Abstract
Toxoplasma gondii is the causative agent of toxoplasmosis in human and animals. In a mouse model, T. gondii strains can be divided into three groups, including the virulent, intermediately virulent, and nonvirulent. The clonal type I, II, and III T. gondii strains belong to these three groups, respectively. To better understand the basis of virulence phenotypes, we investigated mouse gene expression responses to the infection of different T. gondii strains at day 5 after intraperitoneal inoculation with 500 tachyzoites. The transcriptomes of mouse peritoneal cells showed that 1,927, 1,573, and 1,009 transcripts were altered more than 2-fold by type I, II, and III infections, respectively, and that the majority of altered transcripts were shared. Overall transcription patterns were similar in type I and type II infections, and both had greater changes than infection with type III. Quantification of parasite burden in mouse spleens showed that the burden with type I infection was 1,000 times higher than that of type II and that the type II burden was 20 times higher than that of type III. Fluorescence-activated cell sorting revealed that type I and II infections had comparable macrophage populations, and both were higher than the population with type III infection. In addition, type I infection had a higher percentage of neutrophils than type II and III infections. Taken together, these results suggested that there is a common gene expression response to T. gondii infection in mice. This response is further modified by parasite strain-specific factors that determine their distinct virulence phenotypes.
INTRODUCTION
Toxoplasma gondii is the causative agent for toxoplasmosis. T. gondii infects almost all warm-blooded vertebrates and birds. It infects up to one-third of the human population (9). In immunocompromised individuals, infection can cause severe encephalitis. Acute infections in pregnant women can result in a condition known as congenital toxoplasmosis, which may cause blindness, mental retardation, or even death of the fetus (13).
In North America and Europe, T. gondii isolates of animals and humans are divided into the type I, II, and III lineages, with type II strains predominant in infections (6, 11). In mice, the type I lineage is highly virulent (100% lethal dose [LD100], 1 parasite), whereas type II is intermediately virulent (LD50 of 103 to 104 parasites) and type III is nonvirulent (LD50 of >105 parasites) (23, 26). Using reference strains from the clonal type I and II lineages, it was shown that acute virulence was associated with rapid parasite dissemination, high parasite load, and an overstimulation of a Th1 immune response by the type I strain, which eventually kills the mice (3, 10, 19), suggesting that immune pathology plays an important role in mortality.
Gene expression in the host infected with T. gondii has been characterized in vitro. Gene expression analysis of macrophages infected with a type I, II, or III strain revealed that type I infections elicited a stronger immune response than type II and III strains (16). Previous studies also showed that the type II ME49 strain induced translocation of nuclear factor kappa B (NF-κB) to the nucleus but that this was not true of the type I RH strain in mouse splenocytes and mouse bone marrow-derived macrophages (7, 21). NF-κB is known to have a critical role in regulating inflammatory immune and antiapoptotic responses during infection. It was shown that type II strains, but not type I and type III strains, induced increased levels of interleukin-12 p40 (IL-12p40) in a myeloid differentiation primary response gene 88 (MyD88)-dependent and Toll-like receptor 2 (TLR2)/TLR4-independent manner in vitro (21). Altogether, these data indicate that the parasite genetic background is important in inducing host gene expression.
Although knowledge has been gained from these studies, conclusions are limited by the controlled conditions characteristic of in vitro systems. It is unknown how these data will translate to in vivo scenarios. To overcome these limitations, we used a mouse model to study host response to T. gondii infection. This is of relevance to human toxoplasmosis as mouse-virulent T. gondii strains may potentially be associated with severe acquired toxoplasmosis in immunocompetent patients (4). Here, we revealed the differences in gene expression levels in mice infected with three major genotypes of T. gondii.
MATERIALS AND METHODS
Cell culture of parasite strains and infection in mice.
Human foreskin fibroblasts (HFF) were grown in Dulbecco's modified Eagle medium (DMEM) (catalog number MT 10-013 CV PR; Fisher Scientific, Hanover Park, IL) supplemented with 10% of heat-inactivated fetal bovine serum (HyClone-SH30070.03 HI/IR; Fisher Scientific), 1% of 100× nonessential amino acids ([NEAA] catalog number SH3023801; Fisher Scientific), 0.4% of 1 M HEPES buffer, and 0.1% of 10 mg/ml gentamicin (catalog number 15710-64; Invitrogen, Carlsbad, CA). The cell culture was maintained in T25 vented culture flasks at 37°C with 5% CO2. T. gondii strains GT1 (type I, virulent), PTG (type II, intermediately virulent), and CTG (type III, nonvirulent) were expanded and maintained in confluent HFF monolayers to reach consistent 2-day passages prior to use for experiments. The tachyzoites of GT1, PTG, and CTG strains were harvested by filtering through 3-μm-pore-size polycarbonate filters (catalog number 420400; Fisher Scientific, Hanover Park, IL) and counted using hemocytometer. Five hundred tachyzoites were inoculated into four 6- to 8-week-old female outbred CD-1 mice (ICR-Harlan Sprague) by intraperitoneal injection. Mice were euthanized at day 5 postinfection. Two uninfected mice from the same batch were used as negative controls. Day 5 postinfection was selected for microarray analysis based on a tissue burden assay that showed significant changes in parasite load in mice infected with virulent strains compared to that of intermediate and nonvirulent strains when parasite load was monitored on days 3, 5, and 7 postinfection (data not shown). It is likely that at day 5 postinfection significant shifts in host response to infection occur and that type II and III strains start to switch from tachyzoite to bradyzoite growth.
