Abstract
Redox-sensing repressor Rex was previously implicated in the control of anaerobic respiration in response to the cellular NADH/NAD+ levels in Gram-positive bacteria. We utilized the comparative genomics approach to infer candidate Rex-binding DNA motifs and assess the Rex regulon content in 119 genomes from 11 taxonomic groups. Both DNA-binding and NAD-sensing domains are broadly conserved in Rex orthologs identified in the phyla Firmicutes, Thermotogales, Actinobacteria, Chloroflexi, Deinococcus-Thermus, and Proteobacteria. The identified DNA-binding motifs showed significant conservation in these species, with the only exception detected in Clostridia, where the Rex motif deviates in two positions from the generalized consensus, TTGTGAANNNNTTCACAA. Comparative analysis of candidate Rex sites revealed remarkable variations in functional repertoires of candidate Rex-regulated genes in various microorganisms. Most of the reconstructed regulatory interactions are lineage specific, suggesting frequent events of gain and loss of regulator binding sites in the evolution of Rex regulons. We identified more than 50 novel Rex-regulated operons encoding functions that are essential for resumption of the NADH:NAD+ balance. The novel functional role of Rex in the control of the central carbon metabolism and hydrogen production genes was validated by in vitro DNA binding assays using the TM0169 protein in the hydrogen-producing bacterium Thermotoga maritima.
INTRODUCTION
Bacteria can adapt to changes in their environment by utilizing a range of transcription factors that receive an appropriate intra- or extracellular signal and trigger the specific transcriptional response. The Rex regulator is a transcription factor that responds to intracellular redox potential and negatively controls expression of genes involved in energy metabolism and fermentative growth in Gram-positive bacteria, including the actinobacterium Streptomyces coelicolor (4) and various Firmicutes, such as Bacillus subtilis (35), Staphylococcus aureus (25), and Streptococcus mutans (3). NAD plays an important role in different biological processes, including redox cellular balance. During the catabolism of carbohydrates, NAD+ is reduced to NADH by glycolytic enzymes. The NADH formed is reoxidized back either by respiratory electron transport chains or by NADH-linked fermentative enzymes. The DNA-binding activity of Rex is modulated by the intracellular ratio of NADH:NAD+. Under the low NADH:NAD+ ratio, the Rex protein binds to the target sites and represses transcription of genes involved in NADH reoxidation, while the increase of NADH concentration results in the dissociation of Rex from DNA and derepression of its target genes (4, 25, 35).
Rex was initially described in S. coelicolor, where it controls the cytochrome bd terminal oxidase operon cydABCD and the heme biosynthesis genes hemACD, as well as the membrane-bound proton-translocating NADH dehydrogenase operon nuoA-nuoN (4). The Rex-regulated promoters contain a common DNA motif with consensus 5′-TGTGNNCNNNTTCACA-3′, which was further confirmed by site-specific mutagenesis and electromobility shift assays to function as a Rex-binding site (4). Neither NAD+ nor NADP+ affected the DNA-binding affinity of Rex, whereas both reduced nucleotides decreased Rex-DNA complex formation, although NADH was more effective than NADPH. Further investigation of Rex-DNA complex formation using a range of NADH concentrations in the presence of an excess of NAD+ demonstrated that Rex senses the NADH:NAD+ ratio rather than the concentration of NADH per se and that it dissociates from DNA when NADH concentration is more than 2% of the common NAD(H) pool (4).
The Rex ortholog in Bacillus subtilis (also known as YidH) is a repressor of the respiratory oxidase cytochrome bd (cydABCD), an NADH-linked fermentative lactate dehydrogenase (ldh), a type II NADH dehydrogenase (ndh), and formate/nitrite transporter (ywcJ) (11, 15, 32). Seven Rex-binding sites identified upstream of the above-mentioned B. subtilis genes, including several tandem sites in the cydA and ldh promoter regions, were validated by in vitro binding assays (35). The Rex-binding motif in B. subtilis has a consensus 5′-WWTGTGAANTNNTNNNCAAW-3′, where “W” denotes either A or T. In Staphylococcus aureus, Rex is a central regulator of anaerobic metabolism and controls at least 18 target operons that are involved in nitrate/nitrite respiration (nar, nir, nas, hmp), fermentation (adhE, adh1, pflBA, ldh1, lctP), and amino acid metabolism (arc, ald1) (25). The Rex-binding motif, 5′-TTGTGAAWWWWTTCACAA-3′, is highly conserved in S. aureus. The DNA-binding activity of the S. aureus Rex protein is decreased by NADH and enhanced by NAD+, although NADH binds with a much higher affinity to Rex (95 nM) than NAD+ (150 μM) (25). Similarly to the S. coelicolor Rex regulator, the characterized Rex repressors in B. subtilis and S. aureus sense the cellular NADH/NAD+ redox balance (25, 35). Furthermore, Rex orthologs were recently characterized in two other Firmicutes species, the human dental pathogen Streptococcus mutans and the ethanol-producing anaerobic bacterium Thermoanaerobacter ethanolicus. In the latter species, Rex (termed RSP, for redox-sensing protein) binds to a palindromic sequence, 5′-ATTGTTANNNNNNTAACAAT-3′, to repress the transcription of the ethanol fermentation genes adhA, adhB, and adhE, and this binding was inhibited by NADH (26). In S. mutans, a high-throughput microarray analysis of a Rex-deficient mutant revealed that Rex plays an important role in the regulation of the central metabolism, oxidative stress, and biofilm formation (3).
