Abstract
Chlamydiae are obligate intracellular pathogens replicating only inside the eukaryotic host. Here, we studied the effect of human flotillin-1 protein on Chlamydia pneumoniae growth in human line (HL) and A549 epithelial cell lines. RNA interference was applied to disrupt flotillin-1-mediated endocytosis. Host-associated bacteria were detected by quantitative PCR, and C. pneumoniae growth was evaluated by inclusion counts. C. pneumoniae attachment to host cells was unaffected, but bacterial intracellular growth was attenuated in the flotillin-1-silenced cells. By using confocal microscopy, we detected flotillin-1 colocalized with the inclusion membrane protein A (IncA) in the C. pneumoniae inclusion membranes. In addition, flotillin-1 was associated with IncA in detergent-resistant membrane microdomains (DRMs) in biochemical fractioning. These results suggest that flotillin-1 localizes to the C. pneumoniae inclusion membrane and plays an important role for intracellular growth of C. pneumoniae.
INTRODUCTION
Chlamydia pneumoniae, a Gram-negative obligate intracellular bacterium, is a common cause of community-acquired pneumonia (31). It has a tendency to cause persistent infections, which have been suggested to play a role in chronic vascular inflammation and atherosclerosis (12). The exact mechanism by which C. pneumoniae possibly contributes to the development of atherosclerotic lesions has remained elusive, however. Chlamydiae replicate only within eukaryotic cells, within a specialized parasitophorous vacuole termed an inclusion, which avoids fusion with host lysosomes and other endosomal compartments (20). How Chlamydiae form and maintain this privileged intracellular milieu is an important but poorly understood question. To date, only a few host proteins have been identified to be closely associated with Chlamydia inclusion (5, 14, 26, 42, 43, 47, 51, 54).
Flotillin-1 and flotillin-2 (also known as reggie-2 and reggie-1, respectively) are ubiquitously expressed proteins in eukaryotic cells that localize to lipid membranes, including the plasma membrane, Golgi complex, lipid droplets, multivesicular bodies, endosomes, phagosomes, and lysosomes (4, 9, 32, 45). Flotillins reside on the cytoplasmic side of the membrane by myristoyl and palmitoyl residues and a hydrophobic stretch of amino acids (9). They are enriched in sphingomyelin- and cholesterol-rich membrane microdomains (7), and it has been suggested that these proteins could have a structural function in these membrane domains (4, 32, 45, 57). Flotillins contribute in a cell-dependent fashion to a variety of cellular functions, including neuronal regeneration, insulin-dependent glucose uptake, epidermal growth factor signaling, T cell activation, endocytosis, and neutrophil migration (4, 9, 22, 32, 39, 45). Although the exact function of flotillins in these seemingly distinct processes has remained elusive, they are thought to be involved in a targeted transport of membrane proteins to distinct subcellular sites (58). Flotillin-1 has also been found in maturing phagosomes in macrophages infected with Leishmania donovani, Plasmodium falciparum, Coxiella burnetii, and Mycobacterium tuberculosis (17, 27, 34, 46), yet the function of flotillin-1 in these phagosomes has remains uncharacterized.
In this study, we have assessed whether flotillin-1 augments C. pneumoniae attachment and growth. In addition, we have evaluated the association of flotillin proteins with intracellular Chlamydia. The effect of flotillin-1 in C. pneumoniae attachment and growth was studied with a flotillin-1 RNA interference assay. Localization of flotillin proteins in Chlamydia-infected epithelial cells was studied by confocal microscopy. Cofractioning of chlamydial proteins with flotillin-1 in detergent-resistant microdomains (DRMs) was studied by using sucrose density gradient flotation analysis.
MATERIALS AND METHODS
Cell culture.
Experiments were carried out in human line (HL) cells (15, 30) or in A549 cells (ATCC; LGC Standards AB, Sweden), as indicated. HL, McCoy, and A549 cells were grown in Dulbecco modified Eagle medium (DMEM; Sigma), Glasgow minimum essential medium (BHK-21; Invitrogen), or F-12K nutrient mixture (Kaighn's modification; Invitrogen), respectively, supplemented with 10% fetal calf serum (FCS) and 20 μg ml−1 gentamicin and maintained at 37°C under 5% CO2.
Preparation of Chlamydia EBs.
C. pneumoniae isolate Kajaani 6 (K6) was originally obtained from P. Saikku, University of Oulu, Finland (18), and C. trachomatis L2 (VR-902B from ATCC) was propagated in HL cells and in McCoy cells. Chlamydia elementary bodies (EBs) were purified as described in reference 11. In brief, infected cells were harvested and ultrasonically disrupted, after which the cell debris was removed and the bacteria were purified by centrifugation in a meglumine diatrizoate gradient. Aliquots of the purified Chlamydia EBs were stored at −70°C in 0.25 M sucrose, 10 mM sodium phosphate, 4 mM potassium phosphate, 5 mM l-glutamic acid (pH 7.5) (SPG) until used. The infective titers of the preparations were determined in HL cell cultures.
Antibodies.
The following primary antibodies were used in the Western blotting experiments: anti-β-actin antibody (Sigma-Aldrich), anti-Rab5 antibody (BD Biosciences), and anti-transferrin receptor (Zymed). Antibodies against C. pneumoniae IncA (Cpn0186), CopN (Cpn0324), and MOMP (Cpn0695) were raised as described earlier (1). Rabbit antibody against C. trachomatis IncA was a kind gift from Ted Hackstadt (Rocky Mountain Laboratories; reference 25). Goat anti-mouse IgG horseradish peroxidase (HRP)-conjugated (SantaCruz Biotechnology) and goat anti-rabbit IgG HRP-conjugated (BD Transduction Laboratories) antibodies were used to recognize the primary antibodies. Rabbit anti-caveolin-1 (SantaCruz Biotechnology), goat anti-flotillin-1 (Abcam), and mouse anti-flotillin-2 (BD Transduction Laboratories) antibodies were purchased for confocal microscopy. The fluorophore-conjugated secondary antibodies were from Invitrogen. For inclusion counting, Chlamydia inclusions were detected with fluorescein-labeled anti-Chlamydia antibody (Bio-Rad, Hercules, CA).