To collect peritoneal cells, 5 ml of ice-cold phosphate-buffered saline (PBS) was injected into the peritoneal cavity, and then peritoneal lavage fluid was collected. Peritoneal cells were collected by centrifugation at 1,000 × g for 10 min, resuspended in 1 ml of RNAlater solution (Ambion, Austin, TX), incubated in 4°C for 1 h, and then stored at −70°C before RNA extraction. The mouse spleens were collected, weighed, homogenized, and diluted in PBS to 0.02 g/ml for DNA extraction and subsequently to determine parasite burden (see the section “In vivo parasite load determination by RT-PCR” below). In a separate experiment, the above three parasite strains were inoculated to two mice each, and peritoneal cells were collected on day 5 postinfection. Two uninfected mice were used as negative controls. A total of 1 × 106 peritoneal cells for each mouse were plated and stained with conjugated antibodies purchased from eBioscience (San Diego, CA). These antibodies included fluorescein isothiocyanate (FITC)-conjugated anti-mouse F4/80 (catalog number 11-4801) for macrophages, allophycocyanin (APC)-conjugated anti-mouse CD11c (catalog number 17-0114) for dendritic cells (DCs), and phycoerythrin (PE)-conjugated anti-mouse Ly-6G (catalog number 12-5931) for neutrophils. Antibodies were diluted 1/200 in fluorescence-activated cell sorting (FACS) buffer (PBS–0.1% bovine serum albumin [BSA]) prior to staining. The stained samples were counted with a FACSCalibur (BD Biosciences) instrument, and data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Microarray analysis.
Total RNA was extracted from peritoneal cells using a Qiagen RNeasy Plus Mini Kit (catalog number 74131; Qiagen, Valencia, CA) following the manufacturer's instructions. For host gene expression profiling, an Affymetrix Mouse 1.0 ST array containing 28,853 genes was used. The microarray was carried out at the University of Tennessee Affymetrix Core Facility. RNA samples were processed according to the Affymetrix protocol for one-cycle DNA synthesis using a Message Amp II-Biotin Enhanced Kit (P/N AM1791; Ambion, Austin, TX). Five micrograms of fragmented cRNA was hybridized to the Affymetrix GeneChip. Arrays were processed using the Affymetrix Hyb/Stain kit PN900720. Arrays were immediately scanned using the Affymetrix 7G scanner. Array images were visually inspected for anomalies. The individual chip scans were quality checked for the presence of control genes and background signal values.
Data normalization and statistical analysis.
The raw data from the arrays were normalized with the robust multichip algorithm with a GC-content correction (GC RMA) using Partek Genomics Suite software. A two-way analysis of variance (ANOVA) was used to detect genes with statistically significant expression levels between T. gondii-infected mice and the uninfected controls. Gene transcripts were considered to be differentially expressed when there was at least a 2-fold change (up or down) from the uninfected controls, and the ANOVA P value was set at ≤0.05.
Microarray validation by RT-PCR.
RNA samples for microarray analysis were used to quantitate mRNA transcript levels of genes either highly upregulated or highly downregulated in mice. cDNA was synthesized by reverse transcription using a DyNAmo SYBR green two-step quantitative reverse transcription-PCR (qRT-PCR) kit (New England BioLabs, Beverly, MA). The RT-PCR mixture had a total volume of 20 μl containing 8.6 μl of H2O, 10 μl of 2× Master Mix (containing modified Tbr DNA polymerase), 0.2 μl of 50 μM (each) forward and reverse primer (see Table S2 in the supplemental material). The reaction was carried out in a Smart Cycler (version 2.0b; Cepheid, Sunnyvale, CA) under the following conditions: 95°C for 15 min, followed by 40 cycles of 94°C for 10 s, 55°C for 30 s, and 72°C for 30 s. Quantification of the target transcripts was normalized to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH; NCBI accession number NM_008084) reference gene using the 2−ΔΔCT method (where CT is threshold cycle) (24).
Data mining.
Gene functional analysis was conducted for differentially expressed genes using the Onto-Express (OE) annotation database (vortex.cs.wayne.edu/ontoexpress). The input into OE was a list of gene symbols corresponding to genes differentially expressed in T. gondii-infected mice compared to expression in uninfected control samples. The output was the functional category assigned to each gene based on specific experimental evidence or by theoretical inference (8). For visual representation, fold change signal intensity values were natural-log transformed, and values for each gene were plotted. For each functional category of genes, statistical analyses were performed to compare mean fold changes of gene expression in mice infected with the type I, II, and III parasite strains. The following formula was used: mean fold change = (Σ|x|/n), where x is the natural logarithm of fold change/negative control. The |x| is used to remove the negative sign when genes are downregulated. n is the number of genes affected in a functional category. Standard deviation was also calculated for each mean value.