The structures of Rex protein dimers in complex with an NADH, an NAD+, and/or a DNA operator have been determined for Rex proteins obtained from Thermus aquaticus (17, 33), Thermus thermophilus (20), B. subtilis (35, 36), and Streptococcus agalactiae (Protein Data Bank [PDB] accession number 3KET). Each Rex subunit contains two domains, an N-terminal winged-helix DNA-binding domain and a C-terminal Rossmann-like domain involved in dinucleotide binding and subunit dimerization. In the closed conformation, when Rex forms a complex with NADH, its DNA recognition domains are tightly packed, which is incompatible with binding to the Rex operator site. Release of NADH enables DNA binding by triggering a global change in the Rex dimer conformation. The structural studies allowed determination of amino acid residues responsible for DNA recognition and NADH binding in Rex proteins from T. aquaticus and B. subtilis. These two best-studied representatives of the Rex family were recently utilized in the elegant design of fluorescence-based in vivo NADH sensors (12, 39).
The main goal of this study was to refine and expand the knowledge of transcriptional regulation by Rex from a few model species, where it was previously studied, to all bacteria that encode Rex orthologs in their genomes. To identify Rex-binding motifs and reconstruct Rex regulons in diverse bacterial lineages, we used the comparative genomics approach that was previously applied for analysis of other regulatory systems (28). Here, we report the detailed description of Rex regulons in the genomes of 119 bacteria from the Firmicutes, Actinobacteria, Proteobacteria, Chloroflexi, Deinococcus-Thermus, and Thermotogales phyla. The bioinformatic analysis revealed a number of remarkable features of the Rex repressor family, such as (i) conservation of the regulator and its DNA-binding motif over a large phylogenetic distance and (ii) significant variations of the regulated target genes and associated metabolic processes that, in addition to anaerobic respiration and fermentation, include hydrogen production and glycolysis.
To evaluate the accuracy of the genomic reconstruction of the Rex regulon, we performed experimental testing of the predicted regulatory sites in Thermotogales. In vitro binding assays with the purified Thermotoga maritima Rex protein (TM0169) confirmed all 12 Rex-binding sites identified in silico in T. maritima and two candidate Rex sites found upstream of the adhE gene in Thermotoga sp. RQ-2. In addition, we confirmed the similar inhibitory effect of NADH on Rex-DNA complex formation in T. maritima, as it was previously reported in other species. Thermophilic bacteria from the Thermotogales order can produce hydrogen by fermenting a wide range of carbohydrates. The Rex regulon in Thermotogales includes multiple genes involved in the central carbohydrate metabolism and hydrogen production. Thus, the Rex regulon functional context in Thermotogales differs substantially from the Rex regulons previously described in other species.
MATERIALS AND METHODS
Bioinformatics approaches and tools. (i) Regulon reconstruction.
We used the established approach of regulon reconstruction that is based on the comparative genomic analysis of candidate regulator-binding sites in closely related bacterial genomes (reviewed in reference 28). The RegPredict Web server tool (regpredict.lbl.gov) (22) was used for the identification and further comparative analysis of candidate DNA operators using the position weight matrices (PWMs) trained on sets of known Rex-binding sites. Starting from the genomic identification of a reference set of organisms that encodes Rex orthologs, the Rex regulons were reconstructed in each reference genome set separately. Two main workflows were applied for reconstruction of Rex regulons: (i) propagation and expansion of previously experimentally described Rex regulons (in Bacillaceae, Staphylococcaceae, Actinomycetales, and Thermoanaerobacterales) and (ii) ab initio prediction of Rex regulons in the remaining seven taxonomic groups of bacteria for which no experimental knowledge on the sets of Rex-regulated genes was available. In each of 11 studied taxonomic groups, an iterative motif detection procedure implemented in the RegPredict tool was used to identify common regulatory DNA motifs in a set of upstream gene fragments and to construct PWMs that were further used to scan the genomes in this group and to predict novel genes in the regulon. Scores of candidate sites were calculated as the sum of positional nucleotide weights. We used a regulog concept (1) to represent a regulon inferred and projected in a set of closely related genomes. The content of each lineage-specific Rex regulog was reconstructed by application of the comparative genomics approach implemented in the RegPredict tool. Rex regulogs have included regulatory interactions with at least one of the following characteristics: (i) candidate Rex-binding sites that are conserved in two or more genomes, (ii) highly scored Rex-binding sites located upstream of target genes without orthologs in the analyzed taxonomic group, and (iii) regulatory interactions previously validated in experiments. Candidate sites associated with new members of the Rex regulon were added to the training set, and the respective PWM describing a lineage-specific Rex motif was rebuilt to improve search accuracy. The details of the reconstructed Rex regulons are captured and displayed in the RegPrecise database of bacterial regulons inferred by the comparative genomics approach (27) (regprecise.lbl.gov).
(ii) Other bioinformatic resources.
Genome sequences were uploaded from the MicrobesOnline database (8). Orthologs of the previously characterized Rex proteins were identified by a procedure based on the analysis of MicrobesOnline database phylogenetic trees for protein domains and validated by bidirectional genome-wide similarity searches using the Genome Explorer package with a 25% protein sequence identity threshold (18). Rex homologs in distantly related taxa were identified using BLAST searches in the refseq_protein database (2). Multiple sequence alignment of Rex family proteins was performed with MUSCLE (9). A phylogenetic tree of Rex family proteins with bootstrap values was computed with PhyML 3.0 (10) and visualized with Dendroscope (13). Sequence logos for Rex-binding DNA motifs were drawn with WebLogo (7). Correlations between amino acid/nucleotide pairs for Rex proteins and their operators were computed using the Prot-DNA-Korr program (http://bioinf.fbb.msu.ru/Prot-DNA-Korr/main.html). A three-dimen-sional (3-D) structure of the Rex-DNA complex (PDB accession number 3IKT) was analyzed using the WHATIF server (http://swift.cmbi.ru.nl/servers/html/index.html). Functional annotations of the predicted regulon members and the associated metabolic systems were collected from the SEED database (24).
Experimental approaches. (i) 5′ RACE.
Transcription start site (TSS) analysis was performed by the 5′ rapid amplification of cDNA ends (RACE) technique as previously described (27). Briefly, total RNA was extracted from exponentially growing T. maritima MSB8 cells using hot acid (phenol-chloroform) and purified using the SurePrep TrueTotal RNA purification kit (Fisher). Purified RNA was treated with Terminator-5′-phosphate exonuclease (Epicentre) to degrade RNA carrying a 5′-monophosphate followed by RNA 5′-polyphosphatase (Epicentre) to convert RNA with 5′-triphosphate to 5′-monophosphate in preparation for 5′-RNA adaptor ligation. cDNA libraries were synthesized from adaptor-ligated RNA and gel purified in the range of 100 to 300 bp. Final cDNA libraries were amplified, quantified, and then sequenced on the Illumina GAIIx platform. Reads were mapped to the T. maritima MSB8 genome using Mosaik Aligner (http://bioinformatics.bc.edu/marthlab/Mosaik) and filtered for uniquely aligned 5′ transcript loci.
(ii) Cloning and overproduction of Rex.
