Abstract
The Reg3 protein family, including the human member designated pancreatitis-associated protein (PAP), consists of secreted proteins that contain a C-type lectin domain involved in carbohydrate binding. They are expressed by intestinal epithelial cells. Colonization of germ-free mice and intestinal infection with pathogens increase the expression of Reg3g and Reg3b in the murine ileum. Reg3g is directly bactericidal for Gram-positive bacteria, but the exact role of Reg3b in bacterial infections is unknown. To investigate the possible protective role of Reg3b in intestinal infection, Reg3b knockout (Reg3b−/−) mice and wild-type (WT) mice were orally infected with Gram-negative Salmonella enteritidis or Gram-positive Listeria monocytogenes. At day 2 after oral Listeria infection and at day 4 after oral Salmonella infection, mice were sacrificed to collect intestinal and other tissues for pathogen quantification. Protein expression of Reg3b and Reg3g was determined in intestinal mucosal scrapings of infected and noninfected mice. In addition, ex vivo binding of ileal mucosal Reg3b to Listeria and Salmonella was investigated. Whereas recovery of Salmonella or Listeria from feces of Reg3b−/− mice did not differ from that from feces of WT mice, significantly higher numbers of viable Salmonella, but not Listeria, bacteria were recovered from the colon, mesenteric lymph nodes, spleen, and liver of the Reg3b−/− mice than from those of WT mice. Mucosal Reg3b binds to both bacterial pathogens and may interfere with their mode of action. Reg3b plays a protective role against intestinal translocation of the Gram-negative bacterium S. enteritidis in mice but not against the Gram-positive bacterium L. monocytogenes.
INTRODUCTION
In many countries in the industrialized world, food-borne intestinal infections are continuing to increase (11). For example, outbreaks of salmonellosis and listeriosis have been reported for decades, but within the past 25 years, their incidence has increased on many continents (29). To inhibit the colonization (adhesion to the intestinal epithelium) and translocation (invasion of host tissues) of food-borne pathogens and commensals, the intestinal mucosal surfaces are armed with an array of physical and chemical defense mechanisms. The low pH of the gastric compartment and bile acids secreted in the proximal intestine reduce the number of bacteria that will survive within the gut (7, 8). Other important defense mechanisms against pathogens include competition with commensal bacteria for nutrients and adhesion sites, a thick mucus layer on the luminal side of the epithelium, and the rapid innate response of the intestinal epithelium (6, 23).
Studies in rodents indicate that innate recognition of bacteria or bacterial components triggers epithelial expression of secreted C-type lectins Reg3g and Reg3b (3, 12, 22, 25). In rodents, mucosal and fecal levels of the Reg3 protein are associated with the severity of infection (25). The human homologue of this protein (i.e., they are, on the amino acid level, the most identical proteins between the two species [bidirectional best hit]) is pancreatitis-associated protein (PAP). PAP is also detectable in feces, and its use as a biomarker to monitor or discriminate human intestinal disease is the subject of ongoing studies.
Currently, the role and function of the Reg3 family members are of much interest. The murine Reg3g and human PAP have been shown to have antimicrobial activity against Gram-positive bacteria but not Gram-negative Escherichia coli (3). PAP and Reg3g were shown to bind peptidoglycan carbohydrate, which is critical for bacterial killing (14). Expression of Reg3g is greatly reduced in mice deficient in MyD88, an intracellular adaptor protein involved in most Toll-like receptor (TLR)-mediated signaling. MyD88-deficient mice have been shown to be more susceptible to Listeria monocytogenes infection than wild-type (WT) mice (2). Injection of recombinant Reg3g into the lumen of MyD88-deficient ligated ileal loops before inoculation with L. monocytogenes was shown to reduce L. monocytogenes survival in the gut. Other in vivo studies showed that genetic ablation of Reg3b, which is another Reg3 murine isoform, resulted in impaired clearance of the bacterial load in ileal Peyer's patches during Yersinia pseudotuberculosis infection without affecting luminal bacterial levels (5). It remains unclear, however, how Reg3b protects against bacterial translocation and whether this is specific for Gram-negative pathogens.
In this study, we used Reg3b knockout (Reg3b−/−) mice and their WT littermates to investigate the protective role of Reg3b following oral challenge with Gram-negative Salmonella enteritidis or Gram-positive L. monocytogenes. Additional ex vivo experiments were performed to gain insights into the mechanisms involved in the protective function of Reg3b.
MATERIALS AND METHODS
Animals and diet.
The experimental protocol was approved by the animal welfare committee of Wageningen University (Wageningen, The Netherlands). Reg3b−/− mice with a C57BL/6 × 129O1a background were generated as described previously (15). Knockout mice were obtained by breeding heterozygote mice. Genotyping was performed as described previously (15), and Reg3b−/− mice were used as the knockout mice and Reg3b+/+ littermates were used as the WT control. The mice were selected based on genotype and age and were fed a semipurified AIN93-G (21) diet (Abdiets, Woerden, The Netherlands). At 7 to 9 weeks of age, mice with an average body weight of 22 g (range, 16 to 31 g) were housed individually. They received a purified diet containing (per kg of feed) 200 g acid casein, 326 g cornstarch, 174 g glucose, 160 g palm oil, 40 g corn oil, 50 g cellulose, and a vitamin and mineral mix (without calcium) according to AIN93 (21). Diets were supplemented with CaHPO4 · 2H2O (Merck, Darmstadt, Germany) to a final concentration of 30 mmol/kg. To mimic the composition of a Western human diet, the prepared diets were relatively low in calcium and high in fat compared to standard rodent diets (21). Food and demineralized drinking water were supplied ad libitum. Mice were separated into groups, and age and gender were randomized over all groups as much as possible. The power analysis used to calculate the minimum number of animals needed per group was based on infection studies previously performed in this model (our unpublished data). In total, 3 Reg3b−/− and 3 WT groups were formed. From each mouse strain, one group was orally infected with Salmonella (Reg3b−/−, n = 10; WT, n = 12), a second group of each strain was orally infected with Listeria (Reg3b−/−, n = 7; WT, n = 8), and a final group was sham treated (Reg3b−/−, n = 5; WT, n = 6). Body weight was measured every 2 days before infection and daily after infection.
Nramp1 genotype of Salmonella-infected mice.
To monitor susceptibility to Salmonella infection, the natural resistance-associated macrophage protein 1 (Nramp1) genotype was determined in the Salmonella-infected Reg3b+/+ and Reg3b−/− mice. A PCR product was created from the genomic DNA by using primers 5′-GGAATGAATGTCAAGCAGCCAG-3′ and 5′-ATCCCACCTCATAGCCGAAG-3′ in order to sequence the area of the Nramp1 gene that is known to vary between different mouse strains (G or A at position 596) (9). The PCR cycle was 3 min at 95°C, 35 cycles of 30 s at 95°C, 30 s at 58°C, and 1 min at 72°C, and 5 min at 72°C. The PCR product was subsequently sequenced by Baseclear (Leiden, The Netherlands) using the primers 5′-CACCCCATTTCCAGTAGGG-3′ and 5′-CCTGTGACACCTGGATGTTC-3′ to determine the Nramp1 genotype of the Salmonella-infected mice.
Infection.
After adaptation to individual housing and the diet for 8 days, mice were orally infected by gavage with 0.2 ml saline containing 5 × 108 CFU of S. enteritidis (clinical isolate, phage type 4, strain NIZO1241; NIZO food research, Ede, The Netherlands) or L. monocytogenes (animal isolate, strain EGD-e, serotype 1/2a; NIZO food research). The virulence of each strain was sustained by routine oral passage in Reg3b+/− mice and subsequent isolation of the pathogen from extraintestinal organs. S. enteritidis was grown on brilliant green agar modified (BGAM) plus sulfamandalate supplement (Oxoid, Basingstoke, United Kingdom) and quantified as described previously (18). L. monocytogenes was grown and enumerated by similar culturing methods using Palcam agar (28) plus Palcam selective supplement (Oxoid).
Collection of biological samples and bacterial quantification.
A pilot study indicated that the determination of the number of L. monocytogenes and S. enteritidis bacteria in organs of mice was optimal, which means that the numbers of Listeria and Salmonella CFU could be detected in organs at 2 and 4 days after oral infection, respectively (our unpublished data). Noninfected mice were sacrificed 3 days after oral sham treatment. Mice were anesthetized by isoflurane, and blood was collected via an orbital puncture to isolate heparin plasma. After cervical dislocation, the colon and the distal third of the small intestine, representing the ileum, were excised. Approximately 1 cm was cut from the middle of each intestinal segment to quantify the number of viable Salmonella or Listeria cells present in the colonic and ileal tissue. This piece was cut open longitudinally, briefly flushed in sterile saline, and homogenized in 250 μl of saline, and 10-fold dilutions were plated for bacterial growth on selective agar for Salmonella or Listeria (as described above). The remaining parts of the ileum and colon were cut open longitudinally and flushed with saline, and the mucosa was isolated by scraping with a spatula. Mucosal samples were immediately frozen in liquid nitrogen for protein analysis. Furthermore, the mesenteric lymph nodes (MLN), spleen, and liver were removed and, after homogenization in sterile saline, directly used for Salmonella or Listeria quantification as described above. Bacterial counts were expressed as the total log10 CFU per gram of tissue.
