Abstract
The Duffy binding protein (DBP) is a vital ligand for Plasmodium vivax blood-stage merozoite invasion, making the molecule an attractive vaccine candidate against vivax malaria. Similar to other blood-stage vaccine candidates, DBP allelic variation eliciting a strain-specific immunity may be a major challenge for development of a broadly effective vaccine against vivax malaria. To understand whether conserved epitopes can be the target of neutralizing anti-DBP inhibition, we generated a set of monoclonal antibodies to DBP and functionally analyzed their reactivity to a panel of allelic variants. Quantitative analysis by enzyme-linked immunosorbent assay (ELISA) determined that some monoclonal antibodies reacted strongly with epitopes conserved on all DBP variants tested, while reactivity of others was allele specific. Qualitative analysis characterized by anti-DBP functional inhibition using an in vitro erythrocyte binding inhibition assay indicated that there was no consistent correlation between the endpoint titers and functional inhibition. Some monoclonal antibodies were broadly inhibitory while inhibition of others varied significantly by target allele. These data demonstrate a potential for vaccine-elicited immunization to target conserved epitopes but optimization of DBP epitope target specificity and immunogenicity may be necessary for protection against diverse P. vivax strains.
INTRODUCTION
Plasmodium vivax is the most widely distributed human malaria parasite, responsible for about 50% of malaria cases outside Africa (21). Distinct from Plasmodium falciparum, blood-stage infections in vivax malaria are generally restricted to reticulocytes and to persons who are blood group positive for the Duffy antigen receptor for chemokines (DARC) (25, 33). Preference for these red blood cell types is determined by specific parasite ligands that mediate the merozoite invasion process. Recognition of DARC is mediated by the Duffy binding protein (DBP), a member of the Duffy binding-like erythrocyte binding protein (DBL-EBP) family, and is associated with the decisive and irreversible step of junction formation just before invasion (1). This vital event for host cell invasion (25) marks DBP as a prime target for vaccine-mediated immunity against blood-stage infections in vivax malaria.
DBP contains two highly conserved cysteine-rich domains (regions II and VI) that define sequence homology for members of the DBL-EBP family (2). Much work has been focused on DBP region II (DBPII), since it contains residues critical for receptor recognition (2, 8, 32). However, DBPII also contains most of the polymorphic residues that occur within the entire DBP, indicating an active immune selection mechanism used by the parasite to escape from inhibitory antibodies (11, 26, 30). In a recent study, we used naturally acquired antibodies that correlate with anti-DBP inhibition to identify B-cell epitopes within DBPII (10). The dominant B-cell epitopes identified were polymorphic surface-exposed motifs. These variant residues were previously determined to lie adjacent to residues functionally important for receptor recognition (32). Similar patterns of immune selection have been observed with other microbial adhesion molecules such as the influenza hemagglutinin (HA) (15, 35). Therefore, consistent with this immune escape paradigm for microbial pathogen ligands is a concern that variation found in the DBPII may lead to strain-specific immunity, thereby reducing the effectiveness of any anti-DBP immune response (6, 19, 31).
Studies from different populations have demonstrated that naturally acquired anti-DBP antibodies increase with exposure, and these antibodies can block DBP-erythrocyte binding and invasion of erythrocytes in vitro (7, 17, 22, 36). However, relatively few individuals respond with an anti-DBP response broadly inhibitory against multiple allelic variants (10, 19). These limitations pose a great challenge in developing DBP as an effective vaccine against vivax malaria. An effective vaccine for vivax malaria should be able to overcome the problems of immunogenicity and be broadly effective against the different alleles of the DBP. In order to address these issues, we produced a set of monoclonal antibodies against DBPII to determine if we could develop a high-titer inhibitory antibody broadly reactive to different alleles of the DBP. This study leads to a better understanding of the specificity needed for a protective immune response against DBP and designing an effective anti-DBP vaccine against vivax malaria.
MATERIALS AND METHODS
Production of recombinant DBPII.
DNA coding for DBPII was amplified by PCR from five different alleles of Plasmodium vivax DBPII (DBPII-SalI, DBPII-AH, DBPII-O, DBPII-7.18, and DBPII-P) (Table 1) present in different regions of endemicity (12). The amplified products were cloned into an expression vector (pET21a+) with a C-terminal histidine tag. The resulting plasmid (pET21a+-DBPII) was transformed into BL21(DE3) LysE Escherichia coli (Invitrogen). Cells were grown in LB medium in a bioreactor (New Brunswick), induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), collected by centrifugation, and stored at −80°C until needed. Recombinant DBPII was purified from inclusion bodies by standard methods (27, 29, 37), and the recombinant proteins were checked for purity by visualizing with SDS-PAGE. Eluted fractions containing enriched protein were then refolded by rapid dilution as previously described (27). The final product was concentrated to 1 mg/ml using the Amicon ultra centrifugal filter units (Millipore) and then stored at −80°C until needed. Denatured forms of the refolded recombinant proteins were generated as previously described (3, 14), dialyzed against phosphate-buffered saline (PBS), and stored as aliquots at −80°C.