Plasmids and transfection.
Flotillin-1–enhanced green fluorescent protein (EGFP) was originally constructed by Duncan Browman (10) and was kindly provided by Ritva Tikkanen (University of Giessen, Institute of Biochemistry). PEGFP-N1 vector was obtained from Clontech. The plasmids were transfected with Lipofectamine reagent (Invitrogen) 24 h prior to inoculation according to the manufacturer's instructions.
C. pneumoniae attachment and internalization assay.
To evaluate an effect of flotillin-1-dependent microdomains on C. pneumoniae attachment and internalization, the flotillin-1 small interfering RNA (siRNA) or nontargeting oligonucleotide-transfected A549 cells growing confluent in 24-well plates were inoculated with C. pneumoniae K6 at a multiplication of infection (MOI) of 0.1 in serum-free culture medium and centrifuged at 900 × g at 37°C for 1 h. Next, the cells were incubated at 37°C for 1 h and were extensively washed. Subsequently, the cells were incubated in culture medium supplemented with 0.5 μg ml−1 cycloheximide at 37°C for 2 h and were extensively washed. To detect internalized and attached bacteria, cells were resuspended in SPG buffer, and DNA was extracted using MPC nucleic acid isolation kit I and the automated MagnaPure compact instrument (Roche Diagnostics) according to the manufacturer's instructions.
C. pneumoniae growth assay.
To assay bacterial growth, the flotillin-1-silenced A549 cells or control-treated (Scramble) cells growing on glass coverslips on 24-well plates were inoculated with C. pneumoniae K6 at an MOI of 0.1, centrifuged at 900 × g for 1 h at room temperature, and incubated at 37°C for 1 h. The cells were washed and incubated in culture medium supplemented with 0.5 μg ml−1 cycloheximide at 37°C for 72 h. Finally, inclusions were detected by immunofluorescent microscopy.
RNA interference assays.
For flotillin-1 and caveolin-1 mRNA silencing, A549 and HL cells were plated on 24-well plates a day prior to transfections at 30% confluence. The following target sequence was used to design siRNA against flotillin-1: 5′-TAGTTTGTGCCTTGTCTTGAA-3′ (Sigma). This sequence has been targeted in flotillin-1 mRNA silencing in a previous study (22). Predesigned siRNA (Ambion) was used against caveolin-1. A nontargeting sequence, 5′-GCGCGCTTTGTAGGATTCG-3′ (44), was applied to design a negative control (Scramble) oligonucleotide (Sigma). Transfections were carried out with Lipofectamine (Invitrogen) according to the manufacturer's recommendations. For flotillin-1 silencing, cells were incubated at 37°C for 24 h, and the transfection was repeated, after which the cells were incubated at 37°C for 48 h. For caveolin-1 silencing, cells were transfected once with caveolin-1-targeted siRNA and incubated at 37°C for 48 h. Following this, RNA and protein samples were collected for flotillin-1 and caveolin-1 expression analysis, and cells were subjected for internalization and bacterial growth assays.
Quantitative reverse transcription-PCR (qRT-PCR).
RNA from siRNA-transfected cells was isolated at 72 h or 48 h after transfection, by using RNAeasy kit (Qiagen) or MagnaPure LC RNA isolation kit (Roche) according to the manufacturer's protocol. Subsequently, cDNA was generated from purified RNA by using Superscript II (Invitrogen) according to the manufacturer's instructions. The primers used for flotillin-1 (5′-TGCCAGAGAGTGTGGAAAGA-3′ and 5′-GGCTGTTCTCAAAGGCTTGT-3′) and hypoxanthine-guanine phosphoribosyltransferase-1 (HPRT-1) (5′-TGACCTTGATTTATTTTGCATACC-3′ and 5′-CGAGCAAGACGTTCAGTCCT-3′) for PCR were used in a final concentration of 3 μM. The probes (Universal ProbeLibrary probe no. 14 for flotillin-1 and no. 73 for HPRT1; Roche) were used in a final concentration of 1 μM. Caveolin-1 and beta-actin mRNA expression was evaluated by using TaqMan gene expression assay (Applied Biosystems). Reactions were run in a 7900HT real-time PCR (Applied Biosystems) with a thermocycler program of 15 min at 95°C, followed by 15 s at 95°C and 1 min at 60°C for 40 cycles. The threshold cycles (CT) were determined by using 7900HT sequence detection systems (version 2.3; Applied Biosystems). Flotillin-1 and caveolin-1 expression ratios were calculated by the ΔΔCT method (38) by using HPRT1 and beta-actin as internal controls, respectively. Flotillin-1 mRNA expression was expressed relative to the Scramble oligonucleotide-transfected samples.
Quantitative assay for Chlamydia genome equivalents.
The C. pneumoniae genomes were detected as published previously (40). Briefly, the sense primer, 5′-AAGGGCTATAAAGGCGTTGCT-3′, and the antisense primer, 5′-TGGTCGCAGACTTTGTTCCA-3′, amplified a 79-bp fragment of the major outer membrane protein gene (ompA) of C. pneumoniae. Primers were from Amersham Pharmacia Biotech, and the dually labeled (5′-6-carboxyfluorescein [FAM], 3′-6-carboxy-tetramethylrhodamine [TAMRA]) probe for ompA (5′-TCCCCTTGCCAACAGACGCTGG-3′) was synthesized by TAGC (Copenhagen, Denmark). A standard curve was prepared using 10-fold serial dilutions of the ompA plasmid (40). Amplification and detection were done simultaneously in an Applied Biosystems 7500 real-time PCR system according to the manufacturer's instructions. Student's t test was applied to calculate statistical significances from 3 independent assays with triplicate measurements.
Confocal microscopy.
To examine localization of flotillin-1 and flotillin-2 in C. pneumoniae K6- and C. trachomatis L2-infected cells, the cells were fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature, after which cells were permeabilized and blocked with 0.05% saponin (Sigma) in 1% bovine serum albumin (BSA) (Sigma) in PBS for 20 min at room temperature. Cells were rinsed with BSA-PBS and stained with primary antibodies for 2 h and with secondary antibodies for 1 h at room temperature. Bacterial and host DNA were stained with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen), and samples were mounted with Prolong antifade mounting medium (Invitrogen). Confocal images were produced with an LSM510 meta confocal microscope (Zeiss) using <0.1-μm optical slices. To prepare confocal images for publication, signals were gamma corrected, and brightness and contrast were adjusted to a whole image area by using ImageJ software (rsbweb.nih.gov/ij/).