The T. maritima rex gene (TM0169) was amplified by PCR using genomic DNA as the template (forward primer, 5′-CGATCATGGATCCATGGCGGAAAAGATACC-3′; reverse primer, CTGCAGTCGAAGCTTTCAAGAATTCCTCC). The resulting DNA fragment carrying the rex gene was digested with BamHI/HindIII and cloned into the pSMT3 expression vector (19). The obtained vector encodes a fusion between the Rex protein and N-terminal hexahistidine Smt3 polypeptide (a yeast SUMO ortholog), allowing it to enhance protein solubility. The resulting construct was transformed in Escherichia coli BL21/DE3 cells. The cells were grown in 50 ml LB medium containing 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to an optical density at 600 nm (OD600) of 1.2 at 20°C and harvested after 16 h. The recombinant Smt-His6-Rex protein was purified by a rapid Ni-nitrilotriacetic acid (NTA) agarose column as described previously (23). Briefly, harvested cells were resuspended in 20 mM HEPES buffer (pH 7) containing 100 mM NaCl, 0.03% Brij 35, and 2 mM beta-mercaptoethanol supplemented with 2 mM phenylmethylsulfonyl fluoride and a protease inhibitor cocktail (Sigma-Aldrich). Lysozyme was added to 1 mg/ml, and the cells were lysed by a freeze-thaw cycle followed by sonication. After centrifugation, the supernatant was supplemented with Tris-HCl buffer (pH 8) to achieve a final concentration of 50 mM and then was loaded onto an Ni-NTA acid agarose minicolumn (0.3 ml). After the column was washed with the starting buffer containing 1 M NaCl and 0.3% Brij-35, bound proteins were eluted with 0.3 ml of the starting buffer supplemented with 250 mM imidazole. The final Rex protein buffer contains 20 mM Tris-HCl (pH 8.0), 2 mM dithiothreitol (DTT), and 200 mM NaCl. Protein size, expression level, distribution between soluble and insoluble forms, and extent of purification were assessed with 12% SDS-PAGE gels.
(iii) DNA binding assays.
The interaction of the purified recombinant tagged Rex protein with its cognate DNA-binding sites in T. maritima MSB8 and Thermotoga sp. RQ-2 was assessed at 22°C using two techniques: electrophoretic mobility shift assay (EMSA) and fluorescence polarization assay (FPA). Using FPA, we tested 14 DNA fragments containing the predicted Rex binding sites. The 28-bp DNA fragments were obtained by annealing synthesized oligonucleotides 3′-labeled with 6-carboxyfluorescein with unlabeled complementary oligonucleotides at a 1:10 ratio (see Table S1 in the supplemental material). The obtained fluorescence-labeled double-stranded DNA (dsDNA) fragments (1 nM) were incubated with the increasing concentrations of purified Rex (10 to 500 nM) in a 100-μl reaction mixture in 96-well black plates (VWR, Radnor, PA). The binding buffer contained 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.3 mg/ml bovine serum albumin (BSA), and 1 μg poly(dI-dC). Fluorescence measurements were taken on a Beckman DTX 880 multimode plate reader with excitation and emission filters at 495 nm and 520 nm, respectively. The background fluorescence from the buffer was subtracted, and the fluorescence polarization value was defined as follows: FP = (I1 − G · I2)/(I1 + G · I2), where I1 and I2 are the fluorescence intensities measured in the parallel and perpendicular directions respective to the orientation of the excitation polarizer and G is a correction factor (34). Using the EMSA, we tested a single 28-bp DNA fragment with a Rex-binding site from the upstream region of the TM0201 gene. The fragment for EMSA was obtained by the same approach as for FPA, where one of the two oligonucleotides was 3′ labeled with biotin instead of 6-carboxyfluorescein. The biotin-labeled dsDNA fragment (0.1 nM) was incubated with increasing concentrations (5, 12, 25, 50, and 100 nM) of purified Rex protein in a total volume of 20 μl as previously described (29). The binding buffer contained 20 mM Tris-HCl (pH 8.0), 150 mM KCl, 1 mM DTT, 1 mM EDTA, 0.05% NP-40, and 2.5% glycerol. Poly(dI-dC) was added as a nonspecific competitor DNA at 1 μg to reduce nonspecific binding. After 25 min of incubation at 37°C, the reaction mixtures were separated by electrophoresis on a 5% native polyacrylamide gel in 0.5× Tris-borate-EDTA (100 min, 90 V, room temperature). The DNA was transferred by electrophoresis (30 min at 380 mA) onto a Hybond-N+ membrane and fixed by UV cross-linking. Biotin-labeled DNA was detected with the LightShift chemiluminescent EMSA kit. The effect of NAD+ and NADH was tested by their addition to the incubation mixture.
RESULTS
Genomic reconstruction of Rex regulons in bacteria. (i) Phylogenetic distribution of Rex proteins.
Orthologs of the Rex proteins from S. coelicolor, B. subtilis, S. aureus, and S. mutans were identified in the reference protein database (refseq_protein) using the BLASTP tool provided by NCBI. Rex homologs were identified in the genomes of 16 bacterial phyla (see Table S2 in the supplemental material), being most widely distributed in Firmicutes, Actinobacteria, Thermotogales, Chloroflexi, Deinococcus-Thermus, and Bacteroidetes. Additional Rex orthologs were found in Acidobacteria, Dictyoglomi, Elusimicrobia, Fusobacteria, Gemmatimonadetes, Lentisphaerae, Proteobacteria, Spirochaetes, Synergistetes, and Verrucomicrobia. Interestingly, among Proteobacteria, Rex was identified only in the delta subdivision (e.g., in all Desulfovibrionales species). Among Actinobacteria, Rex was found in only 24 out of 35 families of the Actinomycetales order. Thus, Rex proteins are universal but not ubiquitous in bacteria and are absent from archaea. In general, the presence of the Rex regulator correlates with chemoheterotrophy (Rex was not found in chemoautotrophs) but does not correlate with the aerobic/anaerobic lifestyle of microorganisms.
Like some other bacterial repressors (e.g., ArgR, NiaR, NrdR) controlling the central metabolism, Rex regulators are present in a single copy in most genomes with the exception of the Thermotogales group and some Firmicutes species, where we have identified two rex paralogs (termed Rex and Rex1). To get insight into a possible evolutionary history of the duplicates, we constructed the maximum likelihood phylogenetic tree for all Rex proteins analyzed in this study (see Fig. S1 in the supplemental material). A Rex1 paralog in Clostridium bartlettii (locus_tag CLOBAR_01830) appears to group with the Firmicutes/Clostridia branch, whereas Rex1 paralogs in Lactobacillus genomes (e.g., LGG_01260) form a diverged branch that was clustered with Rex proteins from the Firmicutes/Bacilli branch, suggesting their possible appearance by species- or lineage-specific gene duplications. In contrast to Firmicutes, Rex1 paralogs identified in the Thermotogales group (e.g., TM1427 in T. maritima) form a distinct branch, which is not clustered with the main branch of Rex proteins with representatives from all studied Thermotogales genomes (e.g., TM0169 in T. maritima).
(ii) Structural and functional analysis of Rex proteins.
The crystal structures of Rex proteins from T. thermophilus and T. aquaticus (T-Rex), B. subtilis (B-Rex), and S. agalactiae demonstrate a dimeric organization of two modular subunits. All studied Rex proteins have two domains: an N-terminal DNA-binding domain and a C-terminal pyridine nucleotide-binding domain (Rossmann fold) for NAD(H) binding and dimer formation. A multiple alignment of the selected Rex family proteins, including four proteins with known 3-D structures, is provided in Fig. S2 in the supplemental material. Most of the analyzed Rex proteins are well conserved, whereas the Rex1 paralogs from Thermotogales demonstrated the largest number of amino acid substitutions, suggesting their possible functional diversification. Using the solved structure of T-Rex in complex with NAD+ and a DNA operator, we mapped the NAD(H)- and DNA-binding residues on the multiple alignment of Rex sequences. Among 12 residues involved in the T-Rex–DNA contacts, 10 residues (Pro4, Arg10, Ser31, Arg46, Lys47, Tyr51, Gly56, Gly59, Gly61, and Tyr62) are conserved in most Rex family proteins, whereas Phe43 and Arg58 are variable in the Rex family. These observations correlate with the relative conservation of the identified Rex-binding DNA motifs across 11 studied taxonomic groups (see the next section). Within the NAD(H)-binding domain, a key acidic residue (Asp112 in T-Rex) that discriminates NAD(H) from phosphorylated NADP(H) is conserved in all Rex family proteins. The Gly-X-Gly-X-X-Gly signature sequence of the P loop that is important for pyridine nucleotide binding is conserved in all main Rex proteins but has several mutations (Gly-X-Asn-X-X-Ala) in Rex1 paralogs in Thermotoga spp. These findings suggest that the Rex effector molecule NADH experimentally validated in B. subtilis (35), S. aureus (25), S. coelicolor (4), T. aquaticus (17), and T. maritima (this study) is conserved in all Rex orthologs but not in Rex1 paralogs in Thermotoga spp. Potential functional consequences of the observed multiple substitutions in Rex1 paralogs are discussed below.
(iii) Identification of Rex-binding motifs and regulons.
To perform the comparative genomics reconstruction of Rex regulons, we selected the reference set of 119 bacterial genomes from 11 taxonomic groups based on the following considerations (see Table S2 in the supplemental material). First, the analyzed genome has to encode at least one regulator from the Rex family and be present in the MicrobesOnline genomic database and the RegPredict server. To facilitate the comparative genomics analysis, all analