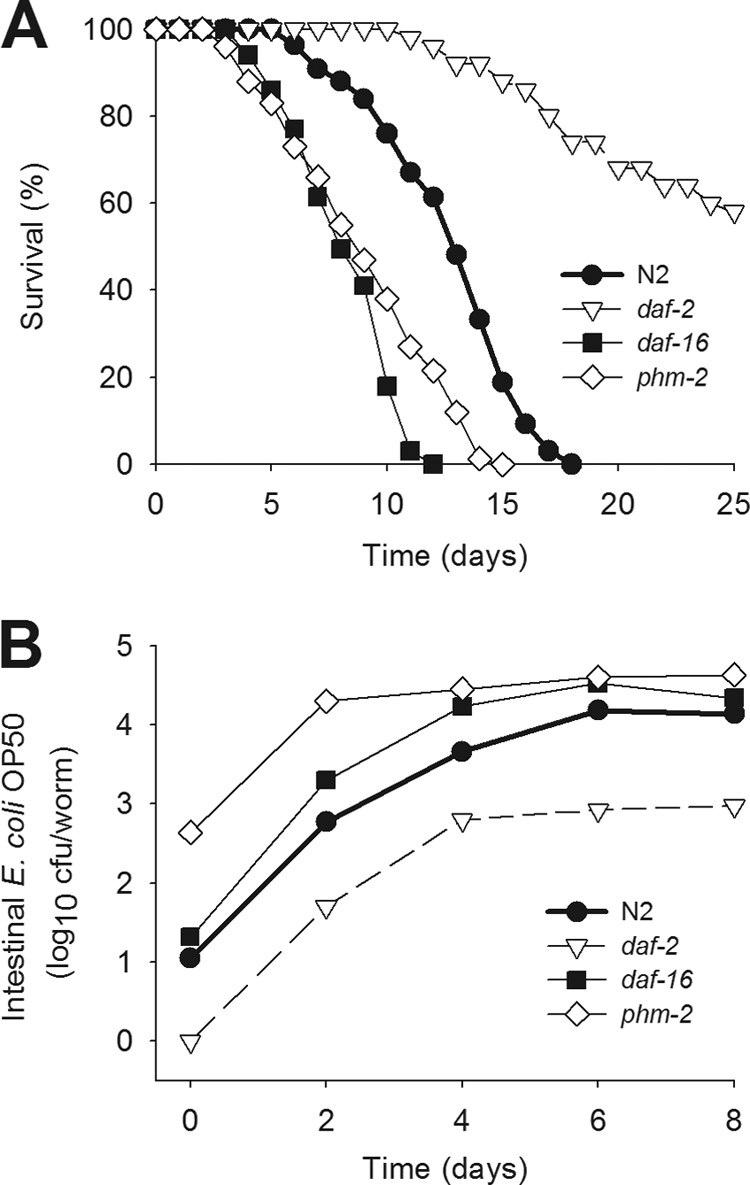
Survival and density of colonizing bacteria in the C. elegans gut. (A) Survival of the C. elegans N2 strain and daf-2, daf-16, and phm-2 mutants when grown on lawns of E. coli OP50. (B) Load of E. coli OP50 within the gut of wild-type C. elegans N2 or in the daf-2, daf-16, and phm-2 mutants for the first 8 days after the L4 stage.
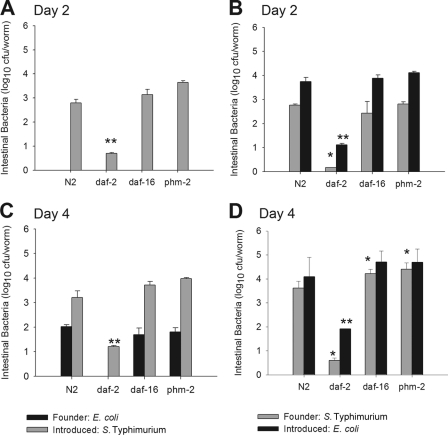
The rise in gut bacterial counts that occurs during the early days of worm aging (Fig. 1B) could reflect new bacterial entry from the lawn, in excess of the grinder capacity, or alternatively, could reflect the persistence and proliferation of bacteria already present. We created model systems to evaluate these two possibilities. First, to determine whether individual bacteria are able to persist within the C. elegans gut, N2 worms were grown on lawns of founder bacteria (E. coli pSMC21) and transferred 48 h later to new plates seeded with S. Typhimurium pGB5 (introduced bacteria). In opposite experiments, worms were grown on lawns of founder S. Typhimurium pSMC21 and then transferred 48 h later to new plates seeded with (introduced) E. coli pGB5. We asked whether the founder bacteria present in the gut lumen could resist the introduction of bacteria from the new lawn that had circumvented the grinder. First, we found that no viable founder E. coli could be recovered from the C. elegans intestine 24 h after feeding with the introduced Salmonella; Salmonella completely replaced E. coli (Fig. 2A). In contrast, founder Salmonella was detected in the intestinal tracts of worms that had been transferred to lawns of introduced E. coli (Fig. 2B). Since the worms no longer were exposed to Salmonella, these observations indicated the resilience of the founder Salmonella cells in the intestinal niche, permitting persistence. As worms age, the founding bacterial populations show greater resilience in the face of introduced organisms (Fig. 3).
Fig 2.
