Abstract
Live attenuated strains of Salmonella enterica have a high potential as carriers of recombinant vaccines. The type III secretion system (T3SS)-dependent translocation of S. enterica can be deployed for delivery of heterologous antigens to antigen-presenting cells. Here we investigated the efficacy of various effector proteins of the Salmonella pathogenicity island (SPI2)-encoded T3SS for the translocation of model antigens and elicitation of immune responses. The SPI2 T3SS effector proteins SifA, SteC, SseL, SseJ, and SseF share an endosomal membrane-associated subcellular localization after translocation. We observed that all effector proteins could be used to translocate fusion proteins with the model antigens ovalbumin and listeriolysin into the cytosol of host cells. Under in vitro conditions, fusion proteins with SseJ and SteC stimulated T-cell responses that were superior to those triggered by fusion proteins with SseF. However, in mice vaccinated with Salmonella carrier strains, only fusion proteins based on SseJ or SifA elicited potent T-cell responses. These data demonstrate that the selection of an optimal SPI2 effector protein for T3SS-mediated translocation is a critical parameter for the rational design of effective Salmonella-based recombinant vaccines.
INTRODUCTION
The use of live attenuated bacterial pathogens is a promising approach for the rational design of new recombinant vaccines. Several attenuated carrier strains have been deployed for expression and delivery of various viral, bacterial, or parasitic antigens for vaccination (16). Live attenuated strains of Salmonella that synthesize and secrete foreign antigens were developed as vaccines for a number of infectious diseases and cancer treatment (19). Moreover, the use of live attenuated Salmonella to deliver recombinant antigens to the immune system is an attractive strategy for the construction of multivalent vaccines (17). Salmonella-based vaccines provide a number of advantages over other antigen delivery strategies, including low cost of production, oral delivery, the absence of animal products, genetic stability, and safety (3). In addition, Salmonella vaccines delivering heterologous antigens elicit efficient immune responses via stimulation of both innate and adaptive immunity (3). The availability of S. enterica serovar Typhimurium for preclinical work in mouse models and favorable clinical experience with the live attenuated vaccines S. enterica serovar Typhi Ty21a, CVD908, and CVD909 (20) further facilitate the development of Salmonella-based vaccination approaches.
Salmonella enterica is a facultative intracellular pathogen that inhabits a unique membrane-bound host cell compartment, termed the Salmonella-containing vacuole (SCV) (reviewed in reference 12). Localization within the SCV prevents delivery of in vivo-expressed foreign proteins to the major histocompatibility complex (MHC) class I-restricted antigen presentation pathway and hinders the use of Salmonella as a vaccine carrier to induce specific CD8 T cells, which are crucial for protection against viruses, intracellular bacteria, and tumors.
The majority of Gram-negative pathogens deploy complex virulence factors termed type III secretion systems (T3SS). T3SS mediate distinct functions that include antiphagocytic and cytotoxic effects on host cells (Ysc/Yop system of Yersinia spp.), invasion of host cells (S. enterica SPI1 system, Shigella Mxi/Spa system), and intracellular pathogenesis (S. enterica SPI2 system, Chlamydia T3SS) (6). T3SS consist of at least 20 different subunits which enable these bacteria to translocate specific effectors directly into the host cell cytoplasm in order to exert a broad range of virulence functions. The T3SS assembles needle-like appendages, which share similarity with the flagellar basal body and some of its proteins, including those which form the core of the central channel and are highly conserved between the two systems (2). The T3SS apparatus, also referred to as the injectisome, spans the inner and outer membranes of the bacterial envelope and secretes translocon and effector proteins. Translocon proteins allow effector proteins access to the eukaryotic cells by forming pores in the host cell membrane and forming a connecting channel-like complex between the bacterium and the eukaryotic membrane. After translocation into eukaryotic host cells, effector proteins subvert various host cell functions and immunity, thereby promoting bacterial virulence (4).
Several important virulence factors of S. enterica are encoded by genes within Salmonella pathogenicity islands (SPI), and two important loci are termed SPI1 and SPI2 (9). SPI1 and SPI2 genes encode distinct T3SS that translocate bacterial effectors during different phases of pathogenesis (5). Salmonella translocates T3SS effector proteins into the host cell cytoplasm mediated by either the SPI1 T3SS from the extracellular stage and from within the SCV during the early stage after entry or the SPI2 T3SS from within the SCV at later stages during intracellular life (9).
Heterologous antigens can be synthesized by Salmonella as fusions with native or recombinant proteins. This approach was mainly used to direct the expression of the desired antigen to a particular location in the bacterial cell and increase the immunogenicity of foreign antigens by fusing them to proteins that could exert a carrier effect (15). Salmonella T3SS-mediated translocation was used for efficient delivery of heterologous antigens into the cytosol of antigen-presenting cells (APCs), leading to prominent CD8 T cell responses (19, 23). Salmonella T3SS-mediated translocation can be used for efficient delivery of heterologous antigen fusions to SPI2 effector proteins to the cytosol of APC, leading to prominent CD8 T cell priming in orally immunized mice (21). SPI2 effector proteins, especially those which are synthesized only when the Salmonella is inside host cells such as dendritic cells (DCs) or macrophages (1), have been thought to be promising candidates for carriers of antigens to the MHC class I pathway.
Our previous studies used the SPI2 T3SS effector protein SseF as a fusion partner for the translocation of heterologous antigens (11, 23). In addition to SseF, 20 or more effector proteins are translocated by the SPI2 T3SS. We hypothesized that the translocation efficacy of effector-antigen fusions might be directly correlated to the immune response triggered by the vaccine. In this study, we used genes for five membrane-associated SPI2 effectors under the control of the PsseA promoter for the generation of expression cassettes with model antigens. We examined the efficacy of antigen translocation into the cytosol of APCs and in vivo immunogenicity of fusion constructs with effector SseJ, SifA, SseL, or SteC in comparison with that of the previously described SseF effector. We observed that the highest immune response in vitro and in vivo was induced by constructs based on the effector SseJ.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella enterica serovar Typhimurium NCTC 12023 was used as the wild-type strain. The purD htrA double-deficient strain MvP728 (23) was used as the attenuated carrier strain. For the generation of recombinant plasmids, E. coli DH5α was used as the host. Plasmids were introduced into Salmonella strains by electroporation, and recombinant strains were cultured in medium containing 50 μg ml−1 carbenicillin in order to maintain the plasmids.
Generation of plasmids.
For generation of expression cassettes with gene fusions consisting of sseJ, sifA, or sseL plus ovalbumin (OVA) and the hemagglutinin (HA) tag under the control of the SPI2 promoter PsseA, a gene fragment comprising codons 196 to 386 of chicken OVA with the CD4 and CD8 epitopes was amplified by PCR using OVA-For-NaeI, OVA-HA-Rev-XbaI, and pOMP as a template, which was later digested with NaeI and XbaI. PsseJ::sseJ::hSurvivin (p3550), PsifA::sifA::hSurvivin (p3551), or PsseL::sseL::hSurvivin (p3552) was digested with EcoRV and XbaI. Digested OVA was ligated to the large fragment of the digested plasmids to obtain plasmids PsseJ::sseJ::OVA::HA (p3554), PsifA::sifA::OVA::HA, (p3556), and PsseL::sseL::OVA::HA (p3555), and these plasmids were used to amplify sseJ::OVA::HA, sifA::OVA::HA, or sseL::OVA::HA using the forward primers SseJ-For-EcoRI, SifA-For-Effector-EcoRI, and SseL-Effector-For-EcoRI and the reverse primer OVA-HA-Rev-XbaI. PCR products were digested later by EcoRI and XbaI. Plasmid p3342 harboring PsseA::sscB sseF::hSurvivin::HA (23) was digested with EcoRI and XbaI, and the large fragment was ligated to digested sseJ::OVA::HA, sifA::OVA::HA, or sseL::OVA::HA fragments, resulting in PsseA::sseJ::OVA::HA (p3631), PsseA::sifA::OVA::HA (p3632), or PsseA::sseL::OVA::HA (p3633). The plasmids obtained were confirmed by colony PCR, diagnostic digestion, and sequencing using T7-Seq and T3-Seq primers. The plasmids used in this study are listed in Table 1 and the sequences of oligonucleotides are indicated in Table 2.
Table 1.
Plasmids used in this study
| Plasmid | Relevant characteristics | Reference |
|---|---|---|
| pWSK29 | Low-copy-no. vector, Ampr | Lab stock |
| pOMP-OVA | lacZ::OVA | Lab stock |
| p2629 | pWSK29 PsseA sscB sseF::OVA::M45 | 11 |
| p2810 | pWSK29 PsseA sscB sseF::lisA51-363::HA | 11 |
| p3550 | pWSK29 PsseJ sseJ::hSurvivin | This study |
| p3551 | pWSK29 PsifA sifA::hSurvivin | This study |
| p3552 | pWSK29 PsseL sseL::hSurvivin | This study |
| p3553 | pWSK29 PsteC steC::hSurvivin | This study |
| p3554 | pWSK29 PsseJ sseJ::OVA::HA | This study |
| p3556 | pWSK29 PsifA sifA::OVA::HA | This study |
| p3555 | pWSK29 PsseL sseL::OVA::HA | This study |
| p3626 | pWSK29 PsteC steC::OVA::HA | This study |
| p3631 | pWSK29 PsseA sseJ::OVA::HA | This study |
| p3632 | pWSK29 PsseA sifA::OVA::HA | This study |
| p3633 | pWSK29 PsseA sseL::OVA::HA | This study |
| p3634 | pWSK29 PsseA steC::OVA::HA | This study |
| p3635 | pWSK29 PsseA sseJ::lisA51-363::HA | This study |
| p3636 | pWSK29 PsseA sifA::lisA51-363::HA | This study |
| p3637 | pWSK29 PsseA sseL::lisA51-363::HA | This study |
| p3638 | pWSK29 PsseA steC::lisA51-363::HA | This study |
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| SseJ-Rev-EcoRV | acggatatcttcagtggaataatgatgagc |
| SseJ-Pro-For-KpnI | tacggtacctcacataaaacactagcac |
| SseJ-For-EcoRI | ccggaattcgtaaggaggacactatgcc |
| SifA-Rev-EcoRV | acggatatcaaaacaacataaacagccgc |
| SifA-Pro-For-Kpn | tacggtacctcataagcgattaattgcgcaac |
| SifA –EcoRI-eff-For | ggcgaattcatttttactccagtataagtg |
| SseL-Rev-EcoRV | tatgatatcctggagactgtattcatatatttg |
| SseL-For-KpnI | attggtaccatcagacatatacccttc |
| SseL-effector-For-EcoRI | ggagaattccagagcaaaatgaatatatgtgt |
| SteC-Rev-NaeI | tatgccggctttttttaattcatcctttaatac |
| SteC-Rev-EcoRI | tatgaattctttttttaattcatcctttaatac |
| SteC-For-KpnI | attggtaccaaggttctgtaggaagcctg |
| SteC effector-For-EcoRI | ggagaattccagaggatgagacatatgccg |
| ProB-For-KpnI | ctaggtaccagaagagaacaacggcaag |
| ProB-Rev-EcoRI | cacgaattcacgataagataattaacgtgc |
| LisA-51-For-EcoRV | ctagatatcacgccaatcgaaaagaaac |
| LisA-363-HA-Rev-XbaI | gagtctagattaagcgtagtctgggacgtcgtatgggtagaggttgccgtcgatgatttg |
| Ova-For-NaeI | atagccggcgcaatgcctttcagagtgac |
| Ova595-For-EcoRI | ggagaattcgcaatgcctttcagagtgactgag |
| OVA-HA-Rev-XbaI | agatctagattaagcgtagtctgggacgtcgtatgggtaaggggaaacacatctgccaaag |
| T3-Seq | aattaaccctcactaaagg |
| T7-Seq | taatacgactcactataggg |
For generation of expression cassettes consisting of the gene fusion SteC::OVA::HA under the control of SPI2 promoter PsseA, the low-copy-number plasmid pWSK29 was digested with EcoRI and XbaI, and OVA was amplified using the primers OVA595-For-EcoRI and OVA-HA-Rev-XbaI and the plasmid pOMP as the template. Amplified OVA digested with EcoRI and XbaI was ligated to digested pWSK29, resulting in plasmid pWSK29::OVA::HA. The sseA promoter was amplified using the primers ProB-For-KpnI and ProB-Rev-EcoRI and the plasmid p3342 as the template. Plasmid pWSK29::OVA::HA was digested with KpnI and EcoRI and ligated to the amplified SseA promoter, which was digested with KpnI and EcoRI, yielding plasmid pWSK29::PsseA::OVA::HA. The gene for the effector protein SteC was amplified using SteC-Effector-For-EcoRI and SteC-Rev-EcoRI. pWSK29 PsseA::OVA::HA and amplified SteC were both digested by EcoRI and ligated, yielding PsseA::steC::OVA::HA (p3634). The resulting plasmid was confirmed by colony PCR for the right orientation and sequenced using T3-Seq and T7-Seq.