Immunofluorescence microscopy.
To assay growth of C. pneumoniae, the infected cells were rinsed twice with PBS and fixed with methanol for 20 min at 72 h postinfection (hpi). Chlamydia inclusions were stained with fluorescein-labeled anti-Chlamydia antibody for 30 min at 37°C and mounted with ImmuMount (Thermo Shandon). Samples were examined with a fluorescence microscope (Leica DMRB, Germany) with standard filters at ×400 magnification. The mean inclusion size was estimated by measuring the diameter of the 30 to 40 inclusions in the immunofluorescence images with using ImageJ software. The number of inclusions was counted in 30 different ×400 fields per coverslip in 3 parallel samples. Data from 3 independent experiments were combined to calculate the mean and standard deviation.
Triton X-100 extraction, sucrose flotation, and fractionation.
To extract detergent-resistant membranes from infected cells, confluent HL cells (50 ×106) growing in 250-ml cell culture bottles were resuspended in serum-free DMEM and were inoculated with C. pneumoniae K6 at an MOI of 1. The cells were centrifuged at 900 × g at room temperature for 1 h and incubated at 37°C for 2 h. Then the cells were washed twice and resuspended in culture medium supplemented with 0.5 μg ml−1 cycloheximide and incubated at 37°C for 48 h. The infected cells were detached with trypsin treatment and washed with ice-cold HEPES-buffered saline (25 mM HEPES, 150 mM NaCl, 1 mM EDTA, pH 7.4) containing protease inhibitor cocktail (Roche). The cells were kept on ice and lyzed with 1 ml of ice-cold 1% Triton X-100 in HEPES-buffered saline for 30 min. Next, the cells were homogenized by passaging 10 times through a 25-guage needle. The cell lysate was adjusted to ca. 40% (wt vol−1) sucrose by adding an equal volume of 85% (wt vol−1) sucrose in HEPES buffer, placed at the bottom of an SW40 centrifuge tube, and layered with 6 ml of 35% and 6 ml of 5% (wt vol−1) sucrose in HEPES buffer. After centrifugation in an SW40 rotor (Beckmann Coulter) at 250,000 × g at 4°C for 18 h, 1-ml fractions were collected from the top of the gradient.
Protein precipitation and immunoblotting.
Proteins from equal volumes of sucrose gradient fractions were precipitated by adding deoxycholate and trichloroacetic acid to 0.02% and 10% final concentrations, respectively, and incubating for 1 h on ice. The precipitates were washed with acetone, the pH was adjusted to alkaline using sodium hydroxide, and the samples were then resuspended in sodium dodecyl sulfate (SDS) sample buffer. For immunoblot analysis, protein samples in the SDS sample buffer were boiled for 10 min and separated using SDS-polyacrylamide gel electrophoresis. The blotted proteins were probed with primary antibodies, followed by HRP-conjugated secondary antibodies. The ganglioside M1 (GM1) was detected by dot-blotting aliquots from each fraction on nitrocellulose membrane (Amersham Pharmacia Biotech) using cholera toxin β-conjugated horseradish peroxidase (Sigma-Aldrich). Signals were detected using an enhanced chemiluminescence (ECL) Western blotting kit (Amersham Pharmacia Biotech). Flotillin-1 protein expression was analyzed from exposed films by using the MCID-M5+ program (version 4.0; Imaging Research Inc.).
RESULTS
Flotillin-1 mRNA silencing does not affect C. pneumoniae association to the host.
Given that recent studies have shown that flotillin-1 and flotillin-2 mediate distinct lipid raft-dependent and clathrin-independent endocytosis (2, 22), we wanted to examine whether flotillin-1 membrane microdomains are important in C. pneumoniae association to the host. To study this, A549 cells were serially transfected with siRNA oligonucleotides targeted against flotillin-1 (siFLO-1), and the degrees of flotillin-1 mRNA and protein downregulation were examined. Flotillin-1 protein expression was analyzed by immunoblotting, and siFLO-1-transfected cells revealed a greater than 90% reduction in flotillin-1 protein expression compared to that of the control cells transfected with nontargeting (scramble) oligonucleotides (Fig. 1A). This result was in concordance with the flotillin-1 mRNA expression, as judged by qRT-PCR analysis (data not shown). The cells were infected and the genomes of host cell-associated C. pneumoniae were measured at 4 hpi by using qPCR to analyze bacterial genomes. An average of 5 × 105 bacterial genomes were quantified in the control-treated cells. There were no significant changes in host cell-associated bacterial genomes in the siFLO-1-transfected cells compared to those in the scramble-transfected cells (Fig. 1B), implying that C. pneumoniae attaches to the host independent of flotillin-1.
Fig 1.