Resilience of bacterial gut colonization depends on the bacterial strain and on C. elegans age and genotype. Studies were done at 48 h (day 2) (A and B) or at 96 h (day 4) (C and D) following shift and competition assays, as described in Materials and Methods. C. elegans N2 or mutant strains were grown on lawns of the founder bacteria E. coli pSMC21 and then transferred to new plates seeded with S. Typhimurium pGB5 at 48 h (A) or at 96 h (C). Alternatively, C. elegans N2 or mutants were grown on lawns of the founder bacteria S. Typhimurium pSMC21 and then transferred to new plates seeded with E. coli pGB5 at 48 h (B) or at 96 h (D). Bacterial density was determined by plating the intestinal contents of lysed worms on selective media. Data represent means ± standard deviations (SD). Asterisks indicate a significant difference (P < 0.05) compared to the density of the founder strain recovered from N2 worms (*) or significant difference compared to the density of the introduced strain (**).
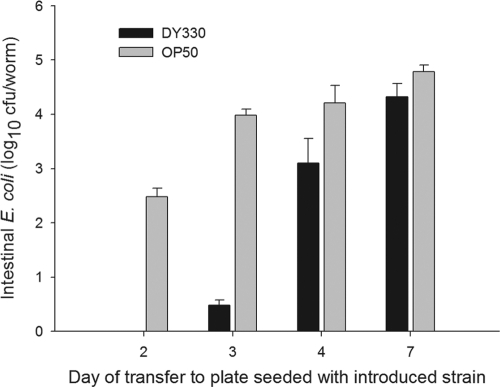
Fig 3.
Effect of aging on E. coli founder strain persistence in C. elegans. C. elegans N2 worms were initially grown on lawns of founder E. coli DY330 (black bars) and then moved at different points in worm maturation (L4 stage plus 2, 3, 4, or 7 days) to plates with lawns of introduced E. coli OP50. Bars show intestinal densities of founder E. coli DY330 (black) and introduced E. coli OP50 (gray bars) recovered 24 h after shifting the worms to lawns of the introduced strain. The older the worms, the more the founder (DY330) strain was able to resist the newly introduced strains; with aging, the concentrations of the introduced (OP50) strain also increased but less dramatically. Data represent means ± SD.
We next asked whether the age of the worm at the time of the transfer affected the resilience of the colonizing populations. When worms were transferred to the new lawns after 96 h instead of after 48 h, overall colonization was at a higher level, as expected (Fig. 1). However, E. coli cells now were able to persist in the gut lumen despite the influx of Salmonella (Fig. 2C). At 96 h, Salmonella also persisted as a founder strain (Fig. 2D), as expected from the 48-h studies, and reached higher levels than did the E. coli founder strain in the reciprocal 96-h studies (Fig. 2C). Thus, the resilience of the founding bacterial population was determined by the specific combination of founder and introduced strains, in the context of host age.
Role of gut environment in persistence.
We next examined how differing C. elegans gut environments affect founding bacterial persistence. First, we studied daf-2 and daf-16 worms, which have the essentially opposite phenotypes of restricting and enhancing bacterial colonization in the gut lumen, respectively (Fig. 1). As with wild-type worms, founder E. coli was not able to persist in the gut of the daf-2 or the daf-16 mutants when Salmonella was introduced at 48 h (Fig. 2A). In contrast, the founder Salmonella cells resisted E. coli introduction at the same time point (48 h) within both mutants (Fig. 2B). However, in the daf-2 mutants, both founder and introduced strains were at significantly lower levels than in N2 worms. Transferring the mutant worms to new lawns at 96 h (instead of at 48 h) showed trends similar to those observed for N2 worms (Fig. 2C and D); however, founder Salmonella cells persisted at significantly higher levels in the daf-16 mutants and at significantly lower levels in the daf-2 mutants (Fig. 2D). In general, the daf-2 environment was the most inhibitory one for founder E. coli or Salmonella in relation to the introduced strains.
We also asked whether the numbers of newly introduced competitors delivered to the gut affect the ability of founder bacterial cells to persist in the lumen. For these studies, we examined phm-2 mutants in which defective grinders (7, 42) allow delivery of higher gut loads (Fig. 1B). With change of the lawn at 48 h, the gut load of the introduced strains was somewhat but not significantly higher than that of N2. With change at 96 h, the founder Salmonella persisted at significantly higher levels than those of the N2 worm (Fig. 2). In total, across the range of gut conditions present in the daf-2, daf-16, and phm-2 mutants, parallel phenomena of strain specificity and worm age dependence in the gut colonization competition were observed. The worm genotypes studied affected overall bacterial load and, to a limited extent, altered the dynamics between the founder and introduced bacteria.
Worm age affects the resilience of the colonizing population.
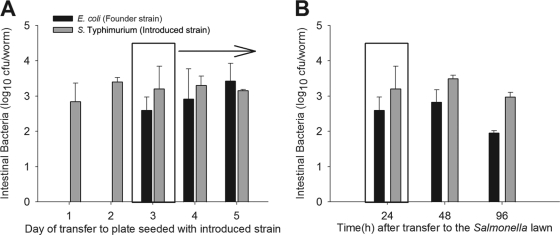
Since founder Salmonella organisms are highly resilient to displacement by introduced E. coli, whereas outcomes are more conditional on worm age for founder E. coli (Fig. 2), the use of founder E. coli provides further experimental opportunities to probe the selective pressures. To independently confirm the hypothesis that worm age determines the persistence of the founder bacteria, we conducted experiments in which Salmonella was introduced progressively later to worms with founder E. coli populations and then sampled the worms 24 h after each switch. As increasingly older E. coli-fed worms were exposed to the introduced Salmonella strain, there was progressively greater persistence of the founder E. coli strain (Fig. 4A), confirming and extending the 48-h and 96-h findings (Fig. 2). Next, to determine whether sampling only 24 h after the switch might reflect only transient events, we conducted experiments in which sampling of the worms after the switch ranged from 24 to 96 h. Over the course of the 96-h sampling after the switch (Fig. 2B), the founder bacteria continue to persist in essentially the same proportion to the introduced bacteria, despite continuing exposure to the latter. This experiment provides further evidence that founder bacterial cells can persist and multiply in the gut lumen and that the niches they establish can substantially resist a continuing influx of introduced and, thus, competing bacteria. In total, these data indicate that founder Salmonella is able to establish persistent gut colonization of N2 and mutant C. elegans that resist introduced E. coli, regardless of host age or worm genotype. However, the resilience of E. coli in the luminal environment has greater dependence on host age, whether in wild-type or mutant worms.
Fig 4.
E. coli OP50 can resist the intrusion of Salmonella in the gut of older worms. (A) Gut densities of E. coli OP50 (black bars) and S. Typhimurium SL1344 (gray bars) recovered from C. elegans N2 after their growth on founder E. coli OP50 lawns and then after the substitution of S. Typhimurium lawns at several points in worm maturation (L4 stage plus 1, 2, 3, 4, or 5 days). Data represent means ± SD. (B) Intestinal bacteria recovered from C. elegans N2 after being switched from lawns of founder E. coli OP50 (black bars) to lawns of S. Typhimurium SL1344 (gray bars), which were transferred on day 3 (L4 plus 3), with worms then maintained on the introduced S. Typhimurium strain with sampling of the worms at 24, 48, and 96 h after the shift.
Competition in vitro.
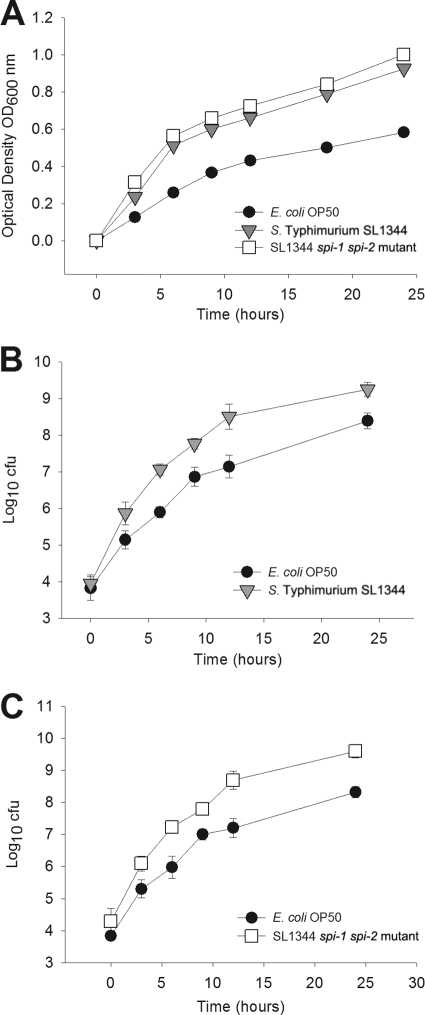
In the above-described studies, we examined the sequential exposure to potential bacterial colonizers, but next we questioned whether introducing the strains simultaneously would change the dynamics. To address this question, we performed experiments in which lawns were mixtures of E. coli and Salmonella in different proportions. First, we assessed whether the E. coli or Salmonella strains used had substantial in vivo growth advantage. We found that at 37°C, E. coli and Salmonella growth rates are almost identical (data not shown). However, at 25°C, the temperature at which the competition assays were conducted and in which the worms were incubated, Salmonella had a growth advantage (Fig. 5A). Similarly, and in an in vitro competition experiment, both wild-type and Salmonella Δspi-1 Δspi-2 outcompeted E. coli by a difference of 1 log10 (Fig. 5B and C). Taken together, these findings could in part explain the greater resilience of both founder and introduced Salmonella in the sequential colonization studies (Fig. 2 and 4). Nevertheless, despite the growth differences, persistence in the gut lumen also could be demonstrated for E. coli (Fig. 2C and D).
Fig 5.
In vitro growth and competition of E. coli and Salmonella. (A) Growth of E. coli, S. Typhimurium SL1344, and the SL1344 spi-1 spi-2 mutant on LB broth supplemented with 60 μg/ml streptomycin and incubated at 25°C; (B) competition between E. coli and S. Typhimurium SL1344 in vitro in broth culture; (C) competition between E. coli OP50 and the SL1344 spi-1 spi-2 mutant, as described in panel B.
Competition for gut colonization.
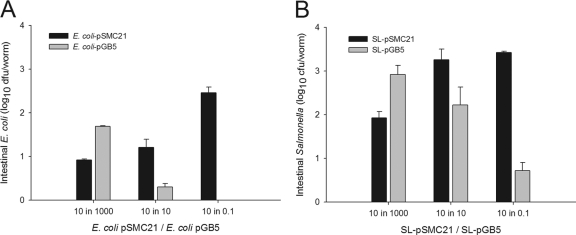
Next, to examine the dynamics of in vivo competition, we created lawns with mixtures of E. coli and Salmonella cells to introduce the two strains simultaneously and then to determine the proportions of each that would be recovered from the worms. To calibrate the system, we first competed two variants of S. Typhimurium strain SL1344, differentiated by their plasmid-encoded antibiotic resistance phenotypes. As expected, the cells were recovered from the worms essentially in proportion to the input ratios; neither strain had any selective advantage (Fig. 6A). That the competition between the two strains was neutral provided an important validation of the assay for examining other strain combinations. Competition between two variants of E. coli was performed as well (Fig. 7); the unequal levels reflected colonization differences between the strains, possibly due to extra genetic cost.
Fig 6.
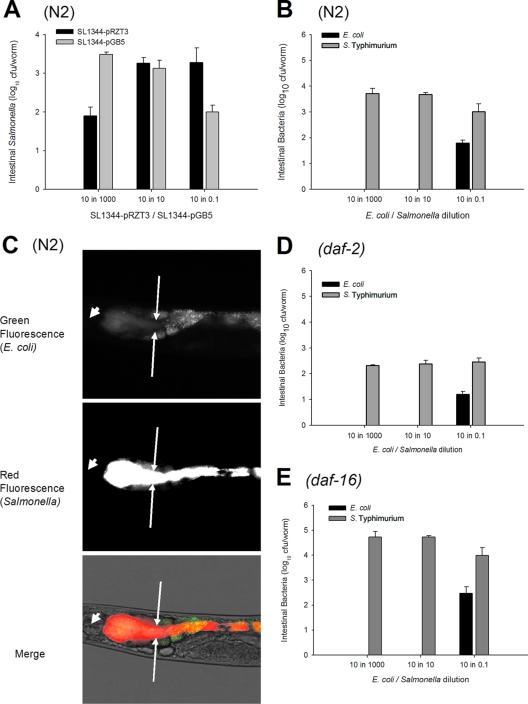
Simultaneous competition between E. coli and S. Typhimurium for gut colonization of C. elegans N2 and mutants. (A) Gut densities at 24 h after exposure of N2 worms to S. Typhimurium SL1344-pRZT3 expressing DsRed (dark bars) and S. Typhimurium SL1344-pGB5 expressing GFP (gray bars) grown for 6 ho on mixed lawns at ratios of 10:1,000, 10:10, and 10:0.1. Data represent means ± SD. (B) Lawns with differing ratios (10:1,000, 10:10, 10:0.1) of E. coli OP50 and S. Typhimurium SL1344 were prepared, and worms fed on these lawns for 6 h. Levels of E. coli (black bars) and S. Typhimurium (gray bars) in the gut were measured 24 h later in wild-type C. elegans N2, as described above. (C) Fluorescence microscopy of C. elegans N2 after feeding for 12 h on mixed lawns of E. coli OP50-pGB5 and S. Typhimurium SL1344-pRZT3 at a ratio of 10:0.1 (100-fold E. coli excess). Arrows demarcate the gut lumen. Arrowheads indicate the pharyngeal grinder. Magnification, ×40. The green fluorescence channel shows E. coli inside the gut lumen and gut autofluorescence, and the red channel shows Salmonella fluorescence. The merged image shows bacterial colocalization (yellow). (D) Same as panel B in daf-2 mutants. (E) Same as panel B in daf-16 mutants.
Fig 7.
Simultaneous competition between two variants of E. coli (A) and two variants of S. Typhimurium (B) for gut colonization of C. elegans N2. (A) Gut densities at 24 h after exposure of N2 worms to E. coli pSMC21 expressing Amp and Km resistance (black bars) and E. coli pGB5 expressing Amp and Tet resistance (gray bars) grown for 6 h on mixed lawns at ratios of 10:1,000, 10:10, and 10:0.1. Data represent means ± SD. (B) S. Typhimurium pSMC21 and pGB5 on lawns, as explained for panel A.
When N2 worms were grown on the mixed lawns, the Salmonella cells substantially outcompeted E. coli even when E. coli was introduced at 100-fold-higher concentrations (Fig. 6B). We confirmed these findings by microscopy (Fig. 6C), visualizing E. coli OP50 expressing green fluorescent protein (GFP) and S. Typhimurium expressing red fluorescent protein (RFP) (DsRed) in worms fed on lawns with 100-fold E. coli excesses. Discrete clusters of green-fluorescing (E. coli) cells were observed predominately in the anterior gut (Fig. 6C, top), whereas red (Salmonella) fluorescence appeared uniformly throughout the gut lumen (Fig. 6C, middle). Overall, red dominates over green (Fig. 6C, bottom). Next, we asked whether the gut environment has a major role in the population dynamics by studying the E. coli/Salmonella competition in the opposing daf-2 and daf-16 mutants. Although the total bacterial levels in the daf-2 and daf-16 worms reflected their overall capacity for colonization (Fig. 2), Salmonella outcompeted E. coli to an extent paralleling that in the wild-type (N2) worms (Fig. 6D and E). Thus, in the setting of simultaneous introduction, bacterial species differences clearly affect the ability of the strains to persist in the worm lumen, whereas the studied host genotypes affect primarily the extent of the lumenal niche but not the competition dynamics.
Role of Salmonella tissue interaction in colonization, competition with E. coli, and C. elegans life span.
We next sought to develop a model to better understand why Salmonella is more efficient than E. coli at colonizing the C. elegans gut. Many of the genes that are involved in the interaction between Salmonella and its vertebrate hosts are located in the Salmonella pathogenicity islands (SPI) (17). SPI-1 and SPI-2 each carry components of type III secretion systems (T3SSs), the former being critical for invasion and intestinal disease in mammals and the latter important for intracellular survival, systemic persistence, and disease (13).
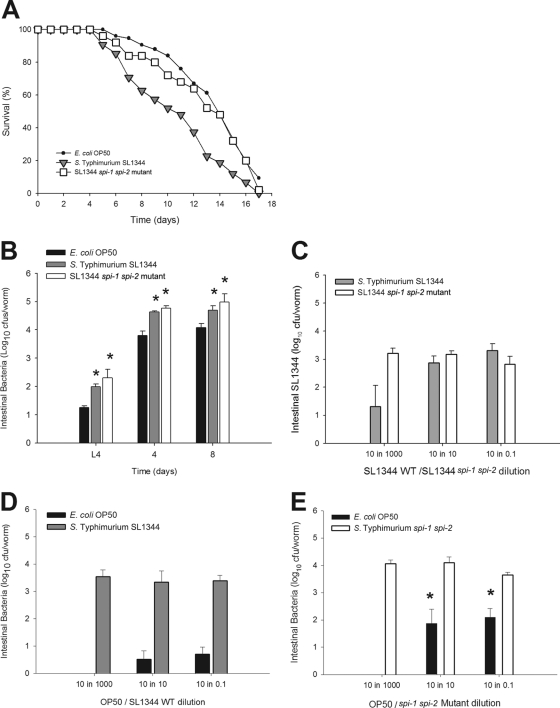
An early step in the establishment of S. Typhimurium infection in the vertebrate host gastrointestinal tract involves attachment and/or invasion of intestinal epithelial cells. To determine the role of SPI-1 SPI-2-carried genes in C. elegans gut colonization, we grew wild-type worms on plates seeded with S. Typhimurium SL1344, isogenic S. Typhimurium SL1344 Δspi-1 Δspi-2, or E. coli. As shown in life span assays for the N2 worms and as expected, Salmonella is more virulent (TD50 = 10.5 ± 0.5 days) than E. coli OP50 (TD50 = 13.8 ± 0.56; P = 0.03) (Fig. 8A). As expected, the Salmonella Δspi-1 Δspi-2 mutant was less virulent (TD50 = 13.1 ± 1.18 days) for C. elegans than isogenic wild-type Salmonella, confirming prior studies (4), and had virulence similar to E. coli OP50 (13.8 ± 0.56 days). However, the density of Salmonella Δspi-1 Δspi-2 in the C. elegans gut was consistently about 10-fold greater than that of E. coli OP50 through the first 8 days of adult life (Fig. 8B). The Salmonella Δspi-1 Δspi-2 mutant also colonized the C. elegans gut at higher levels than did wild-type Salmonella, despite its having a longer life span. Thus, bacterial density in the gut is not the sole determinant in life span, as shown by comparing the two Salmonella strains; host interaction is clearly critical.
Fig 8.
Role of the SPI-1/-2 islands in Salmonella colonization, resilience, and C. elegans longevity. (A) Life span assays of C. elegans raised on lawns of E. coli OP50 (black circles), wild-type S. Typhimurium SL1344 (gray diamonds), or S. Typhimurium SL1344 Δspi-1 Δspi-2 (white squares). (B) For worms grown on lawns of E. coli OP50 (dark bars), S. Typhimurium SL1344 (gray bars), or SL1344 Δspi-1 Δspi-2 (white bars), their gut densities in C. elegans N2 at L4 stage (L4 plus 0), day 4 (L4 plus 4), or day 8 (L4 plus 8) of their adult life span. Data represent means ± SD. An asterisk indicates a significant difference (P < 0.05) compared to growth on E. coli OP50. (C) Gut density of the S. Typhimurium SL1344 wild type (WT) (gray bars) and S. Typhimurium SL1344 Δspi-1 Δspi-2 (white bars), as described in panel D. (D) Gut density of E. coli OP50 (black bars) and S. Typhimurium SL1344 (gray bars) within wild-type C. elegans N2 grown on mixed lawns with ratios of 10:1,000, 10:10, and 10:0.1 of E. coli to Salmonella. Data represent means ± SD. (E) Gut density of E. coli OP50 (black bars) and S. Typhimurium SL1344 Δspi-1 Δspi-2 (white bars), as described in panel B.
To directly examine the dynamics of gut colonization, we performed competition studies using lawns with differing E. coli and Salmonella proportions (Fig. 8). In direct competition studies, the Δspi-1 Δspi-2 strain competed proportionally (or a little more so) in relation to the wild-type strain (Fig. 8C), consistent with the findings from studies in which monocolonization was done (Fig. 8B). Confirming the prior studies, wild-type Salmonella completely outcompeted E. coli, with essentially 4 log10 differentials, summating both the differences in lawn proportions and the observed colonization proportions (Fig. 8D). The Δspi-1 Δspi-2 strain also outcompeted E. coli (Fig. 8E), but the net differential, only 1 to 2 log10, was significantly less than that for wild-type Salmonella (P < 0.05). Since in vitro growth differences of E. coli with both strains are essentially the same (Fig. 5B and C), these experiments provide evidence that the differential colonization by E. coli and Salmonella cannot be explained solely on the basis of differential growth at 25°C but reflects host interaction.
Does worm passage select for better colonizers?
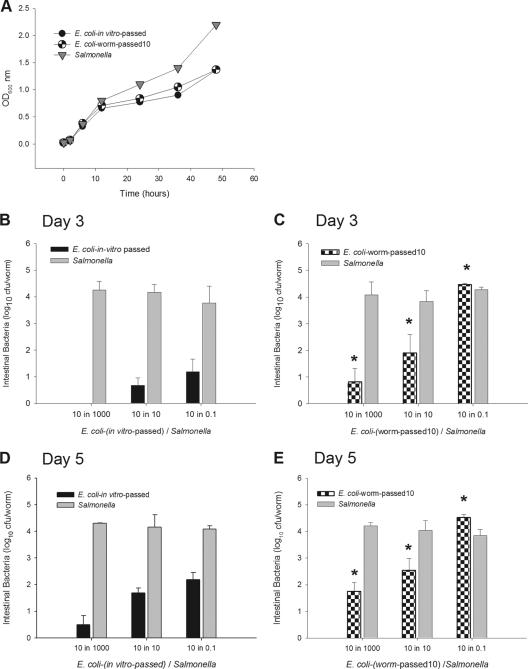
Finally, we asked whether the in vivo milieu selects for better-adapted bacteria or whether the apparent resilience of persisters was merely stochastic and not inherited. To address this question, E. coli OP50 was passed 10 times through the C. elegans gut or, in control procedures, passed in vitro for the corresponding period. These passaged strains then were competed with in vitro-passed Salmonella. Both strains grew similarly in vitro at 25°C and less well than Salmonella (Fig. 9A). When competitions were performed using 3-day-old worms, the ability of E. coli to colonize and compete against Salmonella was greatly enhanced after the in vivo passages, with significantly higher levels (P < 0.05) of colonization than the in vitro-passed strain (Fig. 9B and C). Although Salmonella still outcompeted both of the E. coli strains, the data provide evidence that there had been in vivo adaptation. To further assess the contribution of aging to the colonization process, we carried out the same competition assay using older (5-day-old, or 120-h-old) adult worms. In the older worms, the in vivo-passed E. coli again outperformed the in vitro-passed strain (Fig. 9D and E). We also compared the colonization profiles in the older (5-day-old) and younger (3-day-old) adults to examine the effects of host age on the competition dynamics. Although Salmonella levels were essentially fixed under all conditions studied (Fig. 9B to E), the extent of E. coli colonization was dependent on its both in vivo passage history and host age. In total, these experiments indicate that in vivo selection for bacterial genotypes enables better colonization of worms and that advancing worm age independently and disproportionately favors E. coli colonization.
Fig 9.
Selection for bacteria with improved colonization. (A) Growth of E. coli and Salmonella cells in vitro at 25°C, as shown in Fig. 4A. Cells include E. coli in vivo passed for 10 generations of worms (black-and-white circles), E. coli with parallel in vitro passage (black circles), and wild-type S. Typhimurium SL1344 (gray triangles). Lawns with differing ratios (10:1,000, 10:10, 10:0.1) of in vitro-passed E. coli OP50 and in vitro-passed S. Typhimurium SL1344 were prepared, and 3-day-old worms (B) or 5-day-old worms (D) fed on these for 6 h. Gut levels of E. coli (black bars) and S. Typhimurium (gray bars) were measured 24 h later in wild-type C. elegans N2. An asterisk indicates a significant difference (P < 0.05) compared to panel B. E. coli OP50 was sequentially passed through the worm gut 10 times (see Materials and Methods). Lawns with differing ratios (10:1,000, 10:10, 10:0.1) of worm-passed E. coli OP50 and in vitro-passed S. Typhimurium SL1344 were prepared, and 3-day-old worms (C) or 5-day-old worms (E) fed on these for 6 h. Gut levels of E. coli (black-and-white bars) and S. Typhimurium (gray bars) were measured 24 h later in wild-type C. elegans N2. An asterisk indicates a significant difference (P < 0.05) compared to panel B. OD, optical density.
DISCUSSION
Simplified and genetically tractable models of gut ecosystems are needed to understand how both hosts and bacteria actively collaborate to shape the overall intestinal milieu (27, 28). Earlier studies have shown that bacteria accumulate in the C. elegans intestine as they age (23) and that S. Typhimurium proliferates and establishes a persistent infection in the worm intestine (3). In prior work, we confirmed these findings and found a strong negative correlation between bacterial numbers and life span (Portal-Celhay et al., submitted). We also showed that the bacterial load reaches a strain-specific and host genome-specific plateau that extends until the worm's demise. We attributed this finding to a cohort effect, in which the fraction of worms examined in late worm adulthood constitutes a subpopulation that survived because they maintain the ability to control bacterial proliferation. Alternatively, late in life, the bacterial populations develop specific syntrophic equilibria that are resilient to changes in host milieu. But did we really observe bacterial proliferation, or are the bacteria accumulating in the intestine without dividing? In this study, we established the utility of the C. elegans gut as a model system to study intestinal bacterial colonization and proliferation and identified factors that contribute to microbial persistence. Furthermore, we provide evidence of active competition for colonization niches in the C. elegans gut, as determined by host age, bacterial factors, and in vivo selection.
In our experimental system, we found that there is resilience (defined as the ability to recover quickly from change) of naturally introduced Salmonella cells in the gut niche of young adult worms (L4 plus 48 h), permitting their persistence. In contrast, E. coli as a founder strain was entirely outcompeted by introduced Salmonella. Since Salmonella can disrupt the pharyngeal grinder (3), better resilience could simply be due to a higher number of cells delivered to the gut. However, in the phm-2 mutants, Salmonella persisted despite E. coli introduction (6), and E. coli founders were unable to persist after Salmonella introduction, indicating that grinder function is not a primary determinant of the competition. That founder E. coli also was not able to persist in the daf-2 or daf-16 mutants indicates that gut colonization resilience is bacterial strain specific, whereas the host genotypes studied predominantly determined the total number of bacterial cells that can colonize the gut.
The same experiments using older adult worms (L4 plus 96 h) showed that E. coli now better persists, which shows that worm age is a determinant of the resilience of founder bacteria. These results could be explained by three alternative but not exclusive mechanisms. One possibility is that the gut milieu of older worms is more permissive for bacterial cells in general, and the colonization experiments (Fig. 1 and 2) provide some support, yet colonization generally plateaus by about day 4. A second possibility is that over time within the C. elegans gut, there is selection for E. coli cells, which are better adapted to the gut niche in terms of avoiding killing by antimicrobial defenses, garnering nutrients, or resisting being flushed from the gut. A third possible mechanism is that E. coli is able to persist in the C. elegans gut once cellular and structural damage of the host intestine has been produced by other bacteria, such as Salmonella. Comparison of Salmonella competition with in vitro-passed and in vivo-passed E. coli (Fig. 9) in worms of different ages provided clear evidence for in vivo selection. Thus, this model system can now be used to further identify the selective forces and the relevant phenotypes (and genotypes) of the selected bacterial cells. Future work should include sequencing of the bacteria after several rounds of in vivo passaging.