In vivo parasite load determination by qPCR.
To extract genomic DNA, 500 μl of homogenized spleen tissue at a concentration of 0.02 g/ml in PBS was further diluted with 500 μl of PBS; then 50 μl of 10 mg/ml proteinase K was added, and the mixture was incubated at 55°C for 2 h. DNA was extracted using a Qiagen DNeasy Blood and Tissue Kit (catalog number 69504; Qiagen, Valencia, CA). For quantitative PCR (qPCR) standard controls, the spleen of a noninfected mouse was homogenized and diluted in PBS, and T. gondii tachyzoites were spiked into the homogenized spleen to make a series of concentrations of 1 × 107 to 1 × 103 parasites/ml. All homogenized spleen samples were adjusted to a volume of 0.02 g/ml in PBS, and genomic DNA was purified as above. The concentration of parasites in mouse spleen was estimated by qPCR using a TaqMan probe targeting the ITS1 sequence (GenBank accession number AY143141). The primers for PCR amplification were ITS1-Fx (GAAGGGGCTCAATTTCTGG) and ITS1-Rx (TGTTCCTCAGATTTGTTGTTTGA), which amplify a 117-bp sequence. The ITS1 probe was 5′-5-FAM-CGTGTCTCTGTTGGGATACTGATTTCCAGG-BHQ-1-3′, with the 5′ end labeled with 6-carboxyfluorescein (FAM) and the 3′ end labeled with Black Hole Quencher-1(BHQ-1) (Integrated DNA Technologies, Inc. Coralville, IA). The qPCR mixture had a total volume of 25 μl containing 16.24 μl of H2O, 2 μl of 10× PCR buffer, 2.0 μl of 2.5 mM (each) deoxynucleoside triphosphates (dNTP), 2.0 μl of 25 mM MgCL2, 0.5 μl of 10 mg/ml BSA, 1.3 μl of 50 μM ITS1-Fx and ITS1-Rx primers, 0.2 μl of 50 μM ITS1 probe, and 0.3 μl of 5 U/μl FastStart Taq DNA polymerase (Roche Applied Science, Indianapolis, IN). One microliter of purified DNA was added as a template, and the PCR mix was transferred to 25-μl Cepheid PCR tubes. The reaction was carried out using a Smart Cycler (version 2.0b; Cepheid, Sunnyvale, CA) under the following conditions: 94°C for 60 s, followed by 45 cycles of 92°C for 15 s, 52°C for 30 s, and 72°C for 40 s. The CT value of each sample was compared to values of the standard controls (107 to 103 parasites/ml and negative control) to estimate the concentration of parasite per gram of spleen.
RESULTS
Microarray experiments.
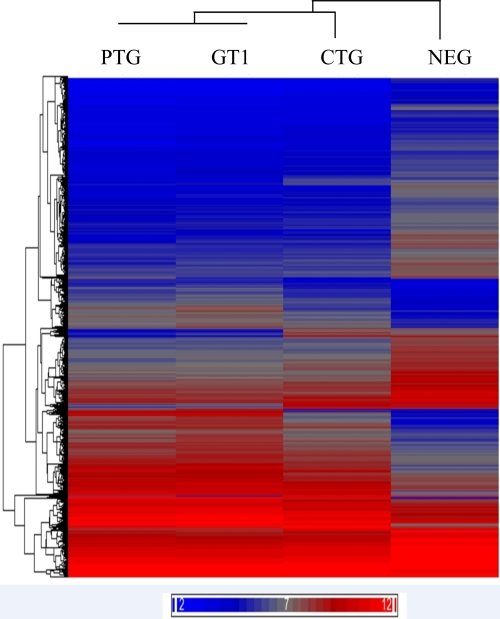
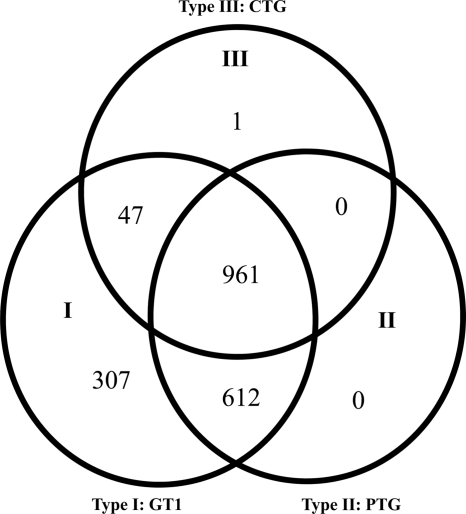
For microarray analysis, total RNAs were extracted from peritoneal cells of mice infected with GT1, PTG, and CTG T. gondii strains. Three of the four RNA samples with high RNA quality (determined by the optical density at 260/280 nm [OD260/280] and OD260/230) were used for gene expression profiling. For PTG strain-infected mice, only two RNA samples were collected for the array study. Differentially expressed genes were identified based on signal intensity values and fold changes compared to values for the uninfected control mice. Mice infected with T. gondii had drastic changes in gene expression compared to uninfected mice, regardless of the parasite genotype (Fig. 1). Among the 28,853 mouse genes investigated, 1,927, 1,573, and 1,009 genes were altered (≥2-fold) in type I, II, and III infections, respectively (Table 1 and Fig. 2). A core set of 961 transcripts was shared among mice infected with the three T. gondii lineages. Type I and II infections shared an additional 612 transcripts, whereas type I and III infections shared an additional 47 transcripts. Type I, II, or III infection each specifically altered 307, 0, and 1 transcript, respectively (Fig. 2).
Fig 1.
Hierarchical clustering analysis of gene expression in CD-1 outbred mice at day 5 postinfection with T. gondii GT1, PTG, and CTG strains. Mice that were not infected with T. gondii were included as negative controls (NEG). Red indicates high expression levels; blue indicates low expression levels. The bars on the left indicate clusters of genes with similar expression levels among different infections.
Table 1.
Numbers of mouse genes affected by T. gondii at day 5 postinfection
| Functional gene class | No. of genes with a change in expression level by straina |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Type I (GT1) |
Type II (PTG) |
Type III (CTG) |
|||||||
| Increased | Decreased | Total | Increased | Decreased | Total | Increased | Decreased | Total | |
| Transcription | 49 | 133 | 182 | 40 | 105 | 145 | 34 | 43 | 77 |
| Transport | 70 | 89 | 159 | 57 | 73 | 130 | 50 | 39 | 89 |
| Immunity and inflammation | 103 | 31 | 134 | 92 | 23 | 115 | 90 | 11 | 101 |
| Signal transduction | 59 | 50 | 109 | 46 | 41 | 87 | 32 | 24 | 56 |
| Oxidation reduction | 32 | 59 | 91 | 30 | 45 | 75 | 30 | 12 | 42 |
| Translation | 9 | 65 | 74 | 7 | 44 | 51 | 5 | 30 | 35 |
| Apoptosis | 28 | 29 | 57 | 22 | 25 | 47 | 21 | 10 | 31 |
| Cell cycle | 36 | 15 | 51 | 34 | 12 | 46 | 29 | 4 | 33 |
| Uncategorized transcripts | 409 | 661 | 1,070 | 350 | 527 | 877 | 243 | 302 | 545 |
| Total | 795 | 1,132 | 1,927 | 678 | 895 | 1,573 | 534 | 475 | 1,009 |
Increased, ≥2-fold change in infected mice versus negative controls (P ≤ 0.05); decreased, ≤ 2-fold change in infected mice versus negative controls (P ≤ 0.05).
Fig 2.
Venn diagram of gene expression in mice infected with GT1, PTG, and CTG strains. Genes with ≥2-fold up- or downregulation in infected mice versus uninfected (NEG) mice were included.
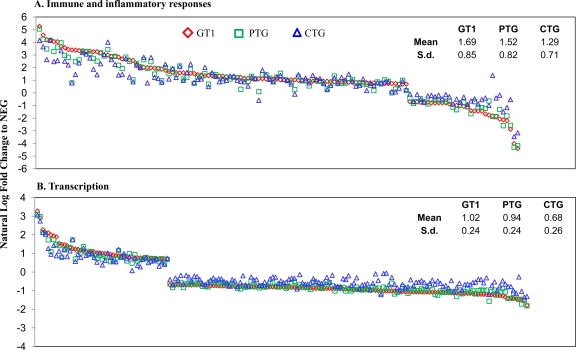
Altered transcripts in infected mice were categorized into classes based on biological processes using OE. Biological processes including transcription, transport, immune and inflammatory response, signal transduction, oxidation reduction, translation, apoptosis, and cell cycle were analyzed, and results are presented in Table 1 and in Table S1 in the supplemental material. Overall, a greater number of genes were downregulated than upregulated in type I (1,132 versus 795) and II (895 versus 678) infections. However, it was the opposite in type III infection, in that fewer genes were downregulated than upregulated (475 versus 534). Among the functional classes, most have higher numbers of downregulated than upregulated genes except the classes of immune and inflammatory response, signal transduction, and cell cycle response. To compare overall expression patterns, fold change values of altered transcripts were natural-log transformed to generate graphs (Fig. 3; see also Fig. S1 in the supplemental material). T. gondii strains elicit different expression levels in mice, with type I having the strongest, followed by the type II, and type III had the least effect on host gene expression. The average fold changes of genes in different functional groups consistently followed the same rank order from high to intermediate to low in type I, II, and III infections, respectively. Individual gene expression levels also largely followed a similar trend (Fig. 3; see also Fig. S1).
Fig 3.
Comparison of mouse gene expression profiles for GT1, PTG, and CTG infection based on biological processes. Mouse genes that were up- or downregulated are sorted by the natural log of the fold change relative to the negative controls.
The immune and inflammatory response had the highest number of upregulated genes by all three parasite strains. It is striking that the majority of genes in this group had increased expression levels, and the number of genes affected were comparable among all three types of infections. There were 103, 92, and 90 transcripts upregulated, and only 31, 23, and 11 downregulated by the type I, II, and III infections, respectively (Table 1). Cytokines including CCL2, CCL3, CCL4, CCL7, CCL8, CCL12, CCL17, CCL22, CXCL9, CXCL10, and CXCL11 were highly expressed, whereas CXCL13 and platelet factor 4 (PF4, also known as CXCL4) were decreased in mice infected by all parasite strains (see Table S1 in the supplemental material). Overall changes in gene expression levels in mice infected with the type I parasite were the highest, closely followed by type II infection, and changes in type III infections were the lowest (Fig. 3A). Type I and II strains had more similar effects on overall expression levels. The type III strain elicited a milder response.
In the functional classes of transcription, translation, and uncategorized transcripts, more genes were downregulated than upregulated in all three types of infections (Table 1). For transcription, 49, 40, and 34 genes were upregulated; however, 133, 105, and 43 genes were downregulated for type I, II, and III infections, respectively (Table 1). The patterns of gene expression followed the same theme as in the immune and inflammatory response described above (Fig. 3B).
In the remaining functional classes (signal transduction, cell cycle response, transport, oxidation reduction, and apoptosis), gene expression patterns varied among different types of infections (see Fig. S1 in the supplemental material). However, in all cases, patterns of type I and II infections were more similar to each other than they were to the type III infection pattern.
Microarray validation via RT-PCR.
In order to validate our microarray data, five upregulated and five downregulated genes were selected. The five upregulated genes were CCL2, CCL7, CCL12, CCL17, and CCL22, and the five downregulated genes were Prg4, serine protease inhibitor 2 (Serpinb2), Serpinb10, ApoE, and Fabp7 (see Table S1 in the supplemental material). The RT-PCR expression data of these 10 genes showed positive correlation with our microarray expression data (r2 = 0.502), with medium level of supporting the results from microarray experiments (see Fig. S2).
Determining parasite load in the spleen tissues of mice by qPCR.
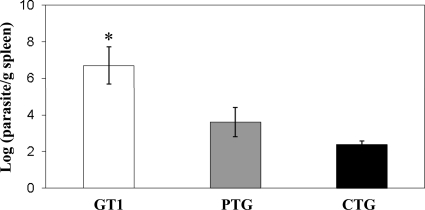
Parasite loads in the spleens of mice infected with 500 tachyzoites at day 5 postinfection were quantified. The log parasites/g of spleen for virulent type I strain GT1 was 6.7 (standard deviation [SD], 1.0). This was more than 1,000 times higher than for the type II strain PTG, which was 3.6 (SD, 0.8), and about 20,000 times higher than that of the Type III strain CTG, which was 2.4 (SD, 0.2). The type II and type III strains differed by a factor of only 20 (Fig. 4).
Fig 4.
Parasite burdens in mice at day 5 postinfection. Parasite load was determined by qPCR. GT1 has a significantly higher parasite burden than PTG and CTG (*, P = 0.001, by ANOVA).
Peritoneal cell population.
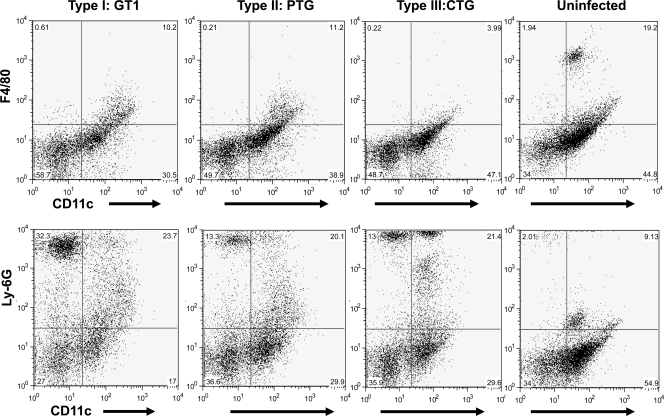
Fluorescence-activated cell sorting (FACS) showed variable cell populations in mice infected with different parasite strains (Fig. 5). The macrophage populations in mice infected with type I and II parasites were comparable, and both were higher than with type III infection. The uninfected mice had a distinct macrophage subpopulation that was not present in T. gondii-infected mice. Type I infection had a lower percentage of dendritic cells (DCs) but a higher percentage of neutrophils than type II and III infection, and percentages were comparable for type II and III infections for both cell types.
Fig 5.
FACS analysis. Mice were infected with GT1, PTG, and CTG strains. Peritoneal cells were collected and stained for markers of dendritic cells (CD11c), macrophages (F4/80), and neutrophils (Ly-6G) and subsequently identified by FACS analysis.