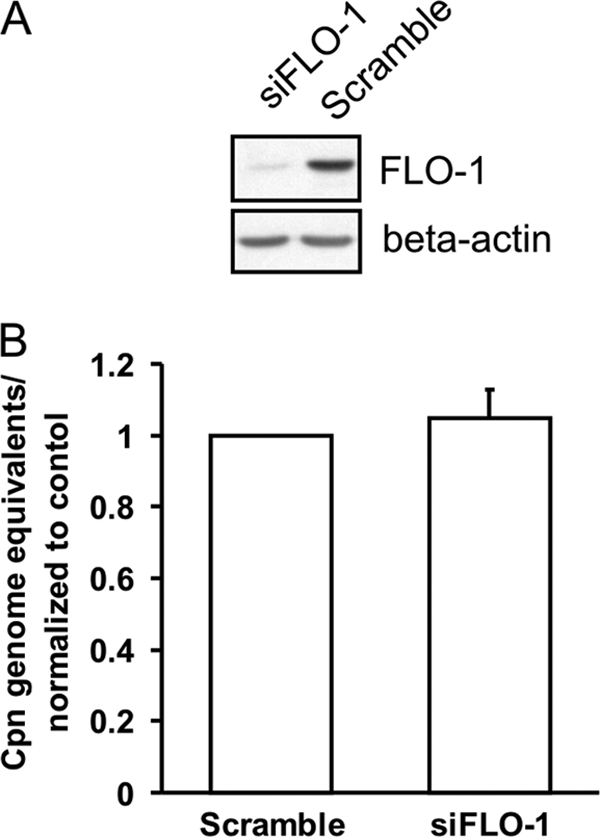
Flotillin-1 RNA silencing does not affect C. pneumoniae host cell association. A549 cells were transfected with siRNA directed against flotillin-1 (siFLO-1) or with nontargeting oligonucleotide (Scramble) for 3 days and inoculated with C. pneumoniae. (A) Representative immunoblot of siFLO-1- and Scramble-transfected A549 cells with flotillin-1 (FLO-1) and beta-actin-specific antibodies. (B) Infected cells were collected and Chlamydia genome equivalents were assayed by qPCR. Chlamydia genome expression was normalized to Scramble-treated cells. Error bars denote the standard errors of the means from 3 independent experiments done in triplicate.
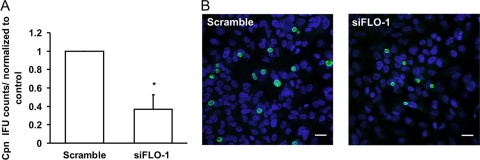
Flotillin-1 mRNA silencing attenuates intracellular growth of C. pneumoniae.
To evaluate the role of flotillin-1 in C. pneumoniae growth, flotillin-1 expression was silenced and bacterial growth was assayed by counting inclusions in the infected A549 cells. An average of 3,600 bacterial inclusions were counted from the coverslips of control-treated cells. The C. pneumoniae growth in the flotillin-1 siRNA-treated (siFLO-1) cells was reduced to approximately 37% compared to that of the control (Scramble) cells at 72 hpi (Fig. 2A). The representative images of siFLO-1 and Scramble-transfected cells infected with C. pneumoniae are shown in Fig. 2B. When the inclusion size was measured from immunofluorescence images, there was no significant difference in the mean inclusion size between the flotillin-1-silenced cells (7.89 ± 1.65 μm) and control-treated cells (7.66 ± 1.47 μm). As flotillin-1 has been reported to stabilize caveolin-1 in epithelial cells (61), we also tested whether caveolin-1 silencing affects Chlamydia growth. Bacterial growth in caveolin-1-siRNA-treated cells was comparable to that in control-treated cells (data not shown), indicating that the suppressive effect of flotillin-1 silencing on Chlamydia growth was not mediated by caveolin-1 depletion. The results suggest an important role for flotillin-1 in the intracellular growth of C. pneumoniae.
Fig 2.
C. pneumoniae growth is attenuated in flotillin-1 mRNA-silenced cells. (A) A549 cells were transfected with siRNA directed against flotillin-1 (siFLO-1) or with nontargeting Scramble siRNA for 3 days and inoculated with C. pneumoniae. Inclusions were stained and counted 48 hpi. Inclusion counts were normalized to Scramble-treated cells. Error bars denote the standard errors of the means of 3 independent experiments done in triplicate. Statistically significant difference is denoted by an asterisk, which indicates P values of <0.05. (B) Representative images of siFLO-1- and Scramble-transfected cells infected with C. pneumoniae at 72 hpi. Inclusions were detected with an antibody specific for IncA and anti-rabbit Alexa Fluor 488-conjugated secondary antibody and are shown in green. Bacterial and host DNA were stained with DAPI and are shown in blue. Bar, 20 μm.
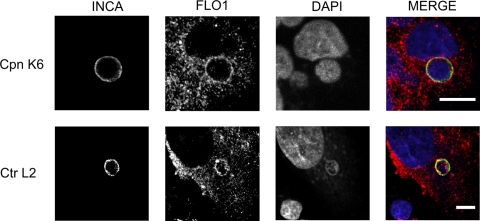
Flotillin-1 colocalizes with Chlamydia inclusions.
Because flotillin-1 was important for C. pneumoniae infection, we analyzed the subcellular localization of flotillin-1 in C. pneumoniae K6- and C. trachomatis L2-infected A549 cells by confocal microscopy at 48 and 24 hpi, respectively. Flotillin-1 colocalized with the inclusion membranes of both C. pneumoniae and C. trachomatis, which were detected with a specific antibody recognizing the inclusion membrane protein IncA of C. pneumoniae and C. trachomatis (Fig. 3).
Fig 3.
Flotillin-1 localizes to Chlamydia inclusions. A549 cells were inoculated with C. pneumoniae K6 (Cpn K6) or C. trachomatis L2 (Ctr L2), and endogenous flotillin-1 was detected at 72 and 48 hpi, respectively, with an antibody against flotillin-1 and anti-goat Alexa Fluor 647-conjugated secondary antibody. Inclusions were detected with antibodies specific for C. pneumoniae IncA (top; Cpn K6) or for C. trachomatis IncA (bottom; Ctr L2) and anti-rabbit Alexa Fluor 488-conjugated secondary antibody. In the merged image, IncA (INCA) is shown in green and flotillin-1 (FLO1) in red. Bacterial and host DNA were stained with DAPI and are shown in blue. Bar, 10 μm.
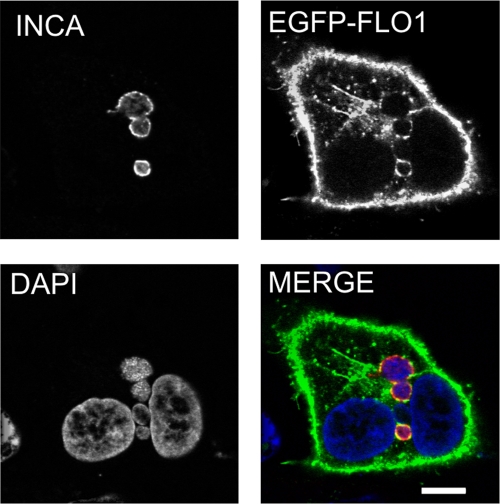
EGFP–flotillin-1 localizes to C. pneumoniae inclusions.