Reg3b and Reg3g protein analysis in the ileal mucosa.
Reg3g and Reg3b expression was determined in the ileum because this is the area where expression is upregulated during infection of rats (22, 25) and upon microbial colonization of germ-free mice (3). Frozen mucosal scrapings of the ileum were pulverized under liquid nitrogen. Approximately two-thirds of the pulverized tissue was suspended in a 0.2-mol/liter sucrose buffer of pH 7.4 containing 20 mmol/liter Tris and a protease inhibitor cocktail (Complete, Roche Diagnostics). To homogenize the samples, they were sonicated on ice for 30 s at level 4 (Sonicator XL2020; Heat Systems, Farmingdale, NY), and protein concentrations were determined using the bicinchoninic acid (BC) assay (Omnilabo, Breda, The Netherlands) according to the manufacturer's protocol. Samples from each group were pooled, and 100 μg protein was denatured at 95°C for 10 min in sample buffer (4× buffer containing 160 mM Tris [pH 6.8], 8% SDS, 40% glycerol, 0.05% [wt/vol] bromophenol blue), subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (4.8% stacking gel [pH 6.8], 12.5% separation gel [pH 8.8]), and transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore, Billerica, MA). After blocking, the membranes were incubated with polyclonal anti-Reg3b (1:50,000) or anti-Reg3g (1:10,000) antibodies (custom made by Eurogentec, Seraing, Belgium). These antibodies were generated in rabbits against synthetically produced peptides by using the peptide sequences GEDSLKNIPSARISC (Reg3b) and EVAKKDAPSSRSSC (Reg3g). The chosen peptide sequences correspond to unique sequences within the Reg3 protein and allow differentiation between the Reg3b and Reg3g proteins. Serum from immunized rabbits was affinity purified using the same peptides. The signal of the secondary horseradish peroxidase (HRP)-conjugated antibody (goat anti-rabbit HRP, 1:100,000; Jackson ImmunoResearch, Suffolk, United Kingdom) was detected by using the ECL Plus chemiluminescent detection kit (GE Healthcare, Den Bosch, The Netherlands).
Myeloperoxidase analysis in the ileal mucosa and serum amyloid A detection in plasma.
A mouse myeloperoxidase (MPO) enzyme-linked immunosorbent assay (ELISA) kit (Hycult Biotechnology, Uden, The Netherlands) was used according to the manufacturer's guidelines to determine the concentration of MPO in ileal mucosal scrapings.
Levels of serum amyloid A (SAA) 2, an acute-phase apolipoprotein present in plasma (24), were determined in order to investigate systemic inflammation. A mouse SAA 2 ELISA kit (Life Diagnostics, Inc., West Chester, Pennsylvania) was used according to the guidelines of the manufacturer.
Pulldown assay.
To investigate the possible mode of action, it was evaluated in a pulldown assay whether Reg3b present in the ileal mucosa binds to S. enteritidis or L. monocytogenes. S. enteritidis and L. monocytogenes strains identical to those used in the infection study were grown overnight (37°C in a horizontal shaker at 250 rpm) in LB and brain heart infusion (BHI) broth, respectively.
Bacteria grown overnight were spun down (3 min at 2,350 × g), and 1 × 108 bacteria were resuspended in 20 μl ileal mucosal scraping extracts from Salmonella-infected Reg3b−/− or WT mice which were diluted 1:5 in the above-described sucrose buffer. The bacteria were incubated for 30 min at 37°C. After the bacteria were spun down (5 min at 1,500 × g), the presence of Reg3b was investigated in the supernatant (15 μl) by SDS-PAGE as described above. Proteins were transferred to an Immobilon-FL membrane (Millipore) and, after blocking, exposed to rat anti-mouse Reg3b antibodies (MAB5110, 1:2,000; R&D Systems, Minneapolis). The secondary antibody used was goat anti-rat IRDye 680CW (1:20,000; Li-Cor, Nebraska), and the blots were scanned using the Odyssey scanner (Li-Cor). Experiments were performed in triplicate.
Statistics.
All data are the means and the standard errors of the means (SEM), and statistical analysis was performed by using Prism 5.0 software (GraphPad, San Diego, California). Our aim was to investigate the role of Reg3b in infection. Therefore, the predefined comparisons were between Reg3b−/− and the WT except for in in vitro investigations to evaluate the binding of Reg3b to Salmonella and Listeria when binding to these pathogens was investigated. Data were tested for normality by the Kolmogorov-Smirnov normality test and the Shapiro-Wilk normality test. If the data were normally distributed, the differences were tested for significance using Student's t test (two-tailed). Data with unequal variances were tested by using the Mann-Whitney U test. Salmonella output in feces was determined at multiple time points, and therefore, these data were analyzed by a repeated-measures two-way analysis of variance (ANOVA) (mixed model). Differences were considered statistically significant when the P value was <0.05.