Table 1.
Panel of DBPII alleles used for protein expression and COS7 cell assaya
| DBPII allele | Accession no. | Amino acid residue |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 308 | 319 | 333 | 369 | 371 | 375 | 384 | 385 | 386 | 390 | 417 | 424 | 437 | 447 | 454 | 467 | 492 | 503 | 511 | ||
| DBPII-SalI | P22290.2 | R | R | L | Y | K | N | D | E | K | R | N | L | W | S | Q | T | K | I | V |
| DBPII-O | AAY79072.1 | S | . | . | . | . | . | G | . | . | H | . | I | . | K | . | . | . | . | . |
| DBPII-27.16 | AAL79076 | S | . | . | . | . | . | G | . | . | H | . | I | . | K | . | . | E | . | . |
| DBPII-D | AAG53621 | S | . | . | . | . | . | G | . | . | H | . | I | . | K | . | P | . | . | . |
| DBPII-7.18 | AAL79051.1 | S | . | . | . | . | . | G | . | Q | . | K | I | R | . | . | . | . | K | . |
| DBPII-AH | AAY34130.1 | S | . | . | . | E | . | G | . | Q | . | K | I | R | . | . | . | . | K | . |
| DBPII-E | AAL79120.1 | S | K | . | . | E | . | G | . | Q | . | K | I | R | . | K | . | . | K | . |
| DBPII-C | AAL79116 | S | . | F | D | . | . | H | K | N | . | K | I | R | . | . | . | . | K | . |
| DBPII-F | AAC47185 | . | . | . | . | E | . | G | K | N | . | K | I | . | . | . | . | . | K | L |
| DBPII-P | AAL79073.1 | S | . | F | . | . | D | G | K | N | H | K | I | R | . | . | . | . | K | . |
Polymorphic residues within DBPII and positions with reference to DBPII-SalI (bold) are indicated. Conserved residues are represented by a dot (.).
Functional analysis.
Recombinant DBPII was tested for functional ligand activity using an in vitro direct erythrocyte binding assay as previously reported (5, 18, 27, 29, 34), with some modifications. Duffy-positive human erythrocytes were washed 3× in incomplete RPMI 1640 (iRPMI 1640) at 500 × g, 5 min. An aliquot of the cells was treated with chymotrypsin to render them Duffy negative by incubating 100 μl of packed cells with 1 ml of chymotrypsin (Sigma-Aldrich) at 1 mg/ml in iRPMI 1640 at 37°C for 1 h in a tube on a rotating wheel. Cells were washed with iRPMI 1640, incubated with 0.5 mg/ml of trypsin inhibitor (Sigma-Aldrich) for 10 min at 25°C, and washed again. One hundred microliters of either chymotrypsin-treated or untreated cells was incubated with 20 μg refolded recombinant DBPII for 4 h at room temperature with constant shaking. The reaction mixture was layered over 500 μl silicone oil (Dow Corning, Midland, MI) and centrifuged for 30 s at 500 × g. Bound recombinant protein was eluted from erythrocytes by resuspending the cell pellet in 10 μl of 1.5 M NaCl for 1 min, followed by 10 μl of 1 M NaCl for 1 min and then 20 μl of 0.3 M NaCl dropwise while shaking, and finally incubated for 10 min at 25°C with agitation every 2 to 3 min. The cells were centrifuged at 500 × g for 5 min, and the supernatant was mixed with SDS-PAGE load buffer and heated at 65°C for 3 min. The samples were separated on SDS-PAGE, transferred onto nitrocellulose membrane, and probed with an anti-DBPII monoclonal antibody (MAb), MAb-3D10, which from preliminary analysis was found to have the same binding specificity to all the recombinant proteins from the different alleles.
Monoclonal antibody production.
Monoclonal antibodies were commercially produced (AG Pharmaceuticals) in BALB/c mice by immunization with purified refolded recombinant DBPII from two alleles, SalI and 7.18. Anti-DBP-positive hybridoma clones were identified by enzyme-linked immunosorbent assay (ELISA) with the homologous antigens and secreted MAbs purified by protein G affinity chromatography. IgG subclasses were determined by an antibody isotyping kit (ThermoScientific, Rockford, IL) according to the manufacturer's instructions. The hybridoma cell lines from the 7.18 allele have been deposited in the MR4 collection as part of the BEI Resources Repository, NIAID, NIH.
Quantification of anti-DBP titer.
Refolded recombinant DBPII in PBS (pH 7.4) was adsorbed onto 96-well microtiter plates at 300 ng/well, incubated overnight at 4°C and washed with PBS-0.5% Tween 20, and unbound surfaces were blocked with 5% (wt/vol) skim milk in PBS-0.5% Tween 20 for 2 h at room temperature. Half-log serial dilutions of each monoclonal antibody in blocking buffer was added to triplicate wells, incubated for 2 h at room temperature, washed, and detected by goat anti-mouse alkaline phosphatase-conjugated antibody according to the manufacturer's protocol (KPL Inc., Gaithersburg, MD). To standardize the optical density (OD) values for plate-to-plate and day-to-day variations, MAb-3D10, which binds equally well with all the recombinant proteins, was used as a standard on each plate. MAb-1F9, a monoclonal antibody with specificity for Plasmodium falciparum AMA-1 (13), was used as a negative control. PBS-coated wells served as background control.
Western blot analysis.
Different variants of recombinant DBPII were separated by SDS-PAGE and electrophoretically transferred onto nitrocellulose membranes (Millipore). Membranes were rinsed in wash buffer (PBS-0.05% Tween 20) and blocked in 5% skimmed milk in wash buffer overnight at 4°C, rinsed in wash buffer, and reacted with 2.5 μg/ml monoclonal antibodies diluted in wash buffer for 2 h at room temperature. After three washes, membranes were incubated in horseradish peroxidase (HRP)-conjugated goat anti-mouse monoclonal antibody (KPL Inc., Gaithersburg, MD) at 0.5 μg/ml in wash buffer. Bound antibody was then detected with ECL substrate (GE Healthcare Life Sciences).
COS7 cell erythrocyte binding-inhibition assay.
An expression plasmid, pEGFP-N1 (where EGFP is enhanced green fluorescent protein) (Clontech), which encodes a red-shifted variant wild-type GFP, was used to target DBPII onto the surface of transfected COS7 cells as a fusion protein to the N terminus of EGFP as previously reported (9, 16, 23, 24). The plasmid constructs (pEGFP-DBPII) contained different alleles of P. vivax DBPII (Table 1). Rosettes (COS7 cells with adherent erythrocytes) were counted as positive when adherent erythrocytes covered ≥50% of the cell surface (20, 22, 32). To evaluate the ability of the MAbs to inhibit binding of erythrocytes to DBP expressed on the surface of transfected COS7 cells, various dilutions of antibodies were preincubated with the transfected COS7 cells for 1 h at 37°C before addition of human erythrocytes. Binding inhibition of each antibody was determined by assessing the percentage of rosettes in wells of transfected COS7 cells in the presence of antibody relative to rosettes in wells of transfected cells in the absence of antibody (% inhibition = 100 − [no. of rosettes in the presence of MAb/no. of rosettes in the absence of MAb] × 100). Differences in inhibitory responses of the MAbs to the different alleles were compared by one-way analysis of variance (ANOVA) and multiple comparison analysis by Tukey's test using SAS software.
RESULTS
Anti-DBPII monoclonal antibody production.
Refolded recombinant DBPII was produced to create and analyze anti-DBP monoclonal antibodies since previous studies demonstrated refolded proteins have the same functional ligand properties as the native DBP. Refolded recombinant DBPII migrates at the expected size of 37 kDa, which is noticeably smaller than nonrefolded or denatured DBPII migrating at 39 kDa, and the mobility shift of the refolded proteins can be used as a simple indicator of native conformation of recombinant DBPII (Fig. 1; see also Fig. S1a in the supplemental material). In vitro erythrocyte binding assays were used as quality control to validate functionality and determine if the refolded recombinant DBPII had native conformation necessary for erythrocyte receptor recognition. Refolded recombinant DBPII was detected binding to Duffy-positive erythrocytes, but negligible or no binding was evident with Duffy-negative erythrocytes (see Fig. S1b in the supplemental material). This binding pattern of the refolded recombinant DBPII was the same as the native DBP, confirming that the antigens were correctly refolded and functionally active. Purified, lipopolysaccharide (LPS)-free recombinant DBPII proteins expressed from alleles 7.18 and SalI were used for immunizations, and a total of 10 anti-DBPII reactive-positive hybridoma clones were selected from the two fusions: 7.18 (2A6, 3A4, 1D2, 2F12, 2C6, 2H2, 3C9, 2D10 represented by ATCC [BEI/MR4] numbers from MRA-967 to MRA-974, respectively) and SalI (3D10 and 3F11). IgG isotyping identified that all the antibodies were IgG1 isotype and carried a kappa light chain, with the exception of MAb-2C6 and MAb-1D2, which were IgG2b (Table 2).
Fig 1.
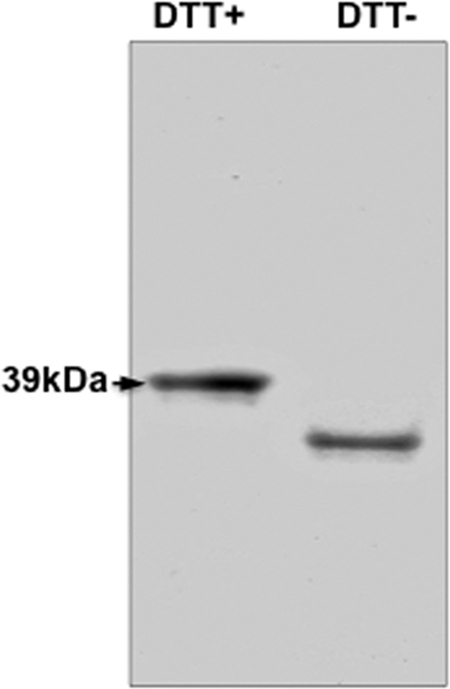
Characteristics of purified and refolded recombinant DBPII-7.18. Recombinant 7.18 was purified from inclusion bodies under denaturing conditions and refolded by rapid dilution. Refolded and reduced forms of the protein were separated on SDS-PAGE gel. Differential mobility of refolded antigen on gel in the presence (+) and absence (−) of dithiothreitol (DTT) indicates the presence of disulfide bonds, a simple indicator of native conformation in refolded antigen. See Fig. S1 in the supplemental material for characteristics of recombinant proteins from all the alleles.
Table 2.
Characteristics of anti-DBPII monoclonal antibodiesa
| Antibody | DBPII allele | IgG isotype |
ELISA titer (ng/ml) |
COS7 assay: IC50 (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H-chain | L-chain | DBPII-AH | DBPII-7.18 | DBPII-O | DBPII-P | DBPII-SalI | DBPII-7.18 | DBPII-SalI | ||
| MAb 3D10 | DBPII-SalI | IgG1 | κ | 2 | 2 | 2 | 2 | 2 | 32 | 25 |
| MAb 2D10 | DBPII-7.18 | IgG1 | κ | 5 | 2 | 2 | 2 | 17 | 0.35 | 0.18 |
| MAb 3C9 | DBPII-7.18 | IgG1 | κ | 50 | 5 | 50 | 17 | 17 | 0.19 | 0.16 |
| MAb 2C6 | DBPII-7.18 | IgG2b | κ | 50 | 17 | 17 | 17 | 17 | 2.25 | 0.75 |
| MAb 2H2 | DBPII-7.18 | IgG1 | κ | 50 | 17 | 17 | 17 | 17 | 0.53 | 1.5 |
| MAb 3A4 | DBPII-7.18 | IgG1 | κ | 50 | 5 | NR | 5 | NR | 24 | 24 |
| MAb 2A6 | DBPII-7.18 | IgG1 | κ | 17 | 17 | NR | 17 | NR | NI | NI |
| MAb 2F12 | DBPII-7.18 | IgG1 | κ | 50 | 5 | 50 | 17 | 17 | NI | NI |
| MAb 1D2 | DBPII-7.18 | IgG2b | κ | 167 | 50 | 50 | 167 | 167 | NI | NI |
| MAb 3F11 | DBPII-SalI | IgG1 | κ | >5,000 | >5,000 | >5,000 | >5,000 | >5,000 | ND | ND |
Monoclonal antibodies and corresponding homologous antigens are indicated. NI, noninhibitory; NR, not reactive; ND, not done; H, heavy; L, light.
Antibody reactivity profiles.
Endpoint ELISA titers revealed that all anti-DBPII monoclonal antibodies except for 3F11 and 1D2 were highly reactive (<20 ng/ml) with recombinant DBPII antigens used for the immunizations, 7.18 or SalI (Table 2; see also Fig. S2 in the supplemental material). To examine potential differences in the reactivity profiles of the monoclonal antibodies for DBPII allelic variants, we analyzed each anti-DBPII monoclonal antibody against refolded recombinant DBPII from three additional heterologous variant alleles genetically distant from SalI and 7.18 (11, 12). The differences in endpoint titers to the variant antigens ranged from 2 to 167 ng/ml (Table 2), except for 2A6 and 3A4, which did not react at all to SalI and O (see Fig. S2). Monoclonal 3F11 reacted poorly to all of the antigens tested and was not further characterized. To directly compare DBP specificity, the reactivity to each recombinant DBPII variant was calculated at a fixed concentration of each monoclonal antibody (Fig. 2). This quantitative analysis was a useful metric to confirm significant differences in antibody specificity.
Fig 2.

Quantitative comparison of anti-DBPII specificity. Binding specificity of each monoclonal antibody was quantified at a fixed concentration (50 ng/ml) to five different DBPII variants. Bars show relative reactivity (ELISA units) of each antibody against the respective DBPII variants, while error bars indicate ± standard deviations for triplicate wells. A P. falciparum apical membrane antigen 1 (PfAMA1)-specific monoclonal antibody, MAb-1F9, was used as a negative-control antibody.
Next, we analyzed by immunoblot analysis the reactivity of each monoclonal antibody to refolded versus denatured homologous antigens (Fig. 3). Except for 3D10, reactivity of all of the antibodies was reduction sensitive, indicating that their target epitopes were disulfide bond dependent. Similar to the ELISA results, the antibodies reacted with all refolded antigens except for monoclonal antibodies 2A6 and 3A4 that failed to recognize DBPII variants SalI and O. Since it was observed that 3D10 retained reactivity with the denatured antigens, ELISA analysis compared this monoclonal antibody's reactivity to refolded and denatured antigens. Reactivity of 3D10 was significantly less (<5-fold) with denatured versus refolded DBPII with native conformation (see Fig. S3 in the supplemental material). Since 3D10 was highly reactive with all the recombinant proteins, it was adopted as a reference standard on each experimental plate.
Fig 3.
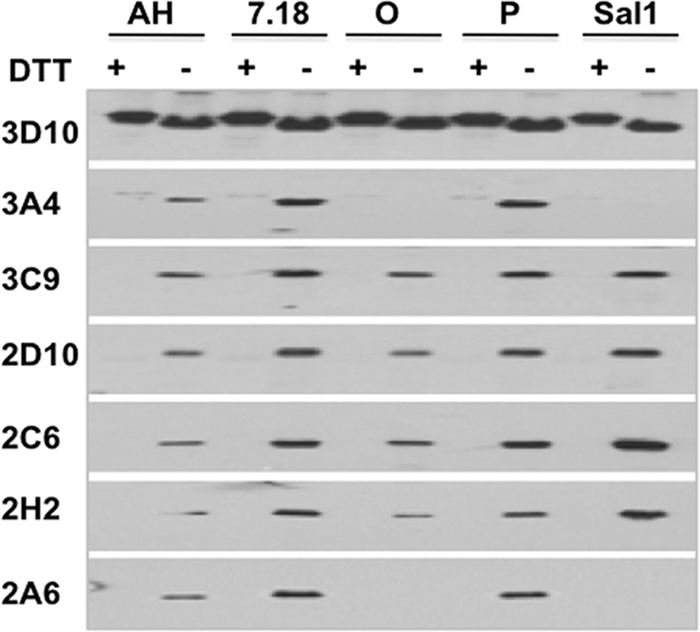
Immunoblot analysis. Fifty nanograms/lane of purified recombinant DBPII, reduced (+) and nonreduced (−) were separated on SDS-PAGE, electrophoretically transferred onto nitrocellulose membrane, and analyzed by Western blotting with the different anti-DBPII monoclonal antibodies at 2 μg/ml. All monoclonal antibodies, with the exception of 3D10, specifically recognized only the refolded antigens. Monoclonal antibodies 3A4 and 2A6 reacted with all antigens except SalI and O. The corresponding Coomassie blue-stained SDS-PAGE gel (2 μg/lane) is shown in Fig. S1.
Anti-DBP monoclonal antibodies inhibit DBPII-erythrocyte binding.
Functional activity of naturally acquired and vaccine-elicited anti-DBPII antibodies that block P. vivax merozoite invasion correlate with levels of inhibition in the in vitro COS7 assay of DBPII-erythrocyte binding (17). Given the limitations of directly measuring inhibition of P. vivax merozoite invasion, the COS7 DBPII-erythrocyte binding assay serves as a useful surrogate to evaluate potential invasion inhibitory effects of anti-DBPII antibodies. Limiting dilutions of each monoclonal antibody were tested in a range of concentrations to give 100% to 0% DBPII-erythrocyte binding inhibition against 7.18 and SalI, which represent the two variants used to produce the monoclonal antibodies. The most inhibitory monoclonal antibodies were 3C9, 2D10, 2C6, and 2H2 (see Fig. S4 in the supplemental material). DBPII binding to the erythrocytes was inhibited in a dose-dependent manner, and we were able to calculate a 50% inhibitory concentration (IC50) for each antibody as a quantitative measure for comparing anti-DBPII efficacy against each DBPII variant (Table 2). For example, 2H2 and 2C6 showed statistical significant differences in IC50s between the two alleles (P < 0.05), but the IC50 of 3C9 and 3A4 were the same for both DBPII SalI and 7.18. Using the respective homologous IC50 concentration (Table 2), each antibody was tested for functional inhibition of DBPII-erythrocyte binding against a panel of seven DBPII variants expressed in the COS7 assay. Functional inhibition by monoclonal antibodies 3D10, 3A4 (not shown), and 2H2 varied little (Fig. 4), indicating the target epitopes are highly conserved in the variants tested. In contrast, significant functional differences in inhibitory efficacy were evident for 3C9, 2C6, 2D10 (P < 0.005) (Fig. 4), and 2A6 (not shown) against the different alleles, indicating these monoclonal antibodies recognized variant epitopes. Interestingly, these significant differences included enhanced as well as reduced inhibitory effects against the heterologous alleles, mirroring differences in increased sensitivity and refractoriness observed with natural immune sera (31).
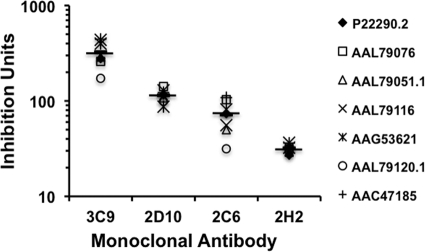
Fig 4.
Inhibition of DBPII binding to human erythrocytes. Monoclonal antibodies were tested for their ability to inhibit binding of COS7 cell-expressed DBPII alleles to Duffy-positive human erythrocytes. Transfected COS7 cells expressing either SalI or 7.18 were preincubated with monoclonal antibodies at different concentrations prior to addition of erythrocytes as described in Materials and Methods. The IC50 of each antibody against the two alleles was determined (see Fig. S4 in the supplemental material). Each monoclonal antibody was then tested at a concentration equal to the IC50 of the homologous antigen for inhibition of DBPII-erythrocyte binding to a panel of homologous and heterologous DBPII alleles (represented by GenBank accession numbers). Chart shows the inhibition ratios for the most inhibitory antibodies (expressed as inhibition units) against the various alleles. The mean inhibition ratios of each monoclonal antibody against all the alleles tested is represented by a horizontal line, and the various symbols above and below the means represent the variability of the inhibitory responses against the different alleles.
DISCUSSION
In P. vivax, DBP is associated with the decisive and irreversible step of junction formation during merozoite invasion and, unlike P. falciparum, there are not any obvious alternate ligand pathways (1, 10). The virtual absence of P. vivax in populations lacking the Duffy blood group presents overwhelming evidence for the vital nature of this interaction and marks DBP as an ideal target for vaccine development. Even though DBP's biological importance justifies its strong consideration as a vaccine candidate, the polymorphic nature of its ligand domain may pose a challenge of effectively developing DBP as a vaccine target against vivax malaria. Previous studies have shown that individuals can acquire robust naturally acquired immunity to P. vivax infections. However, relatively few individuals are capable of developing broadly inhibitory anti-DBP immune antibody responses. Therefore, similar to blood-stage vaccine candidates of P. falciparum, some concern is justified about whether vaccine-induced immunity will be strain specific and limit effectiveness against different alleles of the DBP. Based on this concern, one strategy is to identify conserved neutralizing epitopes that are suitable targets and optimize vaccine development toward those conserved epitopes while avoiding presenting immunogenic variant epitopes in a vaccine. The main objective of this study was to evaluate feasibility for vaccine-elicited immunization to potentially target conserved epitopes of DBP with highly inhibitory antibody desirable for protection against diverse P. vivax strains. Toward that goal, we successfully developed a set of monoclonal antibodies raised against the DBPII that are high titer and broadly inhibitory.
Expressed as a type I membrane protein, DBP is sequestered in the micronemes along with other ligands until required by merozoites for host cell invasion. This “just-in-time” release on the merozoite surface presumably limits exposure to inhibitory antibody targeting the N-terminal 330 amino acid cysteine-rich ligand domain (1). Structural analysis identified six disulfide bonds, which define three subdomains of region II, as critical for maintenance of the native conformation needed for erythrocyte receptor recognition in DBL domains (28). Comparative analysis of the deduced three-dimensional structure with site-directed mutation analysis reveals that many residues important for receptor recognition are surface exposed and lie adjacent to polymorphic residues responsible for DBP allelic variation (4). Together, these data are suggestive of a selection mechanism driven by antibody responses to nonessential variant epitopes adjacent to surface-exposed functionally sensitive areas of the ligand domain required for receptor recognition. Fortunately, this immune escape process does appear to have limits since some individuals exposed to P. vivax in countries where malaria is endemic are capable of producing broadly reactive invasion inhibitory antibodies. Focusing antibody responses on the epitopes recognized by these elite responders requires developing the reagents to validate and characterize the conserved neutralizing epitopes.