In order to confirm the localization of flotillin-1 in Chlamydia-infected cells, A549 cells were transfected with EGFP–flotillin-1 for 24 h, infected with C. pneumoniae for 48 h, and labeled with antibody specific for IncA. Confocal microscopy analysis revealed that EGFP–flotillin-1 colocalized with inclusion membranes labeled indirectly with anti-IncA (Fig. 4). Such an association was not observed in Chlamydia-infected cells transfected with the pEGFP empty vector (data not shown). In addition, we investigated whether another lipid raft-associated protein, caveolin-1, localized to C. pneumoniae inclusion, but the inclusions were negative for caveolin-1 staining (data not shown). Therefore, flotillin-1 localization to inclusion seems to be specific and unrelated to lipid raft-dependent internalization. Together with the results from the RNA interference assays, these findings suggest that flotillin-1 contributes to bacterial intracellular growth by direct interaction with Chlamydia pathogen.
Fig 4.
EGFP-conjugated flotillin-1 localizes to C. pneumoniae inclusions. A549 cells were transfected with EGFP–flotillin-1 for 24 h and inoculated with C. pneumoniae for 48 h. Inclusions were detected with an antibody specific against IncA and anti-rabbit Alexa Fluor 647-conjugated secondary antibody. In the merged image, EGFP–flotillin-1 (EGFP-FLO1) is shown in green and IncA (INCA) in red. Bacterial and host DNA were stained with DAPI and are shown in blue. Bar, 10 μm.
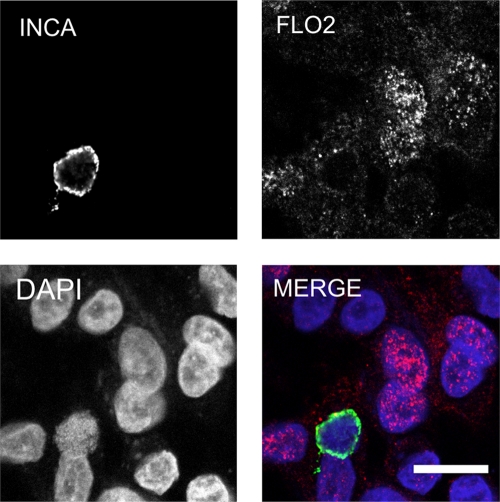
Flotillin-2 does not colocalize with C. pneumoniae inclusion.
Because flotillin-1 and flotillin-2 are able to form homo- or hetero-oligomers (3, 21, 57), we also studied flotillin-2 subcellular localization in Chlamydia-infected cells. Labeling C. pneumoniae-infected cells with antibody specific to flotillin-2 showed prominent nuclear or perinuclear staining with a faint cytosolic staining pattern; however, the localization of flotillin-2 to the chlamydial inclusion could not be confirmed (Fig. 5). Nuclear localization of flotillin-2 was unexpected, since only flotillin-1 has been reported to localize in the nucleus (52). In conclusion, flotillin-1, but not flotillin-2, colocalizes with Chlamydia inclusion.
Fig 5.
Flotillin-2 does not localize to C. pneumoniae inclusion. A549 cells were inoculated with C. pneumoniae K6, and flotillin-2 was detected at 48 hpi with an antibody specific for flotillin-2 and anti-mouse Alexa Fluor 647-conjugated secondary antibody. Inclusions were detected with an antibody specific for IncA and anti-rabbit Alexa Fluor 488-conjugated secondary antibody. In the merged image, IncA (INCA) is shown in green and flotillin-2 (FLO2) in red. Bacterial and host DNA were stained with DAPI and are shown in blue. Bar, 10 μm.
Flotillin-1 associates with IncA in DRMs.
Flotillin-1 belongs to a protein family containing the stomatin/prohibitin/flotillin/HflK/C (SPFH) domain (9). All members of this protein family are cofractioned with cholesterol- and sphingolipid-enriched detergent-resistant membrane microdomains (DRMs) (9, 32, 45). Although the DRMs are not considered to be equal to these cholesterol-enriched microdomains, also termed “lipid rafts,” DRM fractioning can be used to estimate protein association to these ordered membrane domains (35). Because cholesterol-rich microdomains are probably formed in chlamydial inclusion membranes (41), we tested whether chlamydial major outer membrane protein (MOMP), inclusion protein IncA, and CopN were localized to the DRMs together with flotillin-1. To demonstrate this, C. pneumoniae-infected cells were treated with 1% Triton X-100 at 4°C, and DRMs were floated in a sucrose density gradient. Chlamydial proteins IncA and MOMP were partially colocalized with host DRM markers, flotillin-1, and ganglioside M1 (GM1) at 48 hpi (Fig. 6). In contrast, the putative type III-secreted protein CopN was present only in detergent-soluble fractions (Fig. 6). This result was interesting, since both IncA and CopN are secreted and localized to the inclusion membrane (19, 25, 60). Thus, our results imply that CopN, contrary to IncA, is not embedded tightly in the inclusion membrane. This interpretation is consistent with a model suggesting that CopN would be a peripheral inclusion membrane protein (19). Our data suggest that a cholesterol-enriched fraction of an inclusion membrane may act as a platform for the molecular association between flotillin-1 and other inclusion membrane proteins.
Fig 6.
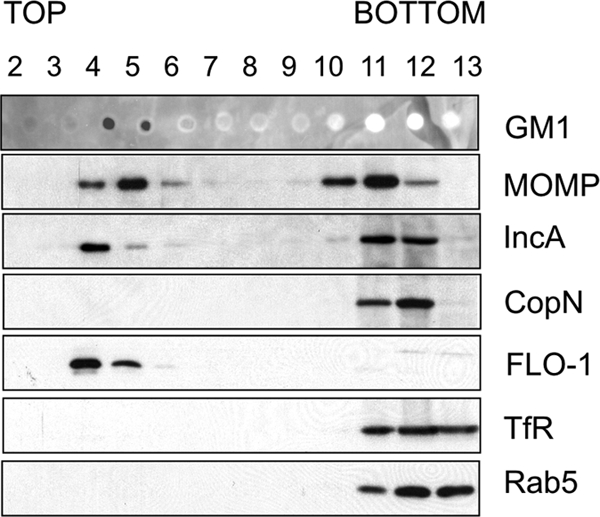
Flotillin-1 and IncA associate with detergent-resistant membranes (DRMs). HL cells were infected with C. pneumoniae for 48 h and treated with 1% Triton X-100 for 30 min at 4°C. The cell extracts were floated in a sucrose gradient, and sucrose fractions were collected from the top to the bottom and analyzed by SDS-PAGE and immunoblotting. GM1 and flotillin-1 (FLO-1) were used as markers for DRM fractions. Transferrin receptor (TfR) and small GTP-ase Rab5 were used to mark detergent-soluble fractions. The figure is representative of results from 3 independent assays.
DISCUSSION
To date, only a subset of host proteins have been identified in chlamydial inclusions, and the mechanisms by which these proteins are translocated to the proximity of the inclusions have remained largely uncharacterized. Here, for the first time, we have demonstrated that flotillin-1 is recruited to the chlamydial inclusion membrane, augmenting intracellular bacterial growth.
In the present study, C. pneumoniae associated to the host independent of flotillin-1, suggesting that bacterial attachment was not dependent on flotillin-1 microdomains on the plasma membrane. Flotillin-1 microdomains mediate a distinct clathrin-independent endocytic pathway (22), and the present study does not exclude the possibility that flotillin-1 contributes to the entry of Chlamydia independent of bacterial attachment or that flotillin-1 in inclusion membranes originates in the host plasma membrane at bacterial internalization. This interpretation would be supported by the fact that flotillin-1 RNA silencing had a significant effect on the number of inclusions formed but an insignificant effect on the mean size of inclusions at 72 hpi. This would imply that those elementary bodies that were internalized developed normally. RNA silencing, however, has a very heterogeneous effect on protein expression from cell to cell and may cause a similar effect on chlamydial growth without impairing bacterial entry in the assay. According to this interpretation, there may be a certain threshold level for endogenous flotillin-1 that enables normal inclusion development. In addition, in an ultrastructural study, it has been shown that host plasma membrane proteins are shed from the chlamydial inclusion at the early infection phase (53). Still, in our study, flotillin-1 was found in association with chlamydial inclusions at the late infection phase, suggesting that flotillin-1 associates with inclusion after internalization. Therefore, we believe that flotillin-1 is transported to inclusion during chlamydial intracellular development. In conclusion, flotillin-1 may have a potential role both in bacterial endocytosis and intracellular development, but in this respect our study remains inconclusive and further studies should be performed to address this.
Flotillin-1 and flotillin-2 are able to form homo- and hetero-oligomers, and hetero-oligomerization seems to be requisite for their endocytosis (3, 21, 57). Although the endocytosis of these proteins appears to be coordinated, a distinct intracellular localization pattern is often observed (32). In a previous study, flotillin-1, but not flotillin-2, was found in the nuclei of PC-3 prostate cancer cells (52). In A549 cells (this study), however, the immunostaining pattern of flotillin-2 suggested prominent nuclear or perinuclear localization, while flotillin-1 localized mainly to the cytosol. We showed that flotillin-1, but not flotillin-2, localizes to chlamydial inclusion, implying that flotillins are not transported in a coordinated fashion to the chlamydial inclusion membrane.
Given that flotillin-1 was demonstrated in the chlamydial infection, one would assume that flotillin-1-associated cargo would also be found in the inclusion. Interestingly, this appears to be the case. Current data suggest that the flotillins are involved in a targeted delivery of specific membrane proteins, especially the glycosylphosphatidyl inositol (GPI)-anchored proteins, to specific regions of the plasma membrane (58). In particular, it has been shown that GPI-anchored protein CD59 is endocytosed from the plasma membrane by a specific flotillin-1-dependent process (2, 22). This complement-protective host protein localizing on the luminal side of a chlamydial inclusion is transported to the C. trachomatis inclusion by a brefeldin A-insensitive, Golgi-independent pathway (26). To date, it is unknown whether the localization of CD59 to the inclusion membrane is important for the chlamydial pathogenesis or host innate immunity response to the infection (26). Future experimental studies should, therefore, investigate whether CD59 is translocated to the chlamydial inclusion by the flotillin-1-dependent pathway and whether CD59 is important in chlamydial intracellular growth.
Sphingomyelin and cholesterol-forming microdomains in lipid membranes (55, 56) are delivered to the chlamydial inclusion membrane (13, 23, 24), and therefore microdomain formation is expected also in inclusion membranes. Indeed, it has been shown that there are cholesterol-rich microdomains in chlamydial inclusion membranes, in which an active Src family kinase, Fyn, is found (41). We showed that both inclusion membrane marker IncA and flotillin-1 were cofractionated with DRMs, suggesting that certain bacterial inclusion membrane proteins, like IncA, may interact with flotillin-1 in cholesterol-enriched inclusion membrane domains. Interestingly, Fyn kinase directly interacts with flotillin-1 and regulates it is activity (36, 49, 59), raising the possibility that Fyn activates flotillin-1 in the inclusion membranes.
When considering the potential contribution of flotillin-1 to the intracellular development of C. pneumoniae, it is notable that flotillin-1 is found in association with lipid droplets (LDs) and multivesicular bodies (MVBs), although the function of flotillin-1 in these endocytic organelles has remained elusive (28, 33, 37, 48). LDs are intracellular storage organelles containing neutral lipids, and translocation of LDs to the inclusion lumen is thought to be a potential source of these lipids for Chlamydiae (16, 29). Although these data are limited to studies with C. trachomatis, an ultrastructural study also showed that C. pneumoniae inclusion fuses with LDs in macrophage foam cells in atherosclerotic plaque specimens (8). Like LDs, MVBs are also involved in normal inclusion development providing essential host lipids for developing Chlamydia (50). MBV markers, including MLN64, LBPA, and a tetraspanin protein CD63, localize to the inclusion membrane and the inclusion lumen (5, 6). Taken together, the evidence reviewed and our observations suggest a compelling possibility that flotillin-1 may have a role in the translocation of LDs to the chlamydial inclusion. Equally, flotillin-1 may have a role in the MVB-mediated molecular transport between the host and chlamydial inclusion. These hypotheses need to be addressed in future studies.
In conclusion, our study adds C. pneumoniae to the growing list of human intracellular pathogens interacting with flotillin-1. Although the molecular function of flotillin-1 in chlamydial inclusion remains to be elucidated, our findings suggest a role for flotillin-1 in the intracellular growth of C. pneumoniae.
ACKNOWLEDGMENTS
This study was supported by Academy of Finland (Microbes and Man Research Program, MICMAN project number 202491, and project numbers 118391, 110340, 217554/ECIBUG, and 130043/CHLAMYTRANS) and Turku University Hospital Fund. J.T.K. was supported in part by Drug Discovery Graduate School and Ministry of Education, Finland, as well as the Siiri Suominen fund.
We thank Outi Rautio and Jouko Sandholm for skillful technical assistance. Ted Hackstadt (Rocky Mountain Laboratories) and Agathe Subtil (Institute Pasteur) are thanked for the antibody against C. trachomatis IncA.
Footnotes
Published ahead of print 3 January 2012
REFERENCES
- 1. Airaksinen U, et al. 2003. Production of Chlamydia pneumoniae proteins in Bacillus subtilis and their use in characterizing immune responses in the experimental infection model. Clin. Diagn. Lab. Immunol. 10:367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ait-Slimane T, Galmes R, Trugnan G, Maurice M. 2009. Basolateral internalization of GPI-anchored proteins occurs via a clathrin-independent flotillin-dependent pathway in polarized hepatic cells. Mol. Biol. Cell 20:3792–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Babuke T, et al. 2009. Hetero-oligomerization of reggie-1/flotillin-2 and reggie-2/flotillin-1 is required for their endocytosis. Cell. Signal. 21:1287–1297 [DOI] [PubMed] [Google Scholar]
- 4. Babuke T, Tikkanen R. 2007. Dissecting the molecular function of reggie/flotillin proteins. Eur. J. Cell Biol. 86:525–532 [DOI] [PubMed] [Google Scholar]
- 5. Beatty WL. 2006. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J. Cell Sci. 119:350–359 [DOI] [PubMed] [Google Scholar]
- 6. Beatty WL. 2008. Late endocytic multivesicular bodies intersect the chlamydial inclusion in the absence of CD63. Infect. Immun. 76:2872–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bickel PE, et al. 1997. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J. Biol. Chem. 272:13793–13802 [DOI] [PubMed] [Google Scholar]
- 8. Bobryshev YV, Killingsworth MC, Tran D, Lord R. 2008. Amalgamation of Chlamydia pneumoniae inclusions with lipid droplets in foam cells in human atherosclerotic plaque. Virchows Arch. 453:69–77 [DOI] [PubMed] [Google Scholar]
- 9. Browman DT, Hoegg MB, Robbins SM. 2007. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 17:394–402 [DOI] [PubMed] [Google Scholar]
- 10. Browman DT, Resek ME, Zajchowski LD, Robbins SM. 2006. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 119:3149–3160 [DOI] [PubMed] [Google Scholar]
- 11. Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Campbell LA, Kuo CC. 2004. Chlamydia pneumoniae—an infectious risk factor for atherosclerosis? Nat. Rev. Microbiol. 2:23–32 [DOI] [PubMed] [Google Scholar]
- 13. Carabeo RA, Mead DJ, Hackstadt T. 2003. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl. Acad. Sci. U. S. A. 100:6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chu HG, Weeks SK, Gilligan DM, Rockey DD. 2008. Host alpha-adducin is redistributed and localized to the inclusion membrane in chlamydia- and chlamydophila-infected cells. Microbiology 154:3848–3855 [DOI] [PubMed] [Google Scholar]
- 15. Cles LD, Stamm WE. 1990. Use of HL cells for improved isolation and passage of Chlamydia pneumoniae. J. Clin. Microbiol. 28:938–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U. S. A. 105:9379–9384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dermine JF, et al. 2001. Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J. Biol. Chem. 276:18507–18512 [DOI] [PubMed] [Google Scholar]
- 18. Ekman MR, et al. 1993. An epidemic of infections due to Chlamydia pneumoniae in military conscripts. Clin. Infect. Dis. 17:420–425 [DOI] [PubMed] [Google Scholar]
- 19. Fields KA, Hackstadt T. 2000. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 38:1048–1060 [DOI] [PubMed] [Google Scholar]
- 20. Fields KA, Hackstadt T. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 18:221–245 [DOI] [PubMed] [Google Scholar]
- 21. Frick M, et al. 2007. Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding. Curr. Biol. 17:1151–1156 [DOI] [PubMed] [Google Scholar]
- 22. Glebov OO, Bright NA, Nichols BJ. 2006. Flotillin-1 defines a clathrin-independent endocytic pathway in mammalian cells. Nat. Cell Biol. 8:46–54 [DOI] [PubMed] [Google Scholar]
- 23. Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15:964–977 [PMC free article] [PubMed] [Google Scholar]
- 24. Hackstadt T, Scidmore MA, Rockey DD. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. U. S. A. 92:4877–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hackstadt T, Scidmore-Carlson MA, Shaw EI, Fischer ER. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell. Microbiol. 1:119–130 [DOI] [PubMed] [Google Scholar]
- 26. Hasegawa A, Sogo LF, Tan M, Sutterlin C. 2009. Host complement regulatory protein CD59 is transported to the chlamydial inclusion by a Golgi apparatus-independent pathway. Infect. Immun. 77:1285–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Howe D, Heinzen RA. 2006. Coxiella burnetii inhabits a cholesterol-rich vacuole and influences cellular cholesterol metabolism. Cell. Microbiol. 8:496–507 [DOI] [PubMed] [Google Scholar]
- 28. Kokubo H, et al. 2003. Ultrastructural localization of flotillin-1 to cholesterol-rich membrane microdomains, rafts, in rat brain tissue. Brain Res. 965:83–90 [DOI] [PubMed] [Google Scholar]
- 29. Kumar Y, Cocchiaro J, Valdivia RH. 2006. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr. Biol. 16:1646–1651 [DOI] [PubMed] [Google Scholar]
- 30. Kuo CC, Grayston JT. 1990. A sensitive cell line, HL cells, for isolation and propagation of Chlamydia pneumoniae strain TWAR. J. Infect. Dis. 162:755–758 [DOI] [PubMed] [Google Scholar]
- 31. Kuo CC, Jackson LA, Campbell LA, Grayston JT. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8:451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langhorst MF, Reuter A, Stuermer CA. 2005. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell. Mol. Life Sci. 62:2228–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langui D, et al. 2004. Subcellular topography of neuronal Abeta peptide in APPxPS1 transgenic mice. Am. J. Pathol. 165:1465–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee BY, et al. 2010. The Mycobacterium bovis bacille Calmette-Guerin phagosome proteome. Mol. Cell. Proteomics 9:32–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lichtenberg D, Goni FM, Heerklotz H. 2005. Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem. Sci. 30:430–436 [DOI] [PubMed] [Google Scholar]
- 36. Liu J, Deyoung SM, Zhang M, Dold LH, Saltiel AR. 2005. The stomatin/prohibitin/flotillin/HflK/C domain of flotillin-1 contains distinct sequences that direct plasma membrane localization and protein interactions in 3T3-L1 adipocytes. J. Biol. Chem. 280:16125–16134 [DOI] [PubMed] [Google Scholar]
- 37. Liu P, et al. 2004. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279:3787–3792 [DOI] [PubMed] [Google Scholar]
- 38. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 39. Ludwig A, et al. 2010. Flotillin microdomains interact with the cortical cytoskeleton to control uropod formation and neutrophil recruitment. J. Cell Biol. 191:771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mannonen L, Kamping E, Penttila T, Puolakkainen M. 2004. IFN-gamma induced persistent Chlamydia pneumoniae infection in HL and Mono Mac 6 cells: characterization by real-time quantitative PCR and culture. Microb. Pathog. 36:41–50 [DOI] [PubMed] [Google Scholar]
- 41. Mital J, Miller NJ, Fischer ER, Hackstadt T. 2010. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell. Microbiol. 12:1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moore ER, Mead DJ, Dooley CA, Sager J, Hackstadt T. 2011. The trans-Golgi SNARE syntaxin 6 is recruited to the chlamydial inclusion membrane. Microbiology 157:830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moorhead AM, Jung JY, Smirnov A, Kaufer S, Scidmore MA. 2010. Multiple host proteins that function in phosphatidylinositol-4-phosphate metabolism are recruited to the chlamydial inclusion. Infect. Immun. 78:1990–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mori S, et al. 2003. PACSIN3 binds ADAM12/meltrin alpha and up-regulates ectodomain shedding of heparin-binding epidermal growth factor-like growth factor. J. Biol. Chem. 278:46029–46034 [DOI] [PubMed] [Google Scholar]
- 45. Morrow IC, Parton RG. 2005. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic 6:725–740 [DOI] [PubMed] [Google Scholar]
- 46. Murphy SC, et al. 2004. Erythrocyte detergent-resistant membrane proteins: their characterization and selective uptake during malarial infection. Blood 103:1920–1928 [DOI] [PubMed] [Google Scholar]
- 47. O'Connell CM, Ionova IA, Quayle AJ, Visintin A, Ingalls RR. 2006. Localization of TLR2 and MyD88 to Chlamydia trachomatis inclusions. Evidence for signaling by intracellular TLR2 during infection with an obligate intracellular pathogen. J. Biol. Chem. 281:1652–1659 [DOI] [PubMed] [Google Scholar]
- 48. Rajendran L, Le Lay S, Illges H. 2007. Raft association and lipid droplet targeting of flotillins are independent of caveolin. Biol. Chem. 388:307–314 [DOI] [PubMed] [Google Scholar]
- 49. Riento K, Frick M, Schafer I, Nichols BJ. 2009. Endocytosis of flotillin-1 and flotillin-2 is regulated by Fyn kinase. J. Cell Sci. 122:912–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robertson DK, Gu L, Rowe RK, Beatty WL. 2009. Inclusion biogenesis and reactivation of persistent Chlamydia trachomatis requires host cell sphingolipid biosynthesis. PLoS Pathog. 5:e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rzomp KA, Scholtes LD, Briggs BJ, Whittaker GR, Scidmore MA. 2003. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect. Immun. 71:5855–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Santamaria A, et al. 2005. PTOV1 enables the nuclear translocation and mitogenic activity of flotillin-1, a major protein of lipid rafts. Mol. Cell. Biol. 25:1900–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scidmore MA, Fischer ER, Hackstadt T. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect. Immun. 71:973–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Scidmore MA, Hackstadt T. 2001. Mammalian 14-3-3beta associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39:1638–1650 [DOI] [PubMed] [Google Scholar]
- 55. Simons K, Ikonen E. 1997. Functional rafts in cell membranes. Nature 387:569–572 [DOI] [PubMed] [Google Scholar]
- 56. Simons K, Ikonen E. 2000. How cells handle cholesterol. Science 290:1721–1726 [DOI] [PubMed] [Google Scholar]
- 57. Solis GP, et al. 2007. Reggie/flotillin proteins are organized into stable tetramers in membrane microdomains. Biochem. J. 403:313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stuermer CA. 2011. Reggie/flotillin and the targeted delivery of cargo. J. Neurochem. 116:708–713 [DOI] [PubMed] [Google Scholar]
- 59. Stuermer CA, et al. 2001. Glycosylphosphatidyl inositol-anchored proteins and fyn kinase assemble in noncaveolar plasma membrane microdomains defined by reggie-1 and -2. Mol. Biol. Cell 12:3031–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Subtil A, Parsot C, Dautry-Varsat A. 2001. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol. Microbiol. 39:792–800 [DOI] [PubMed] [Google Scholar]
- 61. Vassilieva EV, Ivanov AI, Nusrat A. 2009. Flotillin-1 stabilizes caveolin-1 in intestinal epithelial cells. Biochem. Biophys. Res. Commun. 379:460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]