RESULTS
Body weight.
Body weight was determined during the experiment to monitor the general condition of the mice. At the start of the experiment, the average body weight was 22 ± 0.7 g. During the study, there was no significant difference between the body weight of Reg3b−/− mice and that of WT mice (data not shown). The average body weights before infection (average weight during 2 weeks prior to infection) were 22.8 ± 1.4 g and 23.1 ± 1.4 g for the WT and Reg3b−/− groups, respectively. The postinfection body weights of the WT and Reg3b−/− groups were 23.2 ± 1.4 g and 23.2 ± 1.5 g (average of the postinfection period).
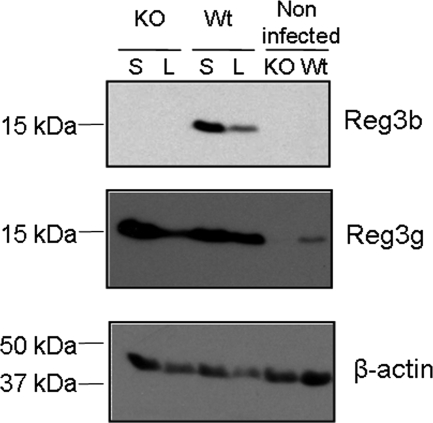
Bacterial infection increases Reg3b and Reg3g in the ileum mucosa.
To evaluate the presence of the Reg3b and Reg3g proteins in the ileal mucosas of WT and Reg3b−/− mice and the subsequent levels upon oral infection, mucosal scrapings were evaluated by immunoblotting using antibodies specific for Reg3b and Reg3g. As expected, Reg3b was not detected in the ileal mucosa of Reg3b−/− animals (Fig. 1). After Salmonella and Listeria infection, the protein level was higher in the mucosal samples of WT mice than it was before infection (Fig. 1). The isoform Reg3g was not detectable in mucosal scrapings of noninfected Reg3b−/− mice (Fig. 1). In contrast, the WT mice had detectable levels of this Reg3 isoform and levels increased during Salmonella and Listeria infection in WT and Reg3b−/− mice.
Fig 1.
Western blot analysis of Reg3b and Reg3g in pooled ileal mucosa samples of Reg3b−/− mice (KO) or WT mice (Wt). Reg3b−/− mice (n = 10) and WT mice (n = 12) were orally infected with S. enteritidis (S), or Reg3b−/− mice (n = 7) and WT mice (n = 8) were orally infected with L. monocytogenes (L). Noninfected mice (n = 5) received saline as a control treatment.
Reg3b−/− mice are more susceptible to salmonellosis, but not listeriosis, than WT mice.
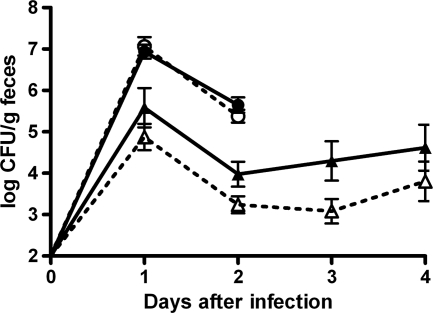
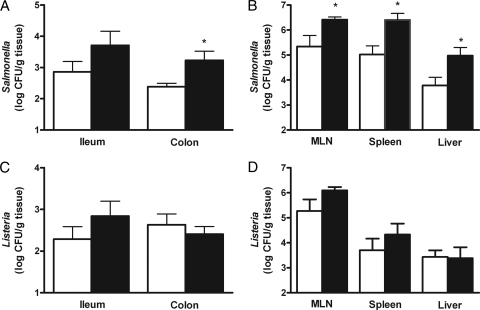
In order to ensure similar Nramp1 genotypes in all Salmonella-infected animals, these mice were sequenced for a previously reported point mutation in this gene which could affect infection susceptibility (9). All mice were shown to be homozygous for the Nramp1 susceptibility allele (data not shown). To investigate the role of Reg3b in intestinal colonization, the numbers of viable Salmonella cells were determined in fresh fecal samples of infected WT and Reg3b−/− mice. Colonization of Salmonella in Reg3b−/− mice was not different from that in WT mice (Fig. 2). In the colonic tissue, which was first flushed with saline to remove intestinal content, higher numbers of Salmonella cells were recovered in Reg3b−/− mice than in WT mice (Fig. 3A) (P < 0.05). In the ileal tissue, Salmonella levels were identical in Reg3b−/− and WT mice. Furthermore, in MLN, spleen, and liver, these levels were higher in Reg3b−/− mice than in their WT counterparts (Fig. 3B) (P < 0.05).
Fig 2.
Salmonella (triangles) and Listeria (circles) excretion in feces. Individually housed mice were orally infected at day 0. Reg3b−/− mice (filled symbols) were infected with 5 × 108 CFU of S. enteritidis (n = 10) or L. monocytogenes (n = 7), and WT littermates (open symbols) were infected with identical numbers of CFU of S. enteritidis (n = 12) or L. monocytogenes (n = 8). Values are the means and SEM.
Fig 3.
Levels of Salmonella (A and B) and Listeria (C and D) in intestinal and extraintestinal tissues at 4 and 2 days after oral infection, respectively. Numbers of CFU of pathogens in the ileal and colonic mucosas (A and C) and in mesenteric lymph nodes (MLN), spleen, and liver (B and D) of Reg3b−/− mice (n = 10; black bars) or WT mice (n = 12; white bars). Values are the means and SEM. An asterisk indicates that the results are significantly different from those of the WT (P < 0.05).
Intestinal colonization of Listeria, as found for Salmonella, was identical in Reg3b−/− and WT mice (Fig. 2). To further investigate the effects of Reg3b on Listeria translocation, the numbers of CFU in ileal and colonic tissue and extraintestinal organs were determined. The number of Listeria cells recovered from the WT did not differ from that recovered from Reg3b−/− mice in ileal and colonic tissues (Fig. 3C) and in MLN, spleen, and liver (Fig. 3D).
Mucosal and systemic inflammation.
The role of Reg3b in inflammation was investigated by measuring the levels of the inflammation marker myeloperoxidase (MPO) in the ileum mucosa at 2 and 4 days after oral infection of Listeria and Salmonella groups, respectively. MPO levels of Salmonella-infected (0.49 ± 0.05 ng/mg protein) and Listeria-infected (0.39 ± 0.06 ng/mg protein) groups did not differ significantly from that of the noninfected groups (0.35 ± 0.05 ng/mg protein) (data not shown). To study systemic inflammation, SAA levels in plasma were determined at the same time points. The levels were elevated from 0.1 ± 0.001 μg/ml (noninfected mice) to 1,164 ± 186 and 502 ± 174 μg/ml upon Salmonella and Listeria infection, respectively (P < 0.05) (data not shown). However, there were no significant differences between WT and Reg3b−/− mice.
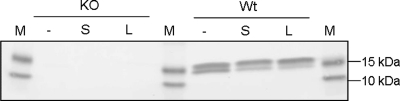
Reg3b binds to Salmonella and Listeria.
To investigate the possible mode of action, it was evaluated in a pulldown assay whether Reg3b present in the ileal mucosa binds to S. enteritidis and L. monocytogenes. Upon ex vivo incubation of mucosal samples from infected mice with these bacteria, it was shown that Reg3b protein levels decreased in the supernatant (Fig. 4). It was mainly the lower-molecular-weight form of the proteolytically processed protein that decreased after incubation with bacteria. This indicated that it is the cleaved form of Reg3b that binds to the pathogens, not the full-length polypeptide. We have shown previously that the Reg3 protein is not precipitated even when centrifuged at 15,000 × g (25). Therefore, the results presented here indicate that ileal mucosal Reg3b is able to directly bind both Salmonella and Listeria.
Fig 4.
Listeria and Salmonella pulldown assays. Presence of Reg3b, detected by Western blot analysis, in supernatant fractions of ileal mucosa samples from S. enteritidis-infected WT (Wt) or Reg3b−/− (KO) mice incubated with Salmonella (S) or Listeria (L). Mucosal samples incubated without bacteria served as the negative control (−). The prestained protein marker is indicated by M.
DISCUSSION
Both microbial infections of conventional mice and intestinal colonization of germ-free mice by commensals increase ileal expression of the secreted C-type lectins Reg3g and Reg3b (3, 12, 22, 25). While Reg3g has been shown to have a bactericidal activity against Gram-positive bacteria (3), the physiological function of Reg3b remains unknown. This study demonstrates a role for Reg3b in protection against infection with Gram-negative S. enteritidis. Despite the fact that similar levels of viable Salmonella cells were recovered from feces, higher numbers were present in MLN, spleen, and liver of infected Reg3b−/− mice than in WT mice, indicating that the translocation and dissemination of Salmonella in host tissues are increased in the absence of Reg3b. As the mice were bred on a mixed background (C57BL/6 and 129O1a) and these strains differ in their susceptibility to Salmonella infection due to a mutation (G to A) in the Nramp1 gene, all mice were genotyped individually (9). All the Salmonella-infected animals were shown to be homozygous for the Nramp1 susceptibility allele, ruling out the possibility that the Nramp allele status would affect the results. In colonic tissue, Salmonella levels were also increased in Reg3b−/− mice compared to those of WT mice. The MLN drain the small intestine, which is a main site of Salmonella invasion (19). Therefore, reduced levels of Salmonella in the MLN of WT mice suggest protective effects of Reg3b on Salmonella translocation in the ileum. Although we did not observe differences in Salmonella counts in the ileal tissues of WT and Reg3b−/− mice, this may be due to a lack of statistical power in this assay, as SEM levels are higher in ileal tissue than in colonic tissue. Moreover, the investigated ileal tissue did not contain Peyer's patches and M cells, which, in addition to absorptive enterocytes, are considered to be important sites of Salmonella translocation (10, 27). All together, the experimental infection data strongly suggest that Reg3b inhibits Salmonella translocation from the gut lumen into intestinal tissues and further extraintestinal tissues but does not kill Salmonella in the gut lumen. These results are supported by the recent findings of Dessein et al., who showed that genetic ablation of Reg3b did not affect the bacterial load of the Gram-negative pathogen Yersinia pseudotuberculosis in the intestinal lumen (5) but significantly increased the bacterial burden in the Peyer's patches, which are the main route of entry for this pathogen. In contrast, Reg3b did not influence resistance to infection with Gram-positive Listeria monocytogenes, as identical numbers of this pathogen were recovered from intestinal and extraintestinal tissues in Reg3b−/− and WT mice, as found in our study.
As murine Reg3g and human PAP are directly bactericidal for Gram-positive bacteria in vitro (3, 17), it seemed possible that Reg3b would have similar antimicrobial activity. However, in this study, there was no effect of Reg3b on the colonization and translocation of L. monocytogenes. The strain of L. monocytogenes used in our infection study was identical to that used previously for in vitro antibacterial assays with Reg3g (3). Previous investigations with rat PAP1, which is the homologue of the murine Reg3b, also showed no direct bactericidal effect against L. monocytogenes in vitro (25). Others also report a lack of bactericidal effect of this protein (5). The coexistence of the two closely related Reg3 proteins in the mouse strongly suggests a different function for each of these proteins. Here we provide evidence that Reg3b has a protective role against Gram-negative Salmonella translocation but not Listeria infection. The exact mechanism by which Reg3b inhibits Salmonella translocation into host tissues is unclear.
The observed difference in Reg3b functionality between listeriosis and salmonellosis might be due to differences in physical appearance and host invasion mechanisms of the two pathogens but might also be related to differences in host responses induced by these pathogens. We can only speculate about the latter, as we did not monitor the exact (immune) responses of the mice. Previous studies by Brandl et al. (2) indicated that the innate immune defense against L. monocytogenes, which requires Toll-like receptor (TLR)-mediated signals, plays a crucial role in Reg3g-related protection against this pathogen. As S. enteritidis and L. monocytogenes may trigger different (TLR-mediated) innate immune signals (1), this might be a cause of differential regulation of Reg3b expression and, therefore, the subsequent protective effects of this protein. On the other hand, it might be that Reg3b modulates the immune response directed against Salmonella but not the response against Listeria. Although measured at a single time point in the current study, Reg3b did not affect the systemic inflammatory response as measured by SAA levels in serum. The local inflammatory response in our infection models was relatively low, as MPO levels did not increase upon infection in the ileal mucosa. This was likely due to the relatively short period that mice were exposed to the orally administered pathogens because our focus was on pathogen colonization and translocation, which normally precede inflammation. Thus, based on our study design, we cannot fully exclude the absence of effects of Reg3b on the inflammatory response. However, previous investigations in our lab with a similar Salmonella infection model in rats indicated that mucosal inflammation in this infection model is relatively low at day 4 postinfection (26). Further analysis of the collected intestinal tissues from the current study might give insight on the role of the innate (immune) response in Reg3b-related protective effects. For example, microarray analysis of intestinal tissues may identify whether and which host defense pathways are associated with protective effects of Reg3b. Performing additional Listeria and Salmonella infection experiments in mice deficient in specific TLRs might reveal the involvement of specific innate immune signaling in Reg3b-mediated protection against intestinal infection.
Besides the proposed association of the host response with Reg3b-related protection of intestinal infection, this lectin might also directly and differentially affect Listeria and Salmonella. The outer membranes of these pathogens are different; for example, Salmonella contains immunoreactive lipopolysaccharides (LPS) which are absent in Gram-positive bacteria, such as Listeria. This influences host signaling pathways, e.g., TLR recognition. Our ex vivo analysis revealed that ileal mucosal Reg3b showed binding affinity to both Salmonella and Listeria. Bactericidal activities of Reg3g depend on binding to cell wall peptidoglycan, a molecule exposed on the Gram-positive bacterial surface (8). This peptidoglycan recognition is determined by the so-called Loop1 EPN tripeptide motif, which is also present in Reg3b (14). This motif is involved in peptidoglycan binding, and binding affinity for carbohydrate ligands depends on carbohydrate chain length (14). Moreover, Reg3b binds to the Gram-positive Bacillus subtilis peptidoglycan, mannose polysaccharides, and chitin, which is a long-chain polymer of N-acetylglucosamine (14). Our results suggest that Reg3b possibly recognizes carbohydrate structures on the bacterial surface of microorganisms which enter the intestinal mucosa. Currently we do not know what the specific target of Reg3b is on the outer membrane of bacteria. It may be related to LPS, which are large molecules consisting of a lipid and a polymeric carbohydrate structure on most Gram-negative bacteria (20). This may be a binding target for the EPN motif in Reg3b. This motif is likely, as shown for Reg3g, also involved in the binding of Reg3b to Gram-positive bacteria. The molecular mechanism by which Reg3b may recognize its binding target remains to be determined. Studies with purified Reg3b are needed to determine what ligands specifically bind to this protein. To address this question, it is necessary to have purified, biologically active Reg3b protein, and efforts are under way to produce and purify this protein in an active form. Although binding may be crucial for the function of Reg3b, it does not necessarily result in a protective effect in vivo, as possible binding of Reg3b to Listeria in the intestinal mucosa does not result in reduced Listeria levels in extraintestinal organs. This might be caused by another important difference between the pathogens, which is that Listeria and Salmonella use complex and very different mechanisms of cellular attachment and invasion (4, 19). Listeria can invade the human intestinal epithelium via the epithelial receptor E-cadherin. A single amino acid change in the murine E-cadherin, however, makes mice relatively resistant to intestinal infection (13). As also shown in the present study, relatively high doses of Listeria do achieve a significant invasive infection in rodents (13). This probably involves a second but less-well-characterized surface protein of L. monocytogenes, internalin B, which enables Listeria to enter and survive within epithelial cells via interaction with three different epithelial membrane receptors (4, 19). In the case of Salmonella, the bacteria are not highly adherent, but their invasion machinery is particularly efficient (19). Salmonella has another mechanism to invade cells which involves binding to epithelial cells by its type III secretory system (TTSS) (10, 16). The TTSS allows direct activation of components of the host-cell cytoskeleton by intracellular delivery of dedicated bacterial effectors, resulting in membrane ruffling and endocytosis. By this event, Salmonella species invade and reside in an atypical acidic compartment called the Salmonella-containing vacuole (SCV) and survive inside the cell (4). It is possible that Reg3b interferes with the expression or function of Salmonella pathogenicity island 1 (SPI-1) TTSS invasion machinery.
In conclusion, Reg3b inhibits intestinal bacterial translocation upon oral infection with Gram-negative Salmonella. In contrast, this protein does not have protective effects against intestinal infection with Gram-positive Listeria. Inhibitory effects of Reg3b on bacterial infection may be linked to its observed binding to pathogens. The protective mechanism of Reg3b in salmonellosis is not associated with direct bactericidal effects but may be related to interference with Salmonella virulence mechanisms or host responses to this pathogen.
ACKNOWLEDGMENTS
We thank the biotechnicians at the Small Animal Centre of Wageningen University (Wageningen, The Netherlands) for excellent assistance. We also thank our colleagues at the Department of Health of NIZO food research and members of the Host-Microbe Interactomics group at Wageningen University for fruitful discussions.
Footnotes
Published ahead of print 17 January 2012
REFERENCES
- 1. Abreu MT, Fukata M, Arditi M. 2005. TLR signaling in the gut in health and disease. J. Immunol. 174:4453–4460 [DOI] [PubMed] [Google Scholar]
- 2. Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. 2007. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 204:1891–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cash HL, Whitham CV, Behrendt CL, Hooper LV. 2006. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313:1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cossart P, Sansonetti PJ. 2004. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304:242–248 [DOI] [PubMed] [Google Scholar]
- 5. Dessein R, et al. 2009. TLR2 is critical for induction of REG3{beta} expression and intestinal clearance of Yersinia pseudotuberculosis. Gut 58:771–776 [DOI] [PubMed] [Google Scholar]
- 6. Duncan HE, Edberg SC. 1995. Host-microbe interaction in the gastrointestinal tract. Crit. Rev. Microbiol. 21:85–100 [DOI] [PubMed] [Google Scholar]
- 7. Giannella RA, Broitman SA, Zamcheck N. 1973. Influence of gastric acidity on bacterial and parasitic enteric infections. A perspective. Ann. Intern. Med. 78:271–276 [DOI] [PubMed] [Google Scholar]
- 8. Gorden J, Small PL. 1993. Acid resistance in enteric bacteria. Infect. Immun. 61:364–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Govoni G, et al. 1995. Genomic structure, promoter sequence, and induction of expression of the mouse Nramp1 gene in macrophages. Genomics 27:9–19 [DOI] [PubMed] [Google Scholar]
- 10. Grassl GA, Finlay BB. 2008. Pathogenesis of enteric Salmonella infections. Curr. Opin. Gastroenterol. 24:22–26 [DOI] [PubMed] [Google Scholar]
- 11. Kaur T, Ganguly NK. 2003. Modulation of gut physiology through enteric toxins. Mol. Cell. Biochem. 253:15–19 [DOI] [PubMed] [Google Scholar]
- 12. Keilbaugh SA, et al. 2005. Activation of RegIIIbeta/gamma and interferon gamma expression in the intestinal tract of SCID mice: an innate response to bacterial colonisation of the gut. Gut 54:623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lecuit M. 2007. Human listeriosis and animal models. Microbes Infect. 9:1216–1225 [DOI] [PubMed] [Google Scholar]
- 14. Lehotzky RE, et al. 2010. Molecular basis for peptidoglycan recognition by a bactericidal lectin. Proc. Natl. Acad. Sci. U. S. A. 107:7722–7727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lieu HT, et al. 2006. Reg2 inactivation increases sensitivity to Fas hepatotoxicity and delays liver regeneration post-hepatectomy in mice. Hepatology 44:1452–1464 [DOI] [PubMed] [Google Scholar]
- 16. Ly KT, Casanova JE. 2007. Mechanisms of Salmonella entry into host cells. Cell. Microbiol. 9:2103–2111 [DOI] [PubMed] [Google Scholar]
- 17. Mukherjee S, et al. 2009. Regulation of C-type lectin antimicrobial activity by a flexible N-terminal prosegment. J. Biol. Chem. 284:4881–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oudenhoven IM, Klaasen HL, Lapre JA, Weerkamp AH, Van der Meer R. 1994. Nitric oxide-derived urinary nitrate as a marker of intestinal bacterial translocation in rats. Gastroenterology 107:47–53 [DOI] [PubMed] [Google Scholar]
- 19. Pizarro-Cerda J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124:715–727 [DOI] [PubMed] [Google Scholar]
- 20. Raetz CR, et al. 1991. Gram-negative endotoxin: an extraordinary lipid with profound effects on eukaryotic signal transduction. FASEB J. 5:2652–2660 [DOI] [PubMed] [Google Scholar]
- 21. Reeves PG, Nielsen FH, Fahey GC., Jr 1993. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 123:1939–1951 [DOI] [PubMed] [Google Scholar]
- 22. Rodenburg W, Bovee-Oudenhoven IM, Kramer E, van der Meer R, Keijer J. 2007. Gene expression response of the rat small intestine following oral Salmonella infection. Physiol. Genomics 30:123–133 [DOI] [PubMed] [Google Scholar]
- 23. Sarker SA, Gyr K. 1992. Non-immunological defence mechanisms of the gut. Gut 33:987–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uhlar CM, Whitehead AS. 1999. Serum amyloid A, the major vertebrate acute-phase reactant. Eur. J. Biochem. 265:501–523 [DOI] [PubMed] [Google Scholar]
- 25. van Ampting MT, et al. 2009. Ileal mucosal and fecal pancreatitis associated protein levels reflect severity of Salmonella infection in rats. Dig. Dis. Sci. 54:2588–2597 [DOI] [PubMed] [Google Scholar]
- 26. van Ampting MT, et al. 2009. Intestinal barrier function in response to abundant or depleted mucosal glutathione in Salmonella-infected rats. BMC Physiol. 9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Asten AJAM, Koninkx JFJG, van Dijk JE. 2005. Salmonella entry: M cells versus absorptive enterocytes. Vet. Microbiol. 108:149–152 [DOI] [PubMed] [Google Scholar]
- 28. van Netten P, Perales I, van de Moosdijk A, Curtis GD, Mossel DA. 1989. Liquid and solid selective differential media for the detection and enumeration of L. monocytogenes and other Listeria spp. Int. J. Food Microbiol. 8:299–316 [DOI] [PubMed] [Google Scholar]
- 29.WHO. Food safety and foodborne illness. 2007. http://www.who.int/mediacentre/factsheets/fs237/en/index.html.