Abstract
Escherichia coli O157:H7 causes food and waterborne enteric infections that can result in hemorrhagic colitis and life-threatening hemolytic uremic syndrome. Intimate adherence of the bacteria to intestinal epithelial cells is mediated by intimin, but E. coli O157:H7 also possess several other putative adhesins, including curli and two operons that encode long polar fimbriae (Lpf). To assess the importance of Lpf for intestinal colonization, we performed competition experiments between E. coli O157:H7 and an isogenic ΔlpfA1 ΔlpfA2 double mutant in the infant rabbit model. The mutant was outcompeted in the ileum, cecum, and midcolon, suggesting that Lpf contributes to intestinal colonization. In contrast, the ΔlpfA1 ΔlpfA2 mutant showed increased adherence to colonic epithelial cells in vitro. Transmission electron microscopy revealed curli-like structures on the surface of the ΔlpfA1 ΔlpfA2 mutant, and the presence of curli was confirmed by Congo red binding, immunogold-labeling electron microscopy, immunoblotting, and quantitative real-time reverse transcription-PCR (qRT-PCR) measuring csgA expression. However, deletion of csgA, which encodes the major curli subunit, does not appear to affect intestinal colonization. In addition to suggesting that Lpf can contribute to EHEC intestinal colonization, our observations indicate that the regulatory pathways governing the expression of Lpf and curli are interdependent.
INTRODUCTION
Infection with enterohemorrhagic Escherichia coli (EHEC) causes abdominal cramping and nonbloody diarrhea that is usually self-limiting but can progress to hemorrhagic colitis and hemolytic uremic syndrome (HUS). EHEC is transmitted through food or water contaminated by feces, usually from cattle, the major reservoir of EHEC. E. coli O157:H7 is the most common EHEC serotype linked to diarrheal disease in the United States, the United Kingdom, Canada, Japan, and Argentina (2, 5, 13, 17, 19). EHEC organisms belong to a group of bacteria that cause attaching and effacing (A/E) lesions on intestinal epithelial cells. Formation of A/E lesions is dependent on the locus of enterocyte effacement (LEE), which encodes a type III secretion system and numerous effector proteins. In particular, tight adherence of EHEC to intestinal cells requires transfer of the bacterial receptor Tir to host cells and expression of the Tir-binding adhesin, intimin (9). However, evidence suggests that an additional factor(s) comprised of both fimbrial and nonfimbrial adhesins may act to promote the initial loose attachment of EHEC to intestinal cells (reviewed in reference 28).
E. coli O157:H7 possesses two tightly regulated operons encoding long polar fimbriae (Lpf), which are induced in vitro in tissue culture media (24) or under iron-restricted conditions (25) and which have been found to influence adherence in vitro and colonization in vivo both for O157 strains and for other pathogenic E. coli strains (4, 6, 8, 26). Mutation of the lpf1 operon in E. coli O157:H7 results in a reduced number of cells adhering to cultured HeLa cells (approximately one-fifth fewer than the number of adhering wild-type cells), as well as a reduction in microcolony formation (24). Similarly, O157:H7 lpf2 mutants show somewhat reduced adherence to Caco-2 cells, and a possible role for lpf2 in early adherence has been suggested (25). In vivo colonization assays using sheep and pigs have also suggested that the absence of Lpf impairs EHEC colonization (8). However, analyses of binding performed with human tissue, using in vitro organ culture, showed that EHEC strains with lpfA mutations colonized intestinal regions that were not normally bound by a wild-type strain (6). Given the relatively subtle effects of lpfA mutations on adherence in vitro, coupled with the divergent findings regarding tissue colonization, the precise role of Lpf in EHEC adherence has remained somewhat ambiguous.
E. coli O157:H7 has the ability to produce, in addition to Lpf, thin aggregative fimbriae termed curli. These appendages may promote adhesion under specific conditions (10), and they have also been implicated in biofilm formation, invasion, and the induction of inflammation (reviewed in reference 1). However, curli are expressed in relatively few E. coli O157:H7 strains mainly in response to certain environmental conditions (29). For example, some strains have been reported to produce curli at 37°C under salt- and nutrient- or oxygen-limiting conditions (reviewed in reference 1). In addition, mutants with altered expression of curli have been described (29). The relationship between the expression of curli and other fimbriae has largely not been explored; however, there is evidence for cross talk in the regulation of other fimbrial structures (22, 30).
In this study, we have further explored the role of Lpf in E. coli O157:H7 adherence to, and colonization of, intestinal tissue. Colonization was assessed using the infant rabbit model of EHEC infection, which reproduces many aspects of intestinal disease seen during human EHEC infection (15). We found that a strain lacking both lpfA1 and lpfA2 (and thus lacking Lpf) is outcompeted in the rabbit intestine by its parental strain, suggesting that Lpf might contribute to intestinal adhesion. In contrast, the ΔlpfA1 ΔlpfA2 mutant showed increased adherence to cultured intestinal epithelial cells, perhaps due to overexpression of curli in this strain. Thus, analyses of the roles of Lpf and curli, both in vivo and in vitro, are likely to be complicated by regulatory cross talk between these adhesins. Nonetheless, our data suggest that both of these factors can promote EHEC interaction with the intestinal epithelium.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains are listed in Table 1 and were grown in Luria broth at 37°C and 225 to 250 rpm. All E. coli O157:H7 strains used in this study are derivatives of strain 86-24 (23). Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 30 μg/ml; kanamycin (Km), 50 μg/ml; streptomycin (Sm), 100 μg/ml; and tetracycline (Tc), 12.5 μg/ml.
Table 1.
Bacterial strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| E. coli | ||
| 86-24 | EHEC O157:H7 strain 86-24 Smr Nalr | 23 |
| 86-24ΔlacZ | 86-24 ΔlacZ Smr | This study |
| CVD468 | 86-24 ΔlpfA1 Smr Cmr | 24 |
| AGT201 | 86-24 ΔlpfA2 Smr Tcr | 25 |
| AGT210 | 86-24 ΔlpfA1 ΔlpfA2 Smr Cmr Tcr | 25 |
| SJL102 | 86-24 ΔcsgA Smr Cmr | This study |
| SJL101 | 86-24 ΔlpfA1 ΔlpfA2 ΔcsgA Smr Cmr Tcr Kmr | This study |
| SM10 (λ pir) | (pCVD442 csgA::kan) Apr Kmr Sucs | This study |
| S. Typhimurium | Strain 2157 | Torres' collection |
Animal experiments.
Litters of 3-day-old New Zealand White infant rabbits were infected as described previously (15, 16). Briefly, the animals were orogastrically inoculated (∼5 × 108 CFU per 90 g of rabbit) with the wild-type strain 86-24, the ΔcsgA mutant, the ΔlpfA1 ΔlpfA2 mutant, or the ΔcsgA ΔlpfA1 ΔlpfA2 mutant or with 1:1 mixtures of 86-24 and the aforementioned mutants. The bacterial inoculum was prepared by growing the strains overnight in L broth at 37°C in the presence of antibiotics, harvesting the cells by centrifugation, and then resuspending the cell pellet in sterile phosphate-buffered saline (PBS; pH 7.2) to give a final cell density of ∼109 CFU/ml. The rabbits were necropsied 5 days postinoculation, tissue samples were taken from the small and large intestines, and the cecum, as well as fecal material, were collected for microbiologic analyses. In the single-infection experiments, the number of EHEC CFU recovered in the samples was determined by plating homogenates on media containing the appropriate antibiotics. In the competition experiments, we used two approaches to determine the number of wild-type and mutant cells present in the intestinal tissue. In some experiments, EHEC was recovered by plating on media containing Sm (to detect total numbers of EHEC), followed by replica plating onto media containing the appropriate antibiotic(s) to select for the mutants. In other experiments, a lacZ mutant of 86-24 was used as the wild-type strain to allow simultaneous detection of colonies based on their ability to utilize bromo-chloro-indolyl-galactopyranoside (also known as X-gal). This second approach was taken in case the use of multiple antibiotics affected the number of mutant cells that were recovered following in vivo passage. Preliminary studies showed that the 86-24 ΔlacZ mutant competed equally with the parental strain in all regions of the intestine (data not shown). Experiments with mutants that exhibited reduced colonization in the rabbit intestine compared to colonization by the wild type were repeated in a second independent litter. In samples where no colonies were detected at the lowest dilution plated, the limit of detection (∼102 CFU/g) was used as a value. Competition indices (CI) were calculated as the ratio of the mutant output to the wild-type output divided by the ratio of the mutant to the wild-type input. The CI ratios for each intestinal segment were compared to a theoretical value of 1 using the one-sample t test. In addition, for each intestinal segment, the CI obtained by the two different detection methods were compared using Student's two-way t test (GraphPad Prism version 5.02).
Cell culture and adherence assays.
T84 cells were maintained in Dulbecco's modified Eagle's medium (DMEM)-F12 at 37°C and 5% CO2. Medium was supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic (Invitrogen/Gibco, Carlsbad, CA). For adhesion assays, 24-well plates were seeded with 105 cells per well and incubated as described above until ∼80% confluent. Monolayers were washed twice with 1 ml of PBS, 1 ml of medium with no supplements was added to each well, and then wells were inoculated with 107 bacterial cells from overnight cultures (multiplicity of infection [MOI], 100). Inoculated monolayers were incubated for 3 h at 37°C and 5% CO2. After incubation, cells were washed twice with PBS, and then 200 μl of 0.1% Triton X-100 in PBS was added to each well. Plates were incubated at 37°C and 5% CO2 until monolayers detached from the bottom of the plate. Monolayers were homogenized by pipetting. Tenfold dilutions were plated onto LB agar using the drop plate technique described by Chen et al. (3). The percentage of bacteria recovered was calculated as the number of CFU/ml recovered divided by the initial number of CFU/ml multiplied by 100. The experiments were repeated 3 times in triplicate. Mean percentages of each mutant recovered were compared to the mean percentage of the wild type recovered using Student's t test. If all group means were compared, a one-way analysis of variance (ANOVA), followed by Holm-Sidak post hoc testing, was used. The in vitro competition assays were performed as described above except that cells were inoculated with 5 × 106 cells each of mutant and wild-type bacteria (the total number of bacteria/well was 107 cells). Numbers of CFU/ml for the mutant and wild type were determined by plating the bacterial mixture onto both LB agar and plates with appropriate antibiotics to select for the mutant. CI were calculated as described above and analyzed using one-way ANOVA and Holm-Sidak post hoc testing.
Gene disruption by allelic exchange.
ΔcsgA mutants were created with E. coli O157:H7 strain 86-24 and the isogenic double lpfA mutant AGT210 by allelic exchange using the E. coli SM10 (λ pir) strain carrying the suicide vector pCVD442 with the csgA gene from E. coli O157:H7 interrupted by a kanamycin resistance gene (pCVD442 csgA::kan). The suicide vector was introduced into the recipient strains by conjugation, and recombinants were selected by plating strains on the appropriate antibiotics and sucrose and then testing for ampicillin sensitivity to confirm the loss of pCVD442, as previously described (24). Interruption of csgA was confirmed by PCR, Congo red binding, and immunoblotting for curli. The lacZ mutant of 86-24 was also constructed by allelic exchange using E. coli SM10λ pir containing pES10 (a kind gift from Elizabeth Shaknovich). Integration of this vector results in deletion of amino acids 359 to 637 of the lacZ gene product.
Congo red binding assay.
Congo red binding assays were performed as previously described (7). Briefly, bacterial cultures were grown overnight in Luria broth at 37°C and 225 rpm. Cultures were diluted 1:1 in sterile distilled water, and then 6-μl drops were spotted onto Congo red indicator (CRI) plates containing LB agar with no salt (LBNS), 40 μg/ml Congo red, and 20 μg/ml Coomassie brilliant blue. Plates were incubated at 30°C or 37°C and photographed at 24 h.
SDS-PAGE and immunoblotting.
Cultures were grown on LB agar with no salt as described above for the Congo red binding assay. At 24 h and 48 h, bacteria were resuspended in 100 μl of 1× PBS. Samples were equilibrated to 109 cells as determined by measurements of optical density at 600 nm (OD600). Cells were spun down and resuspended in 70 μl of formic acid. The formic acid was evaporated off, and the pellets were resuspended in 200 μl 1× sample buffer, 10 μl of which was loaded onto a 15% SDS-PAGE gel. Semidry transfer onto a polyvinylidene difluoride (PVDF) membrane was performed according to standard procedures. The membrane was incubated overnight at 4°C with the primary antibody against purified curli (rabbit anti-curli at 1:7,000; kindly donated by Matt Chapman) and then incubated for 1 h at room temperature with the secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit IgG at 1:10,000). Signal was detected using Amersham ECL Plus Western blotting detection agents (GE Healthcare, Buckinghamshire, United Kingdom) according to the manufacturer's instructions.
Transmission electron microscopy.
Samples were grown statically at 37°C overnight in DMEM-high glucose, spun down, and resuspended in 30 μl 1× PBS. Formvar- and carbon-coated nickel grids (200 mesh) were floated on the cell suspension and stained with 2% potassium-phosphotungstic acid (pH 6.8) to visualize surface structures. For immunogold labeling, grids were prepared as described above but, prior to being stained with phosphotungstic acid, were incubated with rabbit anticurli at 1:1,000 in 1% bovine serum albumin (BSA)-PBS for 30 min at room temperature, washed in 1% BSA-PBS, and then incubated with goat anti-rabbit IgG antibody (1:20 in 1% BSA-PBS) labeled with 15-nm gold particles (Electron Microscopy Sciences, Hatfield, PA) for 30 min at room temperature, washed again with deionized water, fixed in 2% aqueous glutaraldehyde for 10 min, and washed in deionized water. Flagella and curli were visualized using a Phillips 201 electron microscope.
RNA isolation and cDNA synthesis.
Total RNA was obtained from bacteria in contact with monolayers of T84 cells. One ml of DMEM with no supplements was added to each well, followed by infection with 2 × 108 bacterial cells obtained from overnight cultures. Infected monolayers were incubated for 1 and 2 h at 37°C and 5% CO2. The supernatant was pooled from 6 wells; the samples were stabilized with RNAprotect reagent (Qiagen, Valencia, CA) and harvested by centrifugation at 3,500 rpm for 20 min. Then, the samples were resuspended in 10 mM Tris-HCl (in 0.1% diethyl pyrocarbonate [DEPC]-H2O). RNA was purified using the High Pure RNA isolation kit and treated with DNase I (Roche, Mannheim, Germany), quantified, and qualitatively analyzed on 2% agarose gels. Five micrograms of total RNA was used for cDNA synthesis by employing the SuperScript first-strand synthesis system (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. A negative control with no reverse transcriptase was also included. The resulting cDNA was utilized for quantitative real-time reverse transcription-PCR (qRT-PCR).
qRT-PCR.
qRT-PCR was performed by using the iQ SYBR green Supermix on the CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA). The rrsB and rpoS genes were used to normalize our data, and a value of 1 was used to standardize csgA gene expression to the wild-type strain; the primers used are listed in Table 2. For each reaction, 1 μl of reverse-transcribed cDNA was subjected to PCR amplification in a 12.5-μl final volume containing a 500 nM concentration of each primer and 6.5 μl of 2× SYBR iQ SYBR green Supermix. The following conditions were used for amplification: 1 cycle at 95°C for 30 s and then 40 cycles at 95°C for 5 s and 60°C for 30 s. To ensure the specificity of the PCR products, melting curve analysis was performed by heating products from 65°C to 95°C in increments of 0.5°C every 5 s while monitoring the fluorescence. Expression of csgA in each mutant strain at different time points was compared to that of the wild-type strain at 1 h of incubation with T84 cells. These assays were performed in duplicate for each strain. A Student's t test was used for statistical analysis.
Table 2.
qRT-PCR primers used in this study
| Primer name | Sequence (5′–3′) | Target gene | Reference or source |
|---|---|---|---|
| 5RTRRSB | TGCAAGTCGAACGGTAACAG | rrsB | 11 |
| 3RTRRSB | AGTTATCCCCCTCCATCAGG | rrsB | 11 |
| rpoS Fw | AGTCAGAATACGCTGAAAGTTCATG | rpoS | 21 |
| rpoS Rv | AAGGTAAAGCTGAGTCGCGTC | rpoS | 21 |
| csgAFw | TGGTAACAGCGCCACTCTTG | csgA | This study |
| csgARv | GACGGTGGAATTAGATGCAGTC | csgA | This study |
RESULTS
The ΔlpfA1 ΔlpfA2 mutant is outcompeted by wild-type EHEC in an infant rabbit model of infection.
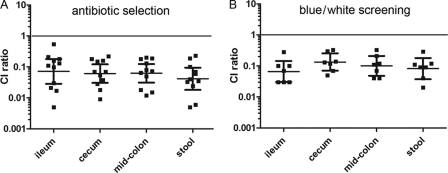
Single-strain infections and competition assays between the wild-type O157:H7 strain 86-24 and mutants lacking either one copy or both copies of lpfA (which encodes the major fimbrial subunits) were performed with infant rabbits. Rabbits inoculated with the wild-type strain or either of the single lpfA mutants (CVD468 and AGT201) or the double lpfA mutant (AGT210) developed diarrhea and were colonized (data not shown). However, when rabbits were coinfected with a 1:1 mixture of the double lpfA mutant AGT210 and wild-type EHEC, there were approximately 10-fold-fewer mutant CFU recovered from all regions of the intestine and in the stool than with the other strains. Thus, the geometric mean CI were around 0.1, regardless of whether antibiotics (Fig. 1A) or blue/white screening (Fig. 1B) was used to determine the relative numbers of bacteria. The CI ratios in all intestinal sections and in the stool were significantly less than 1 (P < 0.001), but there was no significant difference in the CI ratios obtained from the same tissue section using the two detection methods. Deletion of only one of the LpfA subunits was not sufficient to mediate a defect in colonization of the colon (Table 3). The ∼10-fold reduction in intestinal colonization of the strain lacking both Lpf genes in the competition experiment suggests that these fimbriae contribute to the capacity of EHEC to colonize the intestine. The absence of a reduction in colonization or disease for the double lpf mutant in the single-infection experiments may reflect the limited sensitivity of this assay and/or the presence of additional factors that can substitute for Lpf under these conditions.
Fig 1.
The ΔlpfA1 ΔlpfA2 mutant is outcompeted by 86-24 in the infant rabbit intestine. Three-day-old rabbits were orogastrically inoculated with 1:1 mixtures of E. coli O157:H7 strain 86-24 and an isogenic ΔlpfA1 ΔlpfA2 mutant, AGT210 (A), or 86-24ΔlacZ and the ΔlpfA1 ΔlpfA2 mutant (B). Animals were euthanized after 5 days. The number of colonies counted on antibiotic-containing medium or medium containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) was used to calculate competition indexes (CI). A CI is the ratio of the number of mutant CFU to the number of wild-type CFU in output samples divided by the number of mutant to wild-type CFU in the inoculum. A CI value of 1 (shown by the black line) indicates that the mutant competes equally with the wild-type strain. Bars represent the geometric means with 95% confidence intervals. The CI of samples from the same intestinal site but with different selection criteria used were compared by Student's t test and not found to be significantly different.
Table 3.
Competitive index values for mutants obtained following colonization of the large intestines of infant rabbits
| Genotype of mutant |
In vivo colonization |
|
|---|---|---|
| Geometric mean | 95% confidence interval | |
| ΔlpfA1 | 0.70 | 0.38–1.32 |
| ΔlpfA2 | 3.29 | 1.81–5.98 |
| ΔlpfA1 ΔlpfA2 | 0.07 | 0.05–0.12 |
| ΔcsgA | 0.92 | 0.72–1.18 |
| ΔlpfA1 ΔlpfA2 ΔcsgA | 0.02 | 0.007–0.08 |
The ΔlpfA1 ΔlpfA2 mutant shows increased adherence to cultured colonic epithelial cells.
Inactivation of both sets of Lpf genes might impair colonization by disrupting adhesion of E. coli O157:H7 to the intestinal epithelium. To explore this possibility, we assessed the ability of various strains to adhere to T84 cells. Inoculation of T84 monolayers with single strains resulted in recovery of the single lpfA1 and lpfA2 mutants and the ΔlpfA1 ΔlpfA2 mutant (34.4% ± 10%, 27.7% ± 4%, and 32.8% ± 7%, respectively) at levels similar to the recovery of wild-type EHEC (29.5% ± 8%, P = 0.78). However, when competition adhesion assays were performed using the double lpfA mutant AGT210 and wild-type EHEC, the mean CI (1.69 ± 0.12) was significantly greater than 1(P = 0.03). This unexpected finding suggested that the strain lacking all Lpf might produce an additional factor that provided a subtle advantage in the adherence assay and/or might be detrimental in vivo.
Production of curli is increased in the ΔlpfA1 ΔlpfA2 mutant.
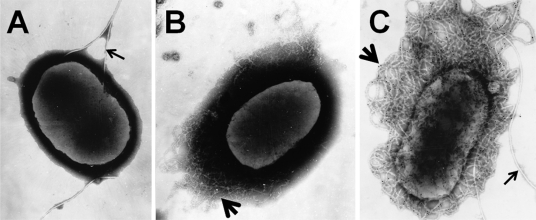
To further explore potential differences in adhesin production by the wild-type strain and the ΔlpfA1 ΔlpfA2 mutant, we used electron microscopy to visualize the surfaces of the two strains. Both negative staining and immunostaining were used to detect the presence of flagella, fimbriae, or other adhesins on bacteria grown in DMEM at 37°C, conditions known to induce Lpf expression. Unexpectedly, micrographs revealed structures like curli on the surface of the ΔlpfA1 ΔlpfA2 mutant strain AGT210 (Fig. 2B). These structures were confirmed to be curli by immunogold labeling using sera generated against purified curli (Fig. 2C). Flagella but no other fimbriae were visible on the surface of wild-type EHEC (Fig. 2A).
Fig 2.
Curli are expressed by the ΔlpfA1 ΔlpfA2 mutant. EHEC 86-24 (A) and the ΔlpfA1 ΔlpfA2 mutant AGT210 (B) were grown statically overnight at 37°C in DMEM. Samples were spun down, resuspended in PBS, and then stained with phosphotungstic acid (A and B) or incubated with anti-CsgA sera and then with a secondary antibody labeled with 15-nm gold particles (C). Thin arrows indicate flagella (A and C), and thick arrows indicate curli-like fibrils (B) or CsgA-labeled fibrils (C).
The production of curli in the wild type and the ΔlpfA1 ΔlpfA2 mutant was investigated using immunoblotting and via growth of EHEC on Congo red indicator (CRI) plates. For both assays, the growth medium was LB with no salt because it has been shown that low salt induces maximal expression of curli (1). Expression was monitored at 30°C as well as at 37°C, as low temperature has also been observed to induce expression (1). As can be seen in the electron micrographs of cells grown at 37°C in liquid culture (Fig. 2), these assays indicated that expression of curli was elevated in the ΔlpfA1 ΔlpfA2 mutant relative to expression in the wild-type strain at 37°C. No production of CsgA (the 17-kDa major constituent of curli) was observed on Western blots of wild-type cells grown at 37°C, while abundant CsgA was evident in samples from the ΔlpfA1 ΔlpfA2 mutant (Fig. 3B). Furthermore, the ΔlpfA1 ΔlpfA2 mutant was pink/red when cultures spotted on indicator plates were grown at 37°C, consistent with binding of Congo red by amyloid-cell-like curli. In contrast, spots of the wild-type strain were a light tan, as were those of a strain lacking csgA and a ΔcsgA ΔlpfA1 ΔlpfA2 triple mutant, which, as expected, lacked detectable CsgA on immunoblots. As seen with other strains, 86-24 had a slightly darker (but not red) color when grown on CRI plates at 30°C, which is consistent with the slight production of CsgA detected on immunoblots for this condition (Fig. 3A). Interestingly, the ΔlpfA1 ΔlpfA2 mutant showed markedly reduced levels of CsgA at 30°C, and on immunoblots, CsgA abundance did not differ from that of the wild-type strain under this condition, suggesting that deletion of lpfA1 and lpfA2 has less effect on csgA regulation at 30°C than at 37°C. However, for reasons that remain unclear, the ΔlpfA1 ΔlpfA2 mutant still appears redder than the wild-type strain on CRI plates at 30°C.
Fig 3.
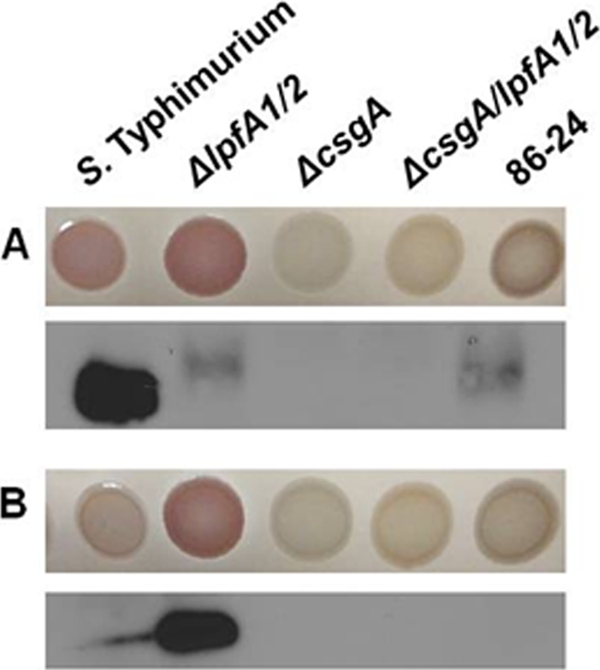
Congo red binding and detection of EHEC curli by immunoblotting. Overnight cultures of wild-type Salmonella enterica serovar Typhimurium and EHEC 86-24 and the isogenic mutants AGT210 (ΔlpfA1 ΔlpfA2), SJL102 (ΔcsgA), and SJL101 (ΔlpfA1 ΔlpfA2 ΔcsgA) were spotted (6 μl) onto LBNS agar containing 40 μg/ml Congo red and 20 μg/ml Coomassie brilliant blue and incubated for 24 h at 30°C (A) or 37°C (B). The positive control for Congo red binding at 30°C was S. Typhimurium.
Quantitative real-time PCR was used to support the immunoblot results and to quantify the expression of csgA in the wild-type, ΔlpfA1 ΔlpfA2 double mutant, and ΔcsgA ΔlpfA1 ΔlpfA2 triple mutant strains while in contact with T84 cell monolayers for 1 or 2 h. Basal expression of csgA in the wild-type strain after 1 h of incubation was set at a value of 1, and all the other values of expression at 1 and 2 h were related to this threshold. The expression of csgA was 1.63-fold higher in the ΔlpfA1Δ lpfA2 double mutant than in the wild-type strain (Fig. 4) (P = 0.01), and threshold expression was observed in the ΔcsgA ΔlpfA1 ΔlpfA2 triple mutant. After 2 h of incubation with T84 cells, the csgA expression level in the ΔlpfA1 ΔlpfA2 mutant remained approximately the same (1.73-fold), while expression by the wild type increased 1-fold compared to the 1-h baseline value (Fig. 4). Expression of csgA in the ΔcsgA ΔlpfA1 ΔlpfA2 mutant remained at baseline levels.