DISCUSSION
In this study, we examined gene expression in mice infected with three distinct T. gondii strains. The results showed several important findings. First, T. gondii drastically altered mouse gene expression in peritoneal cells on day 5 postinfection (Fig. 1). The numbers of altered transcripts were 1,927, 1,573, and 1,009 for type I, II, and III infections, respectively (Table 1). Second, the majority of altered transcripts were shared between the three types of infections. A core set of 961 altered transcripts overlapped among mice infected with the three T. gondii strains. Moreover, type I and II infections shared an additional 612 transcripts, whereas type I and III infections shared an additional 47 transcripts; type I, II, and III infections each specifically altered 307, 0, and 1 transcript, respectively (Fig. 2). These results suggested that commonly shared mechanisms were responsible for modulating most of the host genes by different T. gondii strains. Third, the host immune and inflammatory response was the major class of genes affected by T. gondii infection. It had the highest number of upregulated genes of all functional classes, and the numbers of genes affected were comparable among all three type of infections (Table 1). These data underlined the importance of immune and inflammatory response to T. gondii infection, regardless of parasite genotypes. Fourth, there was a pattern of quantitative difference in modulating genes among the three different infections. The average fold changes of gene expression in different functional classes consistently followed the same order from high to intermediate to low in type I, II, and III infections, respectively. In addition, type II infection was more similar to type I infection than to type III infection. Individual gene expression levels also largely followed a similar trend (Fig. 3; see also Fig. S1 and Table S1 in the supplemental material). Similar quantitative patterns were also observed in a previous study of chemokine response during in vivo infections with type I, II, and III infections (16). The quantitative difference of host transcripts elicited by distinct T. gondii strains may have significant contributions to different levels of immunopathology and the severity of toxoplasmosis in the host.
The results of parasite tissue burden analysis in the spleen showed that the burden in type I infection was more than 1,000 times higher than that of type II, and the type II infection burden was 20 times higher than that of type III (Fig. 4). Interestingly, gene expression patterns showed that types I and II consistently followed similar trends of gene expression among biological processes, with the latter having slightly lower levels, while the type III response was much lower (Fig. 3; see also Fig. S1 in the supplemental material). In addition, both type I and II strains elicited stronger responses in host gene expression, in that host genes were either highly upregulated or highly downregulated, than the type III strain. These results suggested a general expression response common to T. gondii infections. This response may be further modified by parasite strain-specific factors that determine their distinct virulence phenotypes. The factors that control parasite replication are of importance. It was shown that the type I strain RH quickly reached a high tissue load and maintained an upward trajectory until the mice were killed, whereas a type II strain reached a plateau, and the mouse mortality was determined by the initial infection dose (19). Mouse gene expression responses to type I and II infections may be effectively triggered by the presence of a parasite load greater than the threshold value, and the nonvirulent type III parasite does not reach this threshold during infection. This may explain why gene expression patterns were similar between type I and II strains and why both were different from type III infection.
At present, there is limited information on host gene expression in response to T. gondii infection. An in vitro study using a cDNA microarray with 420 expressed sequence tags of a porcine kidney epithelial cell line infected with a temperature-sensitive RH strain (type I) showed that, from a few hours to 3 days after infection, the majority of altered transcripts (263 genes) were induced and that only a small portion of transcripts (48 genes) were downregulated (20). This is different from our study in that, overall, more genes were downregulated than upregulated. A recent study investigated the response of 43,004 human genes in neuroepithelial cells to infection of type I, II, and III T. gondii strains (28). It showed that a type I strain elicited a wide transcriptional response encompassing 1,122 genes, while cells infected with type II and III strains resulted in the alteration of 78 and 344 genes, respectively. In addition, there were higher numbers of downregulated (726 transcripts) than upregulated (396 transcripts) genes in type I infection but a higher number of upregulated (197 transcripts) than downregulated (147 transcripts) genes in type III infection. Similar patterns were observed in our study in mice. However, in the study of Xiao et al. (28), only seven genes were shared from neuroepithelial cells infected with the three T. gondii strains; the majority of altered transcripts were parasite strain specific. This finding was in sharp contrast to our results in that there was large overlap of genes within the three types of infections. In the study of Xiao et al. (28), type II infection had the least effect on human neuroepithelial cell gene expression, which is in contrast to our result that type II was similar to type I infection. The discrepancy among these studies may be due to the different experimental systems employed.
FACS analysis showed different cell populations in mice infected with distinct parasite strains (Fig. 5). The macrophage populations among type I- and type II-infected mice are comparable, and both are higher than that of type III infection. Type I infection has a lower percentage of DCs but a higher percentage of neutrophils (Ly-6G high) than types II and III, which were comparable for both cell types. The high level of neutrophils suggested a strong inflammatory response induced by the type I strain. The uninfected mice have a distinct macrophage subpopulation that is not present in T. gondii-infected mice. This subpopulation might be naïve or have resident macrophages present in the peritoneal cavity. It has been shown that DCs facilitate spread of T. gondii (15); therefore, it is reasonable to suggest that due to the uncontrolled growth of type I parasite, DCs may be recruited from the peritoneal cavity, fulfilling their role as migratory cells aiding in dissemination of parasites, or be destroyed by rapidly replicating parasites. Taken together, the difference of gene expression levels among mice infected with different T. gondii strains is likely a result of the combination of changes in host cell population and in the gene transcription of these cells.
Overreaction of a Th1 proinflammatory response plays a significant part in mortality in mice infected with a virulent type I strain (19). Previous studies of chemokine responses in peritoneal cells during mouse infections with type I, II, and III strains identified highly expressed genes, including CCL2, CCL7, CCL17, CXCL9, CXCL10, and CXCL11 (16). In our study, analysis of genes belonging to the immune and inflammatory response showed that, in addition to previously indentified genes, more cytokines, chemokines, and upstream factors responsible for induction of a proinflammatory response (CCL2, CCL3, CCL4, CCL7, CCL8, CCL12, CCL17, CCL22, CXCL9, CXCL10, CXCL11, NOS2, Myd88, tumor necrosis factor [TNF], and gamma interferon [IFN-γ]) were revealed (see Table S1 in the supplemental material). The proinflammatory cytokines induced during T. gondii infection may serve as important mediators for defense, but due to the uncontrolled response in infections with type I strains, more damage is caused to the host.
The NF-κB pathway is an important host signaling pathway in the regulation of inflammatory, immune, and antiapoptotic responses. It has been shown that both type I and II parasites induced the NF-κB pathway (18, 22). Our microarray analysis revealed that factors in the NF-κB pathway such as TNF and IL-1β were highly expressed in type I and II infections (see Table S1), supporting the conclusions from previous studies.
A few genes were highly suppressed, including 15-lipoxygenase (Alox15), CXCL13, platelet factor 4 (PF4), proteoglycan 4 (Prg4), serine protease inhibitor 2 (Serpinb2), and Serpinb10 (see Table S1 in the supplemental material). Alox15 oxidizes unsaturated lipids in macrophages and regulate IL-12 production, which is critical for resistance to T. gondii during both the acute and chronic stages of infection (17). CXCL13 is required for B1 cell homing to body cavities and for production of a natural antibody which is important for body cavity immunity (2). PF4 is a chemokine that involved in platelet-dependent thrombosis in mice (27). Prg4 functions in lubrication and cytoprotection in the synovial fluid. This protein has large mucin-like repeats which, when replaced with O-linked oligosaccharides, impart lubricating and cytoprotective properties (12). At present, the roles of CXCL13, PF4, and Prg4 in T. gondii infection are not clear.
Serpinb2 and Serpinb10 were highly suppressed in mice infected with the type I and II T. gondii strains (see Table S1 in the supplemental material). Serpinb2 was suppressed by 61.4-, 31.7-, and 1.4-fold in mice infected with type I, II, and III strains, respectively, while Serpinb10 was suppressed by 83.5-, 94.2-, and 6.7-fold in Type I, II, and III infections, respectively. It was suggested that serpins could inhibit host cell apoptosis (1). Some serpins have been shown to inhibit cathepsin and lysosomal enzymes. A previous study showed that when T. gondii serine protease activity was inhibited via the two serine protease inhibitors 3,4-dichloroisocoumarin and 4-(2-aminoethyl)-benzenesulfonyl fluoride, the parasite was unable to successfully invade host cells (5). When T. gondii was treated with chymotrypsin-like serine protease inhibitors Z-Gly-Gly-Phe-chloromethyl ketone (CMK) and tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK), its replication was inhibited, and the viability decreased (25). It was shown that when T. gondii subtilisin-like serine protease 1 (TgSUB1) was disrupted, T. gondii showed decreased attachment and invasion (14). Taken together, these data suggest that proteases are important for survival and transmission of T. gondii. Suppression of host protease inhibitors may be an offensive mechanism developed by highly virulent T. gondii strains. Given that DCs, macrophages, and neutrophils are highly phagocytic cells and are able to release large numbers of proteolytic enzymes, the ability to suppress host protease inhibitors by the type I parasite strain will provide it an advantage in combating host defense mechanisms.
In summary, we showed that distinct parasite strains elicited different responses in the host. The differential host-parasite interaction may ultimately dictate the pathogenesis of T. gondii infection. Future work should be aimed toward identifying gene expression regulators in the host in response to T. gondii infection to better understand the mechanisms of the pathogenesis of toxoplasmosis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the Affymetrix Core Lab Awards at the University of Tennessee Knoxville and by American Heart Association grant 0830134N.
Footnotes
Published ahead of print 5 December 2011
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Ahn HJ, Kim JY, Ryu K, Nam HW. 2009. STAT6 activation by Toxoplasma gondii infection induces the expression of Th2 C-C chemokine ligands and B clade serine protease inhibitors in macrophage. Parasitol. Res. 105:1445–1453 [DOI] [PubMed] [Google Scholar]
- 2. Ansel KM, Harris RBS, Cyster JG. 2002. CXCL13 is required for B1 cell homing, natural antibody production, and body cavity immunity. Immunity 16:67–76 [DOI] [PubMed] [Google Scholar]
- 3. Barragan A, Sibley LD. 2002. Transepithelial migration of Toxoplasma gondii is linked to parasite motility and virulence. J. Exp. Med. 195:1625–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carme B, et al. 2002. Severe acquired toxoplasmosis in immunocompetent adult patients in French Guiana. J. Clin. Microbiol. 40:4037–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conseil V, Soete M, Dubremetz JF. 1999. Serine protease inhibitors block invasion of host cells by Toxoplasma gondii. Antimicrob. Agents Chemother. 43:1358–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Darde ML. 2004. Genetic analysis of the diversity in Toxoplasma gondii. Ann. Ist Super Sanita 40:57–63 [PubMed] [Google Scholar]
- 7. Dobbin CA, Smith NC, Johnson AM. 2002. Heat shock protein 70 is a potential virulence factor in murine Toxoplasma infection via immunomodulation of host NF-κB and nitric oxide. J. Immunol. 169:958–965 [DOI] [PubMed] [Google Scholar]
- 8. Draghici S, et al. 2003. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acid Res. 31:3775–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dubey JP. 2009. Toxoplasmosis of animals and humans, 2nd ed Taylor and Francis Group, Boca Raton. FL [Google Scholar]
- 10. Gavrilescu LC, Denkers EY. 2001. IFN-overproduction and high level apoptosis are associated with high but not low virulence Toxoplasma gondii infection. J. Immunol. 167:902–909 [DOI] [PubMed] [Google Scholar]
- 11. Howe DK, Sibley LD. 1995. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J. Infect. Dis. 172:1561–1566 [DOI] [PubMed] [Google Scholar]
- 12. Ikegawa S, Sano M, Koshizuka Y, Nakamura Y. 2000. Isolation, characterization and mapping of the mouse and human PRG4 (proteoglycan 4) genes. Cytogenet. Cell Genet. 90:291–297 [DOI] [PubMed] [Google Scholar]
- 13. Joynson DH, Wreghitt TJ. 2001. Toxoplasmosis: a comprehensive clinical guide. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 14. Lagal V, et al. 2010. Toxoplasma gondii protease TgSUB1 is required for cell surface processing of micronemal adhesive complexes and efficient adhesion of tachyzoites. Cell Microbiol. 12:1792–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lambert H, Vutova PP, Adams WC, Lore K, Barragan A. 2009. The Toxoplasma gondii-shuttling function of dendritic cells is linked to the parasite genotype. Infect. Immun. 77:1679–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee CW, Sukhumavasi W, Denkers EY. 2007. Phosphoinositide-3-kinase-dependent, MyD88-independent induction of CC-type chemokines characterizes the macrophage response to Toxoplasma gondii strains with high virulence. Infect. Immun. 75:5788–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Middleton MK, et al. 2009. 12/15-Lipoxygenase-dependent myeloid production of interleukin-12 is essential for resistance to chronic toxoplasmosis. Infect. Immun. 77:5690–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Molestina RE, Payne TM, Coppens I, Sinai AP. 2003. Activation of NF-κB by Toxoplasma gondii correlates with increased expression of antiapoptotic genes and localization of phosphorylated IκB to the parasitophorous vacuole membrane. J. Cell Sci. 116:4359–4371 [DOI] [PubMed] [Google Scholar]
- 19. Mordue DG, Monroy F, LaRegina M, Dinarello CA, Sibley LD. 2001. Acute toxoplasmosis leads to lethal overproduction of Th1 cytokines. J. Immunol. 167:4574–4584 [DOI] [PubMed] [Google Scholar]
- 20. Okomo-Adhiambo M, Beattie C, Rink A. 2006. cDNA microarray analysis of host-pathogen interactions in a porcine in vitro model for Toxoplasma gondii infection. Infect. Immun. 74:4254–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robben PM, et al. 2004. Production of IL-12 by macrophages infected with Toxoplasma gondii depends on the parasite genotype. J. Immunol. 172:3686–3694 [DOI] [PubMed] [Google Scholar]
- 22. Rosowski EE, et al. 2011. Strain-specific activation of the NF-kB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J. Exp. Med. 208:195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saeij JP, et al. 2006. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 314:1780–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 25. Shaw MK, Roos DS, Tilney LG. 2002. Cysteine and serine protease inhibitors block intracellular development and disrupt the secretory pathway of Toxoplasma gondii. Microbes Infect. 4:119–132 [DOI] [PubMed] [Google Scholar]
- 26. Sibley LD, Boothroyd JC. 1992. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature 359:82–85 [DOI] [PubMed] [Google Scholar]
- 27. Slungaard A. 2005. Platelet factor 4: a chemokine enigma. Int. J. Biochem. Cell Biol. 37:1162–1167 [DOI] [PubMed] [Google Scholar]
- 28. Xiao J, Jones-Brando L, Talbot C, Jr, Yolken RH. 2011. Differential effects of three canonical Toxoplasma strains on gene expression in human neuroepithelial cells. Infect. Immun. 79:1363–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.