That Salmonella was able to establish persistent gut colonization independent of the order of introduction or age of the host, whereas E. coli resilience depended on host age, establishes the principle of species-specific variation in adaptation for C. elegans gut colonization (18). Further E. coli/Salmonella dilutions and a time course to show proliferation of small inoculums of Salmonella would have been useful to better characterize the resilience of the Salmonella populations. However, that Salmonella grows faster in vitro than E. coli at the temperature (25°C) at which the competition assays are done complicates the analysis. However, the differing dynamics of the two Salmonella strains in relation to E. coli indicate that growth differences are not sufficient to explain the differing findings.
Why is Salmonella better at colonizing the C. elegans gut than E. coli? One possibility is that Salmonella's advantage reflects its more profound interaction with host intestinal cells, garnering an extra resource stream. However, the less virulent Δspi-1 Δspi-2 mutant strain colonized as well or better than the wild-type strain (Fig. 6D). It is not clear whether Salmonella adheres or invades the C. elegans gut epithelial cells (3, 4, 33, 42). Nevertheless, the Δspi-1 Δspi-2 mutant did not compete as effectively as did the wild type against E. coli, making exploration of the role of the SPI-carried genes worthwhile. Another possibility could be that E. coli cannot easily survive in an environment in which an active host response, elicited by wild-type Salmonella, is present. Competition against the Salmonella Δspi-1 Δspi-2 mutant, which elicits less intense host responses than does the wild type (34), should give E. coli the opportunity to fare better, which could explain the experimental observation (Fig. 8). Although the Δspi-1 Δspi-2 strain competes well against its wild-type competitor, it is inferior to the wild type in competing with E. coli and markedly less pathogenic for worms than the wild type (Fig. 8A). Thus, the SPI-1/-2 islands play roles in both intraluminal colonization and in host interactions that affect longevity, and in this case, immunosenescence may result in decreased ability to cope with factors produced by colonizers, in addition to the inability to clear ingested bacteria. Consistent with this hypothesis are the observations that nontoxic low-interaction bacteria (e.g., Enterococcus faecium) may accumulate to high densities in the intestinal environment but with small effects on longevity (24).
The results of these studies provide evidence for bacterial persistence in the C. elegans gut lumen, with ongoing selection dictating interspecies and intrastrain competition for survival. Use of the well-defined C. elegans model to examine bacterial competition within the intestinal tract of animals should enhance a mechanistic understanding of host-microbe interactions underlying commensalism in more complex animals, including humans.
ACKNOWLEDGMENTS
We thank the Caenorhabditis Genetics Center at the University of Minnesota for the strains used in this study, as well as George A. O'Toole, John Rawls, Nicole Iovine, and Heran Darwin for providing bacterial strains and plasmids.
This work was supported in part by NIH grant RO1 GM63270, the Ellison Medical Foundation, the Michael Saperstein Medical Scholars Program, and the Diane Belfer Program in Human Microbial Ecology.
Footnotes
Published ahead of print 19 December 2011
REFERENCES
- 1. Aballay A, Ausubel FM. 2002. Caenorhabditis elegans as a host for the study of host-pathogen interactions. Curr. Opin. Microbiol. 5:97–101 [DOI] [PubMed] [Google Scholar]
- 2. Aballay A, Drenkard E, Hilbun LR, Ausubel FM. 2003. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr. Biol. 13:47–52 [DOI] [PubMed] [Google Scholar]
- 3. Aballay A, Yorgey P, Ausubel FM. 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10:1539–1542 [DOI] [PubMed] [Google Scholar]
- 4. Alegado RA, Tan MW. 2008. Resistance to antimicrobial peptides contributes to persistence of Salmonella typhimurium in the C. elegans intestine. Cell. Microbiol. 10:1259–1273 [DOI] [PubMed] [Google Scholar]
- 5. Apfeld J, Kenyon C. 1999. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402:804–809 [DOI] [PubMed] [Google Scholar]
- 6. Avery L. 1993. The genetics of feeding in Caenorhabditis elegans. Genetics 133:897–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Avery L, Thomas JH. 1997. Feeding and defecation, p. 679–716 In Riddle DL, Blumenthal T, Meyer BJ, Priess JR. (ed), C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 8. Begun J, Sifri CD, Goldman S, Calderwood SB, Ausubel FM. 2005. Staphylococcus aureus virulence factors identified by using a high-throughput Caenorhabditis elegans-killing model. Infect. Immun. 73:872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bloemberg GV, O'Toole GA, Lugtenberg BJ, Kolter R. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl. Environ. Microbiol. 63:4543–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheesman SE, Guillemin K. 2007. We know you are in there: conversing with the indigenous gut microbiota. Res. Microbiol. 158:2–9 [DOI] [PubMed] [Google Scholar]
- 12. Chen CS, et al. 2010. WWP-1 is a novel modulator of the DAF-2 insulin-like signaling network involved in pore-forming toxin cellular defenses in Caenorhabditis elegans. PLoS One 5:e9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coburn B, Sekirov I, Finlay BB. 2007. Type III secretion systems and disease. Clin. Microbiol. Rev. 20:535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Couillault C, Ewbank JJ. 2002. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect. Immun. 70:4705–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Darby C. 6 September 2005, posting date Interactions with microbial pathogens. In The C. elegans Research Community (ed), WormBook. http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- 16. Dierking K, et al. 2011. Unusual regulation of a STAT protein by an SLC6 family transporter in C. elegans epidermal innate immunity. Cell Host Microbe 9:425–435 [DOI] [PubMed] [Google Scholar]
- 17. Ellermeier JR, Slauch JM. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24–29 [DOI] [PubMed] [Google Scholar]
- 18. Engelmann I, et al. 2011. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS One 6:e19055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Evans EA, Chen WC, Tan MW. 2008. The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging Cell 7:879–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Evans EA, Kawli T, Tan MW. 2008. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. 4:e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ewbank JJ. 2002. Tackling both sides of the host-pathogen equation with Caenorhabditis elegans. Microbes Infect. 4:247–256 [DOI] [PubMed] [Google Scholar]
- 22. Felix MA, et al. 2011. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 9:e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garigan D, et al. 2002. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161:1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garsin DA, et al. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U. S. A. 98:10892–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gill SR, et al. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gravato-Nobre MJ, Hodgkin J. 2005. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell. Microbiol. 7:741–751 [DOI] [PubMed] [Google Scholar]
- 27. Hooper LV, Gordon JI. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115–1118 [DOI] [PubMed] [Google Scholar]
- 28. Hooper LV, et al. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881–884 [DOI] [PubMed] [Google Scholar]
- 29. Huffman DL, Bischof LJ, Griffitts JS, Aroian RV. 2004. Pore worms: using Caenorhabditis elegans to study how bacterial toxins interact with their target host. Int. J. Med. Microbiol. 293:599–607 [DOI] [PubMed] [Google Scholar]
- 30. Irazoqui JE, Ausubel FM. 2010. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Caenorhabditis elegans as a model to study tissues involved in host immunity and microbial pathogenesis. Clin. Exp. Immunol. 160:48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Irazoqui JE, et al. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6:e1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Irazoqui JE, Urbach JM, Ausubel FM. 2010. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 10:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jia K, et al. 2009. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc. Natl. Acad. Sci. U. S. A. 106:14564–14569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jones BD, Falkow S. 1996. Salmonellosis: host immune responses and bacterial virulence determinants. Annu. Rev. Immunol. 14:533–561 [DOI] [PubMed] [Google Scholar]
- 35. Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366:461–464 [DOI] [PubMed] [Google Scholar]
- 36. Kim DH, Ausubel FM. 2005. Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans. Curr. Opin. Immunol. 17:4–10 [DOI] [PubMed] [Google Scholar]
- 37. Kim DH, et al. 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297:623–626 [DOI] [PubMed] [Google Scholar]
- 38. Kim DH, et al. 2004. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc. Natl. Acad. Sci. U. S. A. 101:10990–10994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kurz CL, Ewbank JJ. 2000. Caenorhabditis elegans for the study of host-pathogen interactions. Trends Microbiol. 8:142–144 [DOI] [PubMed] [Google Scholar]
- 40. Kurz CL, Ewbank JJ. 2003. Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat. Rev. Genet. 4:380–390 [DOI] [PubMed] [Google Scholar]
- 41. Kurz CL, Tan MW. 2004. Regulation of aging and innate immunity in C. elegans. Aging Cell 3:185–193 [DOI] [PubMed] [Google Scholar]
- 42. Labrousse A, Chauvet S, Couillault C, Kurz CL, Ewbank JJ. 2000. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol. 10:1543–1545 [DOI] [PubMed] [Google Scholar]
- 43. Ley RE, et al. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6:776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848 [DOI] [PubMed] [Google Scholar]
- 46. Mallo GV, et al. 2002. Inducible antibacterial defense system in C. elegans. Curr. Biol. 12:1209–1214 [DOI] [PubMed] [Google Scholar]
- 47. Millet AC, Ewbank JJ. 2004. Immunity in Caenorhabditis elegans. Curr. Opin. Immunol. 16:4–9 [DOI] [PubMed] [Google Scholar]
- 48. Murphy CT, et al. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424:277–283 [DOI] [PubMed] [Google Scholar]
- 49. Rawls JF, Mahowald MA, Goodman AL, Trent CM, Gordon JI. 2007. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc. Natl. Acad. Sci. U. S. A. 104:7622–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Riddle DL, Blumenthal T, Meyre BJ, Priess JR. (ed). 1997. C. elegans II, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 51. Rumney CJ, Rowland IR. 1992. In vivo and in vitro models of the human colonic flora. Crit. Rev. Food Sci. Nutr. 31:299–331 [DOI] [PubMed] [Google Scholar]
- 52. Schulenburg H, Kurz CL, Ewbank JJ. 2004. Evolution of the innate immune system: the worm perspective. Immunol. Rev. 198:36–58 [DOI] [PubMed] [Google Scholar]
- 53. Simon TC, Gordon JI. 1995. Intestinal epithelial cell differentiation: new insights from mice, flies and nematodes. Curr. Opin. Genet. Dev. 5:577–586 [DOI] [PubMed] [Google Scholar]
- 54. Stiernagle T. 11 February 2006, posting date Maintenance of C. elegans. In The C. elegans Research Community (ed), WormBook http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- 55. Styer KL, et al. 2008. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322:460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun J, Singh V, Kajino-Sakamoto R, Aballay A. 2011. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science 332:729–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tan MW. 2002. Identification of host and pathogen factors involved in virulence using Caenorhabditis elegans. Methods Enzymol. 358:13–28 [DOI] [PubMed] [Google Scholar]
- 58. Troemel ER, et al. 2006. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tsolis RM, et al. 1999. Of mice, calves, and men. Comparison of the mouse typhoid model with other Salmonella infections. Adv. Exp. Med. Biol. 473:261–274 [PubMed] [Google Scholar]
- 60. Wray C, Sojka WJ. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139–143 [PubMed] [Google Scholar]