Fig 4.
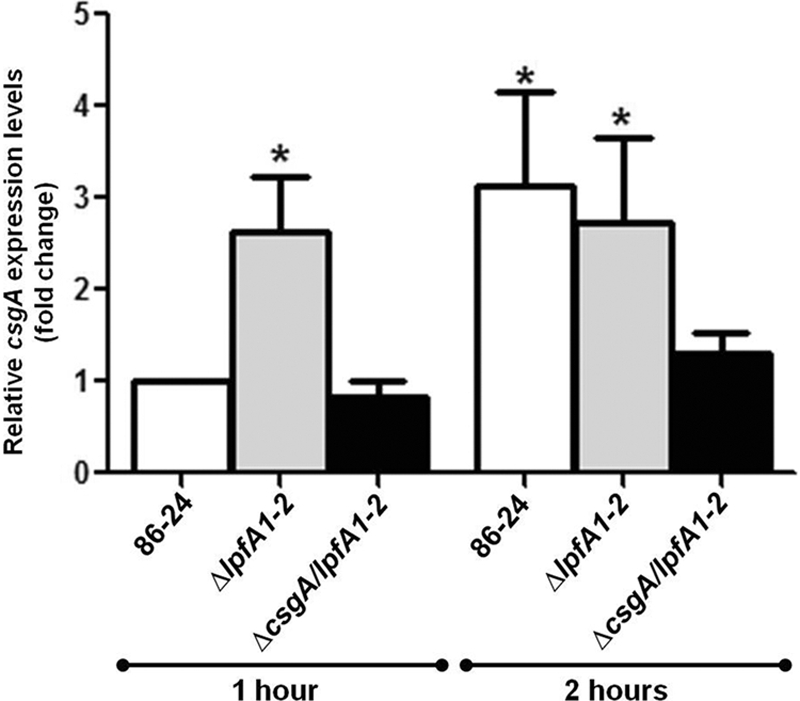
Quantitative real-time RT-PCR analysis of csgA expression. The strains were incubated for 1 h and 2 h with T84 cell cultures at 37°C and 5% CO2. The fold variation of gene expression was obtained by the comparative cycle threshold (ΔΔCT) method. Wild-type EHEC 86-24 csgA expression at 1 h is represented as a value of 1, and all other values of expression at 1 h and 2 h are related to this value. The expression of csgA was 1.63-fold higher (*, P = 0.01) in the ΔlpfA1 ΔlpfA2 mutant than in the wild type at 1 h in contact with T84 cells. After 2 h, the expression of csgA increased 1-fold in the wild type and remained at ∼1.7-fold (*, P < 0.05) in the ΔlpfA1 ΔlpfA2 mutant. Expression levels of csgA in the ΔcsgA ΔlpfA1 ΔlpfA2 triple mutant remained virtually unchanged. Student's t test was used for statistical analysis.
Contribution of curli to adhesion and intestinal colonization.
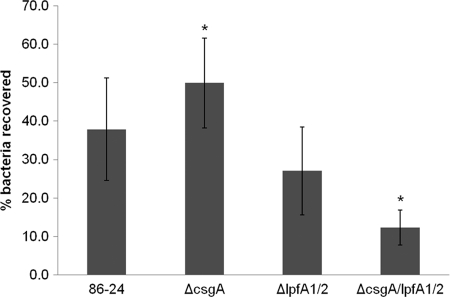
Given the abundance of curli produced by the ΔlpfA1 ΔlpfA2 mutant, we determined whether these might promote its interaction with epithelial cells and thereby account for the enhanced adherence of this strain to T84 cells. As described above, adhesion was assessed using both single-strain and competition assays. In assays of single strains, there was no difference between the levels of adherence of wild-type, ΔcsgA, ΔlpfA1 ΔlpfA2, and ΔcsgA ΔlpfA1 ΔlpfA2 strains to T84 cells (Fig. 5) (P = 0.17). However, a significant difference in adherence for the single ΔcsgA mutant from that of the ΔcsgA ΔlpfA1 ΔlpfA2 triple mutant (two-way Student's t test, P = 0.04) was detected. Thus, the lack of Lpf impairs, rather than aids, adherence in the absence of curli. Together, these observations suggest that the enhanced adherence of the ΔlpfA1 ΔlpfA2 mutant that was observed in initial assays may have been a consequence of the overexpression of curli. However, when expressed at wild-type levels, curli do not appear to play a major role in adherence, based on the results of both the single-strain assays presented above and the competition assays, in which the wild type and ΔcsgA mutant were recovered in equal numbers (CI, 1.32 ± 0.3; P = 0.40). Somewhat unexpectedly, the ΔcsgA ΔlpfA1 ΔlpfA2 triple mutant did not have reduced adhesion relative to that of the ΔlpfA1 ΔlpfA2 mutant (CI, 0.96 ± 0.89; P = 0.89), arguing against a role for curli as the sole adhesin promoting attachment in this strain background. However, unlike the ΔlpfA1 ΔlpfA2 mutant, the ΔcsgA ΔlpfA1 ΔlpfA2 triple mutant did not display enhanced adherence relative to the wild-type strain (CI, 2.73 ± 1.58; P = 0.24), data that support a role for curli in adhesion. Collectively, these data suggest that both Lpf and curli may promote the adherence of EHEC to epithelial cells; however, neither is essential, and interdependence in their expression (and potentially that of other adhesins as well) can complicate analyses of their roles.
Fig 5.
Adherence of E. coli O157:H7 and isogenic lpfA and/or ΔcsgA mutants to T84 cells. Monolayers of T84 cells were inoculated with 107 bacterial cells from an overnight culture of the wild-type EHEC strain 86-24, AGT210 (ΔlpfA1 ΔlpfA2), SJL102 (ΔcsgA), and SJL101 (ΔlpfA1 ΔlpfA2 ΔcsgA) and incubated at 37°C and 5% CO2 for 3 h. T84 cell monolayers were lysed, and 10-fold serial dilutions were plated on LB agar to determine numbers of CFU/ml. Percentages of bacteria recovered were calculated as (final number of CFU per ml/initial number of CFU per ml) × 100. The single ΔcsgA mutant and the triple mutant were compared by Student's t test (*, P = 0.04).
Given that curli appear to enhance the adhesion of the ΔlpfA1 ΔlpfA2 mutant in vitro, we assessed whether the absence of curli influences EHEC intestinal colonization in infant rabbits. In a wild-type background, deletion of csgA had no effect upon colonization of the colon (Table 3) (geometric mean [95% confidence interval], 0.92 [0.72 to 1.18]), suggesting that basal levels of curli do not make a major contribution to colonization. Furthermore, consistent with our findings above, the CI of the ΔcsgA ΔlpfA1 ΔlpfA2 triple mutant was similar to that of the ΔlpfA1 ΔlpfA2 mutant (Table 3) (triple versus double mutant CI geometric mean [95% confidence interval], 0.02 [0.007 to 0.08] versus 0.07 [0.05 to 0.12]), suggesting that even in the ΔlpfA1 ΔlpfA2 background, where curli may be overexpressed, this appendage does not contribute to colonization. The single lpfA mutants were not outcompeted by the wild type in vivo (Table 3), and none of the mutants were outcompeted by the wild type in in vitro growth experiments (data not shown), suggesting that the in vivo intestinal competition deficits of the double and triple mutants are not due to general growth defects.
DISCUSSION
We have investigated the importance of Lpf and curli in the colonization of and adherence to intestinal epithelial cells by E. coli O157:H7, using in vivo and in vitro assays. We found that a strain lacking both lpfA1 and lpfA2 had an impaired ability, relative to the wild-type strain, to colonize the infant rabbit intestinal tract. This result is consistent with previous colonization analyses performed in pigs (8). Unexpectedly, we found that the ΔlpfA1 ΔlpfA2 mutant did not exhibit reduced adhesion to cultured intestinal epithelial cells; instead, it showed enhanced adhesion in competition assays against the wild-type strain. Notably, the ΔlpfA1 ΔlpfA2 mutant produced elevated amounts of curli, both in liquid culture and on solid media, raising the possibility that curli can compensate for the absence of Lpf under certain conditions. A connection between expression of curli and Lpf in E. coli O157:H7 has not previously been described.
Although defects in colonization and adherence were reproducibly observed for the ΔlpfA1 ΔlpfA2 mutant in competition assays against a wild-type strain, such deficiencies were not evident in single-strain infections. It is likely that the competition assays enable better detection of relatively subtle colonization defects, as the presence of an internal control (i.e., the wild-type strain) allows compensation for variability between animals, which in a single-infection study may mask the effects of the mutation of interest. Given that the effects of lpfA1 and lpfA2 deletion upon colonization are relatively small (∼10-fold versus the 10,000-fold effects of a tir or an eae gene deletion [15]), it is not particularly surprising that the deficiencies of this strain could not be detected in the single-strain analyses. However, it should be noted that competition analyses are not always preferable; for example, trans-complementation by a wild-type strain can mask a true deficit in the mutant (14).
Our observation that deletion of lpfA1 and lpfA2 results in augmented expression of curli, at least under a subset of growth conditions, may be emblematic of a paradigm for much of fimbrial expression (22, 30). It is clear that E. coli O157:H7 strains often encode numerous putative fimbrial adhesins; however, many of these do not appear to be widely expressed in wild-type strains (12). In fact, electron micrograph analyses suggest that some EHEC strains do not synthesize any fimbriae when grown in LB, and therefore, alternate growth conditions are needed to induce expression (20). Overall, the current work strongly supports our previous reports indicating that the production of fimbriae in E. coli O157:H7 is tightly regulated (18, 26, 27). However, given that a variety of fimbriae and afimbrial surface structures can promote adhesion, it is possible that the absence of an adhesin often induces EHEC strains to synthesize an alternate structure. Such a compensatory mechanism has also been observed in uropathogenic E. coli (UPEC); strains that do not express type 1 and P fimbriae instead use F1C fimbriae to adhere to mammalian kidney cells (22). The possibility of cross talk playing a role in the regulation of fimbrial structures complicates genetic analyses of their roles, as one structure may compensate for the absence of another.
It remains to be determined why the double deletion of lpfA1 and lpfA2 alters the production of curli, at least in E. coli O157:H7 strain 86-24. However, the absence of Lpf is not sufficient to induce the production of curli, as CsgA production in the ΔlpfA1 ΔlpfA2 mutant grown at 30°C was not different from that of the wild type. Thus, it appears that other stimuli also contribute to the production of curli, i.e., after exposure to T84 cells. It is plausible to propose that curli are induced under cell culture conditions or that csgA is transcribed as detected (Fig. 4) but that curli are not produced due to posttranscriptional regulation. It would also be interesting to explore whether induction of curli via means other than lpfA deletion influences Lpf production, i.e., whether cross talk in the production of these fimbriae is bidirectional. Finally, additional studies to determine the factors and mechanisms that control their expression, the components within epithelial cells that are recognized by these adhesins, and the ways in which adhesion and adhesins change during the course of an intestinal infection are all warranted.
ACKNOWLEDGMENTS
The work in the A.G.T. laboratory was supported by NIH/NIAID grant 5-R01-AI079154-03. Work in the Waldor laboratory was supported by grant R37-AI-42347 and HHMI.
The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID or NIH.
Thanks go to Mauricio Farfan for technical assistance, Matt Chapman for kindly donating the CsgA antibody, Yizhou Zhou for technical advice, and Doug Botkin, Shane Massey, and Roberto Cieza for technical assistance and helpful discussions. We are grateful to Brigid Davis for many helpful suggestions on the manuscript.
Footnotes
Published ahead of print 9 January 2012
REFERENCES
- 1. Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. CDC 2009. Summary of notifiable diseases—United States, 2007. MMWR Morb. Mortal. Wkly. Rep. 56(53):1–94 [PubMed] [Google Scholar]
- 3. Chen CY, Nace GW, Irwin PL. 2003. A 6 × 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli. J. Microbiol. Methods 55:475–479 [DOI] [PubMed] [Google Scholar]
- 4. Doughty S, et al. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70:6761–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. European Food Safety Authority 2010. The community summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in the European Union in 2008. EFSA J. 8:1496 http://www.efsa.europa.eu/en/efsajournal/pub/1496.htm [Google Scholar]
- 6. Fitzhenry R, et al. 2006. Long polar fimbriae and tissue tropism in Escherichia coli O157:H7. Microbes Infect. 8:1741–1749 [DOI] [PubMed] [Google Scholar]
- 7. Jonas K, et al. 2007. Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol. 7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jordan DM, et al. 2004. Long polar fimbriae contribute to colonization by Escherichia coli O157:H7 in vivo. Infect. Immun. 72:6168–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kenny B, et al. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520 [DOI] [PubMed] [Google Scholar]
- 10. Kim SH, Kim YH. 2004. Escherichia coli O157:H7 adherence to HEp-2 cells is implicated with curli expression and outer membrane integrity. J. Vet. Sci. 5:119–124 [PubMed] [Google Scholar]
- 11. Leverton LQ, Kaper JB. 2005. Temporal expression of enteropathogenic Escherichia coli virulence genes in an in vitro model of infection. Infect. Immun. 73:1034–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Low AS, et al. 2006. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ. Microbiol. 8:1033–1047 [DOI] [PubMed] [Google Scholar]
- 13. Public Health Agency of Canada 2009. Canadian integrated surveillance report: Salmonella, Campylobacter, verotoxigenic E. coli and Shigella, from 2000 to 2004. Can. Commun. Dis. Rep. 35:1–50 [PubMed] [Google Scholar]
- 14. Ritchie JM, et al. 2008. EspFU, a type III-translocated effector of actin assembly, fosters epithelial association and late-stage intestinal colonization by E. coli O157:H7. Cell. Microbiol. 10:836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ritchie JM, Thorpe CM, Rogers AB, Waldor MK. 2003. Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect. Immun. 71:7129–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ritchie JM, Waldor MK. 2005. The locus of enterocyte effacement-encoded effector proteins all promote enterohemorrhagic Escherichia coli pathogenicity in infant rabbits. Infect. Immun. 73:1466–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rivas M, et al. 2008. Risk factors for sporadic Shiga toxin-producing Escherichia coli infections in children, Argentina. Emerg. Infect. Dis. 14:763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rojas-Lopez M, et al. 2011. Regulatory control of the Escherichia coli O157:H7 lpf1 operon by H-NS and Ler. J. Bacteriol. 193:1622–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sakuma M, Urashima M, Okabe N. 2006. Verocytotoxin-producing Escherichia coli, Japan, 1999–2004. Emerg. Infect. Dis. 12:323–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Samadder P, et al. 2009. The Escherichia coli ycbQRST operon encodes fimbriae with laminin-binding and epithelial cell adherence properties in Shiga-toxigenic E. coli O157:H7. Environ. Microbiol. 11:1815–1826 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Shamir ER, et al. 2010. Nitazoxanide inhibits biofilm production and hemagglutination by enteroaggregative Escherichia coli strains by blocking assembly of AafA fimbriae. Antimicrob. Agents Chemother. 54:1526–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Snyder JA, et al. 2005. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect. Immun. 73:7588–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tarr PI, et al. 1989. Genotypic variation in pathogenic Escherichia coli O157:H7 isolated from patients in Washington, 1984–1987. J. Infect. Dis. 159:344–347 [DOI] [PubMed] [Google Scholar]
- 24. Torres AG, et al. 2002. Identification and characterization of lpfABCC'DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:5416–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Torres AG, Kanack KJ, Tutt CB, Popov V, Kaper JB. 2004. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol. Lett. 238:333–344 [DOI] [PubMed] [Google Scholar]
- 26. Torres AG, et al. 2007. Environmental regulation and colonization attributes of the long polar fimbriae (LPF) of Escherichia coli O157:H7. Int. J. Med. Microbiol. 297:177–185 [DOI] [PubMed] [Google Scholar]
- 27. Torres AG, Slater TM, Patel SD, Popov VL, Arenas-Hernandez MMP. 2008. Contribution of the Ler- and H-NS-regulated long polar fimbriae of Escherichia coli O157:H7 during binding to tissue-cultured cells. Infect. Immun. 76:5062–5071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Torres AG, Zhou X, Kaper JB. 2005. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73:18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uhlich GA, Keen JE, Elder RO. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:2367–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xia Y, Gally D, Forsman-Semb K, Uhlin BE. 2000. Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the PapB protein. EMBO J. 19:1450–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]