For generation of expression cassettes consisting of fusions of sseJ, sifA, sseL, or steC to a fragment of lisA and the HA tag under the control of SPI2 promoter PsseA, the fragments PsseA::sseJ, PsseA::sifA, PsseA::sseL, and PsseA::SteC were amplified using ProB-For-KpnI as the forward primer and SseJ-Rev-EcoRV, SifA-Rev-EcoRV, SseL-Rev-EcoRV, or SteC-Rev-NaeI as the reverse primer. PCR products were digested with KpnI and EcoRV or NaeI (Table 2). Plasmid p2810 harboring PsseA::sseF::lisA::HA (25) was digested with KpnI and EcoRV, and the large fragment was ligated to amplified digested PsseA::sseJ, PsseA::sifA, PsseA::sseL, or PsseA::steC, yielding plasmids PsseA::sseJ::lisA::HA (p3635), PsseA::sifA::lisA::HA (p3636), PsseA::sseL::lisA::HA (p3637), and PsseA::steC::lisA::HA (p3638). All the plasmids were confirmed with colony PCR, diagnostic digestion, and sequencing using T3-Seq and T7-Seq.
Analysis of synthesis and translocation of recombinant vaccine antigens.
In order to quantify the amounts of recombinant protein produced by the various strains, Western blots were analyzed using the Odyssey detection system, and signal levels for model antigens were normalized to the levels of the constitutively synthesized control protein DnaK.
For analyses of translocation, HeLa cells were infected with various Salmonella strains harboring plasmids for the expression of effector::lisA::HA or effector::OVA::HA under the control of PsseA. As a carrier strain, the double mutant strain MvP728 was used. At 16 h after infection, the cells were fixed and processed for immunostaining of Salmonella LPS (rabbit anti-Salmonella O1,4,5; Difco, BD) and the HA epitope tag (Roche) The cells were analyzed by microscopy using a Zeiss LSM700 confocal laser scanning microscope.
Dendritic cell infection.
The preparation and culture of bone marrow cells from C57BL/6 mice for generation of bone marrow-derived dendritic cells (BM-DC) was described previously (13). After 6 days of culture in RPMI 1640 medium (PAA, Colbe, Germany) containing 10% heat-inactivated fetal calf serum (Gibco BRL, Grand Island, NY) and granulocyte-macrophage colony-stimulating factor (GM-CSF) at 37°C in an atmosphere of 5% CO2, the BM-DC were suspended in small flasks for experiments at a density of 8 × 106 cells per flask and allowed to adhere for at least 12 h.
Bacterial strains were grown overnight in LB, and the optical density at 600 nm (OD600) of the cultures was adjusted to 0.2 in 1 ml of phosphate-buffered saline (PBS). Aliquots of this suspension were added to flasks in order to yield the multiplicities of infection (MOI) indicated in the legends to Fig. 3, 4, and 5. The flasks were centrifuged at 500 × g for 5 min to synchronize the infection and subsequently incubated at 37°C in an atmosphere of 5% CO2 for 30 min. Thereafter, the cells were washed three times with RPMI 1640 medium and incubated with RPMI 1640 medium containing 100 μg ml−1 gentamicin for 1 h, followed by RPMI 1640 medium containing 25 μg ml−1 gentamicin for the rest of the experiment.
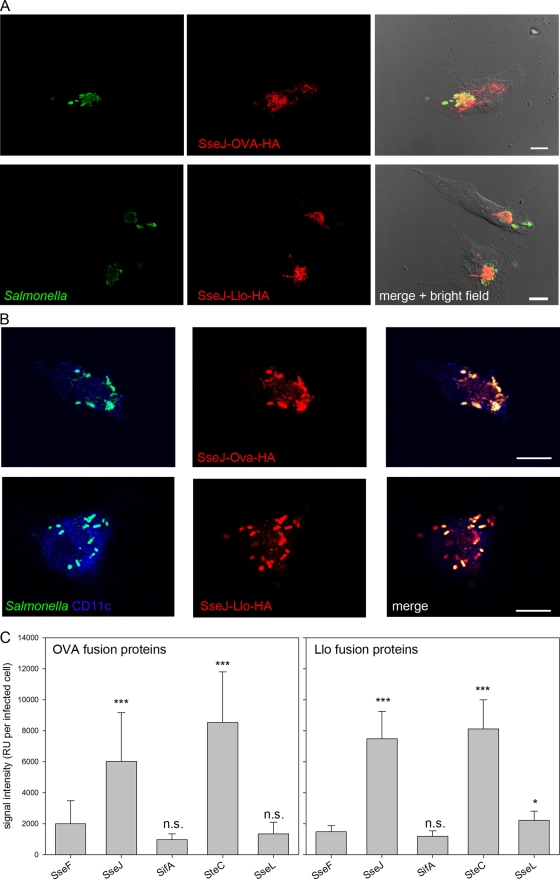
Fig 3.
Translocation of fusion proteins by intracellular Salmonella. Wild-type S. enterica serovar Typhimurium harboring plasmids with cassettes for the expression of fusion proteins consisting of the SPI2 effector protein SseJ, SifA, SseL, or SteC and the model antigen Llo with a C-terminal HA epitope tag under the control of PsseA were used to infect epithelial HeLa cells at an MOI of 10 or BM-DC at an MOI of 25. (A) HeLa cells were fixed 16 h after infection and stained for intracellular Salmonella (green) and translocated fusion protein SseJ-OVA-HA or SseJ-Llo-HA (red). (B) For BM-DC, the DC marker CD11c was labeled (blue) in addition to Salmonella (green) and translocated fusion proteins (red). Cells were analyzed by confocal laser scanning microscopy using the ZEN software package (Zeiss). Representative infected HeLa cells (A) and BM-DC (B) are shown. Scale bars correspond to 10 μm. (C) For quantification, the attenuated purD htrA carrier strain (MvP728) harboring plasmids with expression cassettes for fusion proteins consisting of SPI2 effector protein SseF, SseJ, SifA, SseL, or SteC plus OVA and the HA tag under the control of PsseA were used to infect HeLa cells at an MOI of 10. The cells were fixed 16 h after infection and processed for immunostaining. Infected cells with similar amounts of intracellular Salmonella were selected for the various conditions, and the signal intensities of the Cy3 channel for the anti-HA stain were measured with identical exposure times. The mean signal intensities per cell and standard deviations for at least 50 infected cells per condition are shown. n.s., not significant; ***, P < 0.001.
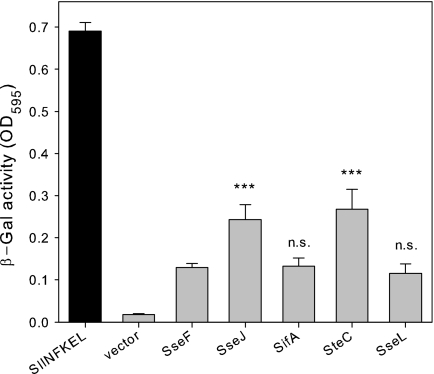
Fig 4.
Efficiency of SPI2 effector fusion proteins in stimulation of T cells. Murine BM-DC were infected at an MOI of 25 with the purD htrA-deficient carrier strain MvP728 harboring the empty plasmid vector or plasmids for expression of the indicated SPI2 effector proteins fused to OVA and the HA tag. As a positive control, BM-DC were infected with Salmonella and stimulated with the SIINFEKL peptide or infected with Salmonella only (vector). Infected BM-DC were incubated with B3Z reporter cells, and after coculture for 24 h, the β-galactosidase substrate chlorophenyl red β-galactopyranoside was added. After additional incubation for 6 h, the reaction was stopped, and the β-galactosidase product (β-gal) was quantified colorimetrically by measurement of extinction at 595 nm. T cell stimulation was analyzed at a ratio of infected BM-DC to T cells of 1:4. Means and standard deviations for triplicate samples are shown, and the data sets are representative of four independent experiments. n.s., not significant; ***, P < 0.001.
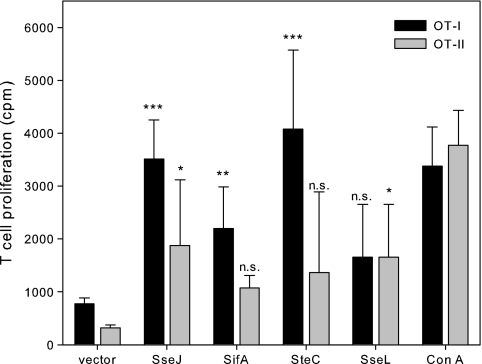
Fig 5.
Efficiency of SPI2 effector fusion proteins in stimulation of OT-I or OT-II cells. Murine BM-DC were infected as described for Fig. 4. Infected BM-DC were incubated with T cells obtained from the transgenic mouse strain OT-I or OT-II. T cell proliferation was quantified by measuring [3H]thymidine incorporation and expressed as counts per minute (cpm). As controls, T cells were stimulated with 1 μg ml−1 concanavalin A (ConA). The means and standard deviations for triplicate samples are shown, and the data sets are representative of three independent experiments. Statistical significance of stimulation by SseJ, SifA, SteC, or SseL fusion proteins compared to the vector control was determined by one-way ANOVA. n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
At 16 h after infection, the cells were fixed and processed for immunostaining of Salmonella LPS (rabbit anti-Salmonella O1,4,5; Difco, BD), the HA epitope tag (Roche), and the dendritic cell-specific marker CD11c (Armenian hamster anti-CD11c; BD). The cells were analyzed by microscopy using a Zeiss LSM700 confocal laser scanning microscope.
Quantification of T cell stimulation.
The SL-H2-Kb-specific murine CD8+ T cell hybridoma B3Z expresses the lacZ reporter gene under the control of the NFAT enhancer (13). The cell line was kindly provided by Nilabh Shastri at the University of California at Berkeley. Briefly, BM-DC from C57BL/6 mice at 105 per well in 96-well plates were infected with bacterial strains grown to stationary phase. Infection was performed for 1 h at MOI of 25 and 100 for wild-type and MvP728 strains, respectively. The plate was centrifuged for 5 min at 500 × g to synchronize the infection. After the infection period, noninternalized bacteria were removed by two washes with phosphate-buffered saline (PBS). To kill remaining extracellular bacteria, infected cells were incubated for 1 h in medium containing 100 μg ml−1 of gentamicin. After a washing step, medium containing 25 μg ml−1 gentamicin was added. B3Z T cells were added to the plates and cocultured at a DC:T cell ratio ranging from 1:8 to 1:0.125 in a total volume of 200 μl per well for 24 h. Cells were recovered by centrifugation at maximal speed and lysed by the addition of 100 μl substrate solution (0.15 mM chlorophenyl red β-galactopyranoside, 0.5% [vol/vol] Nonidet P-40 in PBS). After incubation for 6 to 8 h at 37°C, the absorbance was determined at 595 nm.
Stimulation of T cell proliferation was analyzed as described before (25). Briefly, BM-DCs from C57BL/6 mice were seeded at 105 per well in a 96-well plate. The cells were infected with bacterial strains at an MOI of 25. All experiments were performed in parallel for 1 h. The cells were washed twice with PBS and incubated with RPMI 1640 medium containing 100 μg ml−1 gentamicin for 1 h. After infection, plates were gamma-irradiated (3,600 rad) prior to coculture with 2 × 105 spleen cells from OT-I or OT-II mice (10) at a final ratio of 4:1. These mice express a transgenic T cell receptor (Vα2/Vβ5) specific for the OVA-derived peptides presented either in the MHC class I or MHC class II context. The transgene status was confirmed by fluorescence-activated cell sorting (FACS) with anti-mouse CD4 (clone RM4-5), anti-mouse CD8α (clone 5H10), and anti-mouse Vα2 (clone B20.1) from BD Biosciences. As a control, spleen cells were stimulated with 1 μg ml−1 concanavalin A (Sigma, Deisenhofen, Germany) without the addition of DC. After 48 h of coincubation, the proliferation was measured by 3H uptake for an additional 24 h. All experiments were performed in triplicate and at least three biological replicates were performed.
Vaccination experiments.
The immune responses to vaccination with live Salmonella carriers translocating fusion proteins of listeriolysin (Llo) to various effectors were analyzed as previously described (25). Briefly, BALB/c female mice 6 weeks of age (Jackson Laboratory) were maintained under standard conditions at the Baylor College of Medicine animal facility and were treated according to an IACUC-approved protocol. For immunization, cohorts of seven mice were immunized by orogastric application of 1 × 109 CFU per mouse of Salmonella MvP728 or MvP728 harboring plasmid p2810 (sseF::lisA), p3635 (sseJ::lisA), p3636 (sifA::lisA), p3637 (sseL::lisA), or p3638 (steC::lisA). The inocula were applied in 200 μl of 5% sodium bicarbonate using 20-gauge gastric gavage needles (Popper & Sons, Inc.). Booster immunizations were applied at days 14 and 28 after primary vaccination using the same conditions. The inoculum used was always verified by serial dilution and plating on LB agar plates in the presence of the appropriate antibiotic.
MHC pentamer staining and FACS analysis.
Blood samples were collected 10, 21, and 35 days after immunization from vaccinated mice or control groups. After erythrocyte lysis with ammonium chloride solution, cells were stained with a phycoerythrin (PE)-labeled H-2Kd/Llo91–99 (GYKDGNEYI)-specific Pro5 pentamer (ProImmune, Inc.), rat anti-mouse fluorescein isothiocyanate (FITC)-conjugated CD8 monoclonal antibody (MAb) KT15 (Proimmune, Inc.), allophycocyanin-conjugated hamster anti-mouse CD3e (BD Biosciences), and peridinin chlorophyll protein (PerCP)-conjugated rat anti-mouse CD45 (BD Biosciences). The fluorochrome and isotype-matching MAbs suggested by BD Biosciences were used as negative controls. The analysis was performed on an LSR-II flow cytometer (BD Biosciences) using BD FACDiva software, v. 6.0.
Statistical analyses.
Analyses for unpaired groups and the control group were performed by the one-way analysis of variance (ANOVA) with a Bonferroni t test using SigmaPlot 11.
RESULTS
Generation and evaluation of expression cassettes.
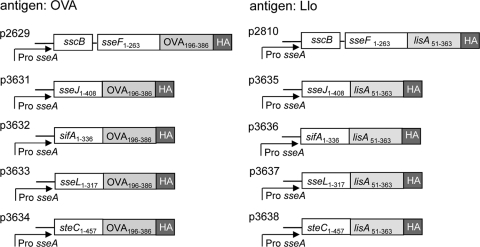
In order to test the efficacy of various SPI2 effector antigen fusions in vaccination approaches with live attenuated carrier strains, we generated expression cassettes containing the sseA promoter, a gene encoding a hybrid protein consisting of one of the SPI2 T3SS translocated effector proteins SseJ, SseL, SteC, and SifA, and portions of the model antigens ovalbumin (OVA196–386) and listeriolysin (Llo51–363). For either model antigen, the fragments encode CD4- and CD8-restricted epitopes, and a large panel of tools is available for the characterization of immune responses. The expression cassettes were placed on the low-copy-number plasmid pWSK29, which has previously been shown to be compatible with SsrAB-regulated in vivo expression (8, 11). The modular design of the various expression cassettes is presented in Fig. 1.
Fig 1.
Generation of expression cassettes for heterologous vaccine antigens. Expression cassettes consist of hybrid genes for the expression of fusion proteins comprising the full-length SPI2 effector protein SseJ, SifA, SseL, or SteC, portions of the model antigen Llo or OVA, and a C-terminal HA epitope tag for the standardized detection of the amounts of fusion protein. Expression cassettes with fusions to sseF were described previously (11). In all cassettes, the expression is controlled by the in vivo-activated promoter PsseA of the SsrAB regulon. All plasmids were generated on the basis of the low-copy-number vector pWSK29, and p3631, etc., are plasmid designations.
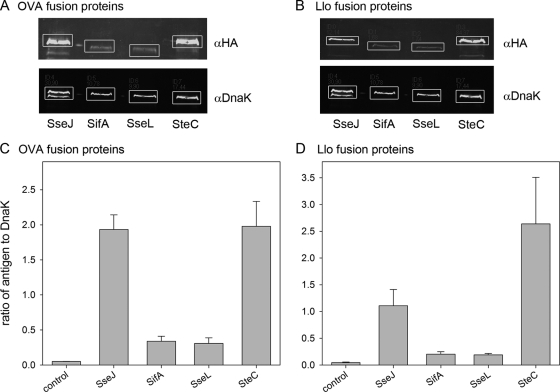
Next, strains harboring the expression cassettes were tested for the levels of recombinant fusion proteins. We first used in vitro culture conditions known to induce the expression of genes of the SsrAB regulon and the synthesis of SPI2 effector proteins. The synthesis of SPI2 effector-OVA-HA (Fig. 2A and C) and SPI2 effector-Llo-HA (Fig. 2B and D) fusion proteins was observed for all constructs analyzed here. In order to quantify the amounts of recombinant protein produced by the various strains, Western blots were analyzed using the Odyssey detection system, and signals for model antigens were normalized to the levels of the constitutively synthesized control protein DnaK. For both model antigens, SseJ and SteC fusions were expressed at significantly higher levels than SseL or SifA fusions. SteC fusions provided the highest level of recombinant-protein synthesis.
Fig 2.
Synthesis of fusion proteins with model antigens. The S. enterica serovar Typhimurium wild-type strain without plasmid (blank) or harboring plasmids encoding fusion proteins consisting of SPI2 effector protein SseJ, SifA, SseL, or SteC and model antigen OVA (A) or Llo (B), each with a C-terminal HA epitope tag. The strains were grown in SPI2-inducing minimal medium (phosphate, carbon, nitrogen minimal medium with 0.4 mM inorganic phosphate; pH 5.8), and bacteria were harvested after 6 h of culture under inducing conditions. Equal amounts of bacteria, adjusted by the OD600, were lysed and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analyses for the detection of the HA epitope tag was performed. Blots were probed with fluorescently labeled secondary antibodies, and signal intensities were quantified using the Odyssey system (Li-Cor). As a loading control, the cytosolic heat shock protein DnaK was detected on the same blot, and signals were quantified. Representative blots are shown. White boxes indicate the areas used for signal quantification. (C and D) The ratios of the HA to DnaK signals were calculated, and means and standard deviations for three samples are shown for OVA (C) and Llo (D) fusion proteins. The experiment was performed at least three times, and means and standard deviations of the three experiments are shown.
Since all fusion constructs were expressed by episomal genes, we tested plasmid stability in the absence of antibiotic selection and under conditions that mimic the intracellular environment of Salmonella in host cells (see Fig. S1 in the supplemental material). These experiments indicated that 80 to 90% of the bacterial cells maintained the plasmids over 7 days of repeated subculture in the absence of antibiotics. Similar numbers of plasmid-harboring bacteria were obtained by subculture in noninducing minimal medium without antibiotics, while 60 to 70% plasmid-harboring cells were obtained after subculture in SPI2-inducing minimal medium without antibiotics. Under these conditions, the greatest loss of plasmids was observed for fusion constructs with SseJ and SseF, while constructs based on SseL were maintained in more than 80% of the cells. These data suggest that the maintenance of expression cassettes is sufficiently stable for potential in vivo applications.
Next, we analyzed the translocation of the model antigens by intracellular Salmonella in mammalian cells. We investigated the translocation of OVA-HA (Fig. 3) and Llo-HA fusion proteins using expression cassettes with the PsseA promoter. All expression cassettes mediated translocation of fusion proteins into HeLa cells (Fig. 3A) and BM-DC (Fig. 3B). However, the intensities of the HA tag immunofluorescence staining of infected BM-DC varied considerably, indicating different amounts of the translocated protein (data not shown). In contrast, infected HeLa cells showed more uniform signal intensities and were used for quantification. Infected HeLa cells harboring similar numbers of intracellular Salmonella were selected, and the signal intensities of the HA tag of the examined constructs were compared. The SPI2 effectors were ranked in the following order depending on the level of the fusion protein expression: SteC > SseJ > SseF > SseL > SifA (Fig. 3C).
Quantification of T cell responses to antigens presented by intracellular Salmonella.
We next compared the antigen-dependent stimulation of T cells after uptake of recombinant Salmonella strains with expression of recombinant antigens under the control of PsseA. BM-DC were infected with the attenuated S. enterica serovar Typhimurium carrier strain MvP728, which is deficient in purD and htrA (21) and harbors plasmids for the expression of SPI2 effector::OVA::HA under the control of PsseA. Subsequently, infected BM-DC were incubated with B3Z T cells. B3Z is a T cell hybridoma that recognizes the OVA-derived SIINFEKL epitope in the context of H2Kb and expresses the lacZ reporter gene under the control of the NFAT enhancer. The β-galactosidase activity thus is a measure of the antigen-dependent stimulation (25). Very low stimulation was observed with the vector controls without expression cassettes, while addition of the SIINFEKL peptide was used as positive control, resulting in maximal stimulation. The use of the Salmonella purD htrA strain with plasmids harboring various expression cassettes resulted in highly increased stimulation of the T cell hybridoma. The highest stimulation was obtained with expression cassettes with SseJ and SteC fusion proteins (Fig. 4). These findings were compatible with the quantification of signals for protein translocation, which also showed that levels of SteC and SseJ fusion proteins were highest.
As a second approach, we used the OVA-specific T cells isolated from OT-I or OT-II transgenic mice (25). OT-I mice are transgenic for CD8+ T cells that recognize the OVA epitope SIINFEKL (OVA257–276) in the context of MHC class I molecules, while OT-II mice are transgenic for CD4+ cells that respond to the OVA323–339 peptide ISQAVHAAHAEINEAGR in the context of the MHC class II I-A2 molecule. BM-DC were infected with various combinations of carrier strains and expression cassettes, and the proliferation of OT-I- and OT-II-derived T cells was determined as a measure of antigen-dependent stimulation (Fig. 5). Compared to the control stimulation of T cells by concanavalin A, high levels of stimulation of OT-I cells were obtained with the Salmonella purD htrA carrier strain with the various expression cassettes for SPI2 effector::OVA::HA. In contrast, stimulation of OT-II cells was rather low under the various conditions investigated in this assay. This indicates that peptides derived from translocated fusion proteins are predominantly presented in a MHC class I-restricted manner. The highest stimulation was observed with strains harboring SteC and SseJ fusion proteins.
Fusion proteins with SseJ and SifA efficiently induce T cell responses after vaccination in mice.
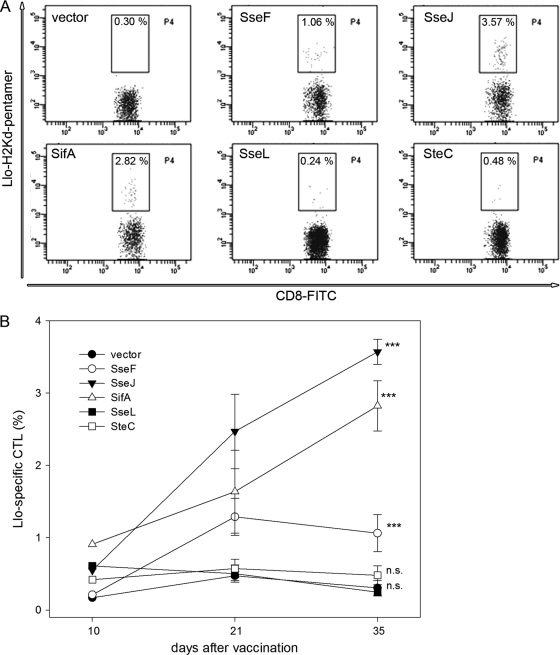
To examine the effect of SPI2 effectors on the in vivo immunogenicity of the recombinant vaccines, we used the purD htrA double-deficient strain MvP728, which harbored expression cassettes for the SPI2 effector-Llo fusion proteins under the control of PsseA. Mice were immunized by oral administration of the vector or vaccine strains followed by booster immunizations on days 14 and 28. In order to quantify the immune response to vaccination, blood samples were analyzed on days 10, 21, and 35 after the initial immunization. The elicitation of Listeria monocytogenes antigen-specific cytotoxic T lymphocytes (CTL) is a reliable correlate of protective immunity against L. monocytogenes infections in murine models (11, 14, 18, 25). The proportion of Llo-specific CTL was determined using specific pentamers (Fig. 6). As previously reported, the immunization with SseF-Llo fusion induced specific CTL responses (25). The quantification of Llo-specific CTL at the various time points indicted that strains translocating SteC-Llo or SseL-Llo fusion proteins did not trigger specific immune responses in mice. In contrast, immunization with strains translocating SseJ-Llo or SifA-Llo fusion proteins induced increased numbers of CTL compared with SseF-Llo. The highest CTL frequency was detected in the group immunized with the SseJ fusion protein. These results indicate that effector proteins SteC and SseL are suboptimal for the use in recombinant vaccines. In contrast, SseJ and SifA are superior to the previously characterized effector protein SseF and can be used for the translocation of heterologous antigens by Salmonella-based recombinant vaccines.
Fig 6.
Efficiency of SPI2 effector fusion proteins in vaccination. Cohorts of BALB/c mice were vaccinated with the purD htrA-deficient carrier strain MvP728 harboring plasmids for the synthesis of fusion proteins consisting of various SPI2 effector proteins and a listeriolysin fragment. Blood samples were collected at days 10, 21, and 35 after initial vaccination. The proportion of Llo-specific cytotoxic T cells (CTL) was determined by staining with the PE-labeled H-2Kd/Llo91–99-specific pentamer and flow cytometry. (A) Representative dot plots obtained on day 35 after initial vaccination. (B) Means and standard deviations of Llo-specific CTL counts from blood samples at days 10, 21, and 35 after vaccination. Statistical significance of stimulation by SseF, SseJ, SifA, SteC, or SseL fusion proteins is indicated. n.s., not significant; ***, P < 0.001.
DISCUSSION
Previous work from others and our labs has demonstrated the broad potential applicability of bacterial T3SS for the translocation of heterologous vaccine antigens by live attenuated bacterial carrier strains (19). Among multiple factors that contribute to the vaccine efficacy, the choice of a T3SS translocation signal or a T3SS effector protein as a fusion partner for the target antigens is of particular importance. Here, we compared the ability of five SPI2 effectors (SseJ, SifA, SteC, SseL, and SseF) to translocate model antigens in APCs and to elicit antigen-specific immune response in vivo. We observed that the amount of translocated protein, as well as the stimulation of T cell responses under in vitro conditions, correlated well with the translocation efficacy of the various effectors. However, the T cell responses triggered in a vaccination experiment were only in partial agreement with the effects observed in vitro. SseJ consistently demonstrated the strongest activities both in vitro and in vivo. While stimulation of T cell responses by fusion proteins based on SseJ was efficient in all assay systems applied here, SseL did not appear to be useful for the generation of the fusion proteins with heterologous antigens in the same settings. The results from in vitro and in vivo experiments were only in partial agreement for fusion proteins based on SteC or SifA. While fusion proteins with SteC were efficiently translocated and stimulated T cell responses in β-Gal- or OVA-responsive T cells, the stimulation of Llo-responsive T cells in vaccinated mice was not detectable.
The reasons for the different levels of immune stimulation observed for the studied effector proteins are still poorly understood. The level of synthesis of the effector proteins under intracellular conditions may vary considerably, but this feature is difficult to quantify for bacterial strains in the intracellular environment. Furthermore, the recognition of effector proteins by dedicated chaperones may vary, as well as the efficacy of translocation by the SPI2 T3SS. Other parameters that will likely affect the efficacy of translocated recombinant antigens are the subcellular localization of the translocated protein in APCs and the route of processing for presentation. The proteins of the subgroup of SPI2 T3SS effectors investigated in this study are all characterized by their association with endosomal membranes after translocation into host cells and their rather long half-life in host cells. Further work is required to test if SPI2 T3SS effectors that are predominantly localized in the cytosol and more rapidly degraded by the host confer different characteristics to vaccine fusion proteins.
Dendritic cells as professional APCs are the key link between innate and adaptive immunity. Immature DC can internalize and process S. enterica for peptide presentation on MHC class II as well as MHC class I, initiating an immune response (22). Interestingly, although S. enterica remains confined to vacuolar compartments, it can be processed for MHC class I presentation of bacterial antigens to CD8+ T cells (26). Because of the cytotoxicity of S. enterica to infected cells, DC can be either direct or indirect presenters of S. enterica antigens. Direct presentation of bacterial antigens to T cells follows phagocytic processing of S. enterica, which does not induce cytotoxicity. Indirect presentation of bacterial antigens to T cells is a consequence of uptake by professional APCs of antigenic material from neighboring cells that have undergone Salmonella-induced apoptotic death (22). Our results of the stimulation of OT-I and OT-II T cells indicate that translocation of all fusion constructs results in MHC class I- as well as MHC class II-restricted presentation. However, it is still unknown how SPI2 T3SS effectors differ in their subcellular localization and route of processing of heterologous antigens in DC. A recent study indicated that a subgroup of SPI2 T3SS effector proteins has DC-specific functions, such as interference with MHC class II presentation (7).
The expression levels of genes encoding SPI2 effector proteins are highly divergent, and this is likely to affect the amount of the translocated effector protein (24). By comparison of various promoters of the SsrAB regulon for expression of heterologous antigen fusions to SPI2 T3SS effector protein SseF, we recently identified PsifB as a promoter with superior stimulation of immune responses to vaccination (25). In this study, we compared various SPI2 effector proteins with high levels of synthesis for the translocation of fusion proteins. Here, the expression of the vaccine antigen fusions was controlled by PsseA, a promoter that has been utilized in our previous approaches (11, 23). The comparison of the effector proteins SifA, SseJ, SteC, and SseL to the previously deployed effector SseF clearly revealed high levels of stimulation of immune responses by SifA and SseJ. Ongoing studies in our lab are examining combinations of selected SPI2 promoters and effectors for optimal immunogenicity of the recombinant vaccines. In addition to the selection of promoters for the controlled in vivo expression of heterologous antigens and the SPI2 T3SS effector protein, other parameters may affect vaccination efficacy. Recent work showed that adaptation of the codon usage of foreign antigens to the codon usage of Salmonella as a carrier strain can strongly affect levels of protein synthesis and in turn the immune response to the antigen (23). Another important aspect is the selection of a suitable attenuated carrier strain. In the previous analysis and in this study, S. enterica serovar Typhimurium strain MvP728 harboring deletions of purD and htrA was used. Other attenuated mutant strains are yet to be tested for their compatibility with SPI2 T3SS-dependent translocation of heterologous antigens.
From results of this study we conclude that the choice of a translocated SPI2 effector is an important parameter for the construction of efficient recombinant vaccines. However, the in vitro efficacy of the various SPI2 effectors only partly correlated with the immunogenicity observed in vivo. This discordance may be explained by several factors. (i) Carrier strains with high levels of expression of heterologous antigens (e.g., with SteC effector) could be more rapidly eliminated in vivo, resulting in reduced immune responses. (ii) The levels of synthesis of certain effectors are different under the in vivo and in vitro conditions applied here. (iii) The subcellular location of the fusion protein under in vivo conditions is not favorable for efficient stimulation of immune response.
In conclusion, our analysis showed that the choice of SPI2 effector protein as a vehicle for intracellular delivery of a target antigen is a critical parameter for the optimal design of Salmonella-based recombinant vaccines. The in vivo vaccination efficacy of such SPI2 effectors did not completely match their in vitro performance. These results reflect the complex interplay between expression level, attenuation of live carrier strains, and properties of the recombinant antigen in vaccination of host organisms. Further mechanistic studies are required for the comprehensive evaluation of these critical parameters for the rational design of recombinant vaccines
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant HE1964 of the Deutsche Forschungsgemeinschaft to M.H., a fellowship of the DAAD to W.A.H.H., and grants from the National Institutes of Health (P50 CA126752) and The Caroline Wiess Law Foundation to L.M. We thank Andreas Goldwich (University Hospital Erlangen) for support with OT-I and OT-II cell preparation and analysis.
Footnotes
Published ahead of print 17 January 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Abrahams GL, Hensel M. 2006. Manipulating cellular transport and immune responses: dynamic interactions between intracellular Salmonella enterica and its host cells. Cell. Microbiol. 8:728–737 [DOI] [PubMed] [Google Scholar]
- 2. Aizawa SI. 2001. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 202:157–164 [DOI] [PubMed] [Google Scholar]
- 3. Cheminay C, Hensel M. 2008. Rational design of Salmonella recombinant vaccines. Int. J. Med. Microbiol. 298:87–98 [DOI] [PubMed] [Google Scholar]
- 4. Cornelis GR. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811–825 [DOI] [PubMed] [Google Scholar]
- 5. Gerlach RG, Hensel M. 2007. Salmonella pathogenicity islands in host specificity, host pathogen-interactions and antibiotics resistance of Salmonella enterica. Berl. Munch. Tierarztl. Wochenschr. 120:317–327 [PubMed] [Google Scholar]
- 6. Ghosh P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Halici S, Zenk SF, Jantsch J, Hensel M. 2008. Functional analysis of the Salmonella pathogenicity island 2-mediated inhibition of antigen presentation in dendritic cells. Infect. Immun. 76:4924–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hansen-Wester I, Stecher B, Hensel M. 2002. Type III secretion of Salmonella enterica serovar Typhimurium translocated effectors and SseFG. Infect. Immun. 70:1403–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53–66 [DOI] [PubMed] [Google Scholar]
- 10. Hogquist KA, et al. 1994. T cell receptor antagonist peptides induce positive selection. Cell 76:17–27 [DOI] [PubMed] [Google Scholar]
- 11. Husseiny MI, Wartha F, Hensel M. 2007. Recombinant vaccines based on translocated effector proteins of Salmonella pathogenicity island 2. Vaccine 25:185–193 [DOI] [PubMed] [Google Scholar]
- 12. Ibarra JA, Steele-Mortimer O. 2009. Salmonella—the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell. Microbiol. 11:1579–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karttunen J, Sanderson S, Shastri N. 1992. Detection of rare antigen-presenting cells by the lacZ T-cell activation assay suggests an expression cloning strategy for T-cell antigens. Proc. Natl. Acad. Sci. U. S. A. 89:6020–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lara-Tejero M, Pamer EG. 2004. T cell responses to Listeria monocytogenes. Curr. Opin. Microbiol. 7:45–50 [DOI] [PubMed] [Google Scholar]
- 15. Mastroeni P, Chabalgoity JA, Dunstan SJ, Maskell DJ, Dougan G. 2001. Salmonella: immune responses and vaccines. Vet. J. 161:132–164 [DOI] [PubMed] [Google Scholar]
- 16. Medina E, Guzman CA. 2001. Use of live bacterial vaccine vectors for antigen delivery: potential and limitations. Vaccine 19:1573–1580 [DOI] [PubMed] [Google Scholar]
- 17. Mollenkopf H, Dietrich G, Kaufmann SH. 2001. Intracellular bacteria as targets and carriers for vaccination. Biol. Chem. 382:521–532 [DOI] [PubMed] [Google Scholar]
- 18. Panthel K, et al. 2005. Salmonella pathogenicity island 2-mediated overexpression of chimeric SspH2 proteins for simultaneous induction of antigen-specific CD4 and CD8 T cells. Infect. Immun. 73:334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Panthel K, Meinel KM, Sevil Domenech VE, Trülzsch K, Rüssmann H. 2008. Salmonella type III-mediated heterologous antigen delivery: a versatile oral vaccination strategy to induce cellular immunity against infectious agents and tumors. Int. J. Med. Microbiol. 298:99–103 [DOI] [PubMed] [Google Scholar]
- 20. Pasetti MF, Simon JK, Sztein MB, Levine MM. 2011. Immunology of gut mucosal vaccines. Immunol. Rev. 239:125–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rüssmann H. 2003. Bacterial type III translocation: a unique mechanism for cytosolic display of heterologous antigens by attenuated Salmonella. Int. J. Med. Microbiol. 293:107–112 [DOI] [PubMed] [Google Scholar]
- 22. Sundquist M, Rydstrom A, Wick MJ. 2004. Immunity to Salmonella from a dendritic point of view. Cell. Microbiol. 6:1–11 [DOI] [PubMed] [Google Scholar]
- 23. Xiong G, et al. 2010. Novel cancer vaccine based on genes of Salmonella pathogenicity island 2. Int. J. Cancer 126:2622–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu X, Hensel M. 2010. Systematic analysis of the SsrAB virulon of Salmonella enterica. Infect. Immun. 78:49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu X, Husseiny MI, Goldwich A, Hensel M. 2010. Efficacy of intracellular activated promoters for generation of Salmonella-based vaccines. Infect. Immun. 78:4828–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yrlid U, Wick MJ. 2002. Antigen presentation capacity and cytokine production by murine splenic dendritic cell subsets upon Salmonella encounter. J. Immunol. 169:108–116 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.