Our study developed a panel of murine monoclonal antibodies that reacted to conserved and variant epitopes. To determine the binding specificity of the various monoclonal antibodies to different DBPII alleles, we performed ELISA studies with five refolded recombinant variant DBPII alleles previously identified as genetically or antigenically distinct. The results demonstrated differential reactivity profiles among the antibodies for different DBP variants, confirming the existence of conserved epitopes as well as antigenic variability in the different DBP alleles, which has previously been suggested with naturally acquired human anti-DBP antibodies (10, 12). Reactivity of all monoclonal antibodies was reduction sensitive, indicating conformation of both types of epitopes were disulfide bond dependent. Only the weakly inhibitory monoclonal antibody 3D10 was able to recognize both denatured and refolded antigens, although there was >5-fold reduction in reactivity of the reduced antigens compared to the refolded antigens.
The COS7 in vitro binding assay was used to determine the efficacy of the anti-DBPII monoclonal antibodies to inhibit DBPII-erythrocyte interaction. As observed in the ELISA, some of the monoclonal antibodies (3C9, 2D10, 2C6, 2A6) showed significant differential inhibitory responses to the different alleles of the COS7-expressed DBPII, indicating that these antibodies might be binding to different epitopes within the different alleles. Monoclonal 3C9 showed the highest level of inhibition (IC50 = 0.19 and 0.16 μg/ml), followed by 2D10 (0.35 and 0.18 μg/ml), 2H2 (0.53 and 1.5 μg/ml), and 2C6 (2.25 and 0.75 μg/ml) for 7.18 and SalI, respectively. Interestingly, 2C6 and 2D10, which were produced by immunizing with 7.18, showed a higher anti-SalI inhibitory response compared to that of the homologous 7.18 allele (3-fold for 2C6 and ∼2-fold for 2D10). This is in line with previous studies, which observed that some changes in variant residues on DBPII enhanced sensitivity to heterologous anti-SalI antibodies and other amino acid changes in the same residues increased refractoriness to antibody inhibition (31). Similarly, some human immune antisera have been found to have an enhancing effect on DBP-erythrocyte binding (10). Whether it is by binding to the receptor site or by steric hindrance, the mechanism by which these monoclonal antibodies inhibit binding is yet to be determined. However, it is clear from our results that optimizing antibody specificity is critical for effective inhibition of DBP-erythrocyte binding and a high-titer antibody alone is not sufficient for protection.
The data presented here represent the first study to use monoclonal antibodies to evaluate binding specificity to different DBPII alleles and inhibition of erythrocyte binding to DBP. The development and characterization of antibody reagents, especially those capable of binding and inhibiting parasite-erythrocyte interaction and subsequent invasion, are critical and are powerful tools for identifying specific targets on the DBP ligand to inhibit invasion. Optimizing the design of DBP immunogenicity to target such conserved epitopes will be important for development of a broadly effective vaccine against P. vivax.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by the U.S. National Institutes of Health grant R01AI064478 (to J.H.A.) and Veteran's Affairs Research Service (C.L.K.).
We have no commercial or other association that poses a conflict of interest.
Footnotes
Published ahead of print 3 January 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Adams JH, et al. 1990. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell 63:141–153 [DOI] [PubMed] [Google Scholar]
- 2. Adams JH, et al. 1992. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. U. S. A. 89:7085–7089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anders RF, et al. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240–247 [DOI] [PubMed] [Google Scholar]
- 4. Batchelor JD, Zahm JA, Tolia NH. 2011. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat. Struct. Mol. Biol. 18:908–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Camus D, Hadley TJ. 1985. A Plasmodium falciparum antigen that binds to host erythrocytes and merozoites. Science 230:553–556 [DOI] [PubMed] [Google Scholar]
- 6. Ceravolo IP, et al. 2009. Naturally acquired inhibitory antibodies to Plasmodium vivax Duffy binding protein are short-lived and allele-specific following a single malaria infection. Clin. Exp. Immunol. 156:502–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ceravolo IP, et al. 2008. Inhibitory properties of the antibody response to Plasmodium vivax Duffy binding protein in an area with unstable malaria transmission. Scand. J. Immunol. 67:270–278 [DOI] [PubMed] [Google Scholar]
- 8. Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, Miller LH. 1996. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J. Exp. Med. 184:1531–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chitnis CE, Miller LH. 1994. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 180:497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chootong P, et al. 2010. Mapping epitopes of the Plasmodium vivax Duffy binding protein with naturally acquired inhibitory antibodies. Infect. Immun. 78:1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cole-Tobian J, King CL. 2003. Diversity and natural selection in Plasmodium vivax Duffy binding protein gene. Mol. Biochem. Parasitol. 127:121–132 [DOI] [PubMed] [Google Scholar]
- 12. Cole-Tobian JL, et al. 2009. Strain-specific Duffy binding protein antibodies correlate with protection against infection with homologous compared to heterologous Plasmodium vivax strains in Papua New Guinean children. Infect. Immun. 77:4009–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coley AM, et al. 2006. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect. Immun. 74:2628–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crewther PE, Matthew ML, Flegg RH, Anders RF. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fitch WM, Leiter JM, Li XQ, Palese P. 1991. Positive Darwinian evolution in human influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 88:4270–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fraser TS, Kappe SH, Narum DL, VanBuskirk KM, Adams JH. 2001. Erythrocyte-binding activity of Plasmodium yoelii apical membrane antigen-1 expressed on the surface of transfected COS-7 cells. Mol. Biochem. Parasitol. 117:49–59 [DOI] [PubMed] [Google Scholar]
- 17. Grimberg BT, et al. 2007. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med. 4:e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haynes JD, et al. 1988. Receptor-like specificity of a Plasmodium knowlesi malarial protein that binds to Duffy antigen ligands on erythrocytes. J. Exp. Med. 167:1873–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. King CL, et al. 2008. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc. Natl. Acad. Sci. U. S. A. 105:8363–8368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mayer DC, et al. 2004. Polymorphism in the Plasmodium falciparum erythrocyte-binding ligand JESEBL/EBA-181 alters its receptor specificity. Proc. Natl. Acad. Sci. U. S. A. 101:2518–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mendis K, Sina BJ, Marchesini P, Carter R. 2001. The neglected burden of Plasmodium vivax malaria. Am. J. Trop. Med. Hyg. 64:97–106 [DOI] [PubMed] [Google Scholar]
- 22. Michon P, Fraser T, Adams JH. 2000. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect. Immun. 68:3164–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michon P, et al. 2001. Duffy-null promoter heterozygosity reduces DARC expression and abrogates adhesion of the P. vivax ligand required for blood-stage infection. FEBS Lett. 495:111–114 [DOI] [PubMed] [Google Scholar]
- 24. Michon PA, Arevalo-Herrera M, Fraser T, Herrera S, Adams JH. 1998. Serologic responses to recombinant Plasmodium vivax Duffy binding protein in a Colombian village. Am. J. Trop. Med. Hyg. 59:597–599 [DOI] [PubMed] [Google Scholar]
- 25. Miller LH, Mason SJ, Clyde DF, McGinniss MH. 1976. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295:302–304 [DOI] [PubMed] [Google Scholar]
- 26. Ntumngia FB, et al. 2009. Genetic variation among Plasmodium vivax isolates adapted to non-human primates and the implication for vaccine development. Am. J. Trop. Med. Hyg. 80:218–227 [PMC free article] [PubMed] [Google Scholar]
- 27. Singh S, et al. 2001. Biochemical, biophysical, and functional characterization of bacterially expressed and refolded receptor binding domain of Plasmodium vivax Duffy-binding protein. J. Biol. Chem. 276:17111–17116 [DOI] [PubMed] [Google Scholar]
- 28. Tolia NH, Enemark EJ, Sim BK, Joshua-Tor L. 2005. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell 122:183–193 [DOI] [PubMed] [Google Scholar]
- 29. Tran TM, et al. 2005. Detection of a Plasmodium vivax erythrocyte binding protein by flow cytometry. Cytometry A 63:59–66 [DOI] [PubMed] [Google Scholar]
- 30. Tsuboi T, et al. 1994. Natural variation within the principal adhesion domain of the Plasmodium vivax Duffy binding protein. Infect. Immun. 62:5581–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. VanBuskirk KM, et al. 2004. Antigenic drift in the ligand domain of Plasmodium vivax Duffy binding protein confers resistance to inhibitory antibodies. J. Infect. Dis. 190:1556–1562 [DOI] [PubMed] [Google Scholar]
- 32. VanBuskirk KM, Sevova E, Adams JH. 2004. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc. Natl. Acad. Sci. U. S. A. 101:15754–15759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Welch SG, McGregor IA, Williams K. 1977. The Duffy blood group and malaria prevalence in Gambian West Africans. Trans. R. Soc. Trop. Med. Hyg. 71:295–296 [DOI] [PubMed] [Google Scholar]
- 34. Wickramarachchi T, Devi YS, Mohmmed A, Chauhan VS. 2008. Identification and characterization of a novel Plasmodium falciparum merozoite apical protein involved in erythrocyte binding and invasion. PLoS One 3:e1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilson IA, Cox NJ. 1990. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 8:737–771 [DOI] [PubMed] [Google Scholar]
- 36. Xainli J, et al. 2003. Epitope-specific humoral immunity to Plasmodium vivax Duffy binding protein. Infect. Immun. 71:2508–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yazdani SS, Shakri AR, Chitnis CE. 2004. A high cell density fermentation strategy to produce recombinant malarial antigen in E. coli. Biotechnol. Lett. 26:1891–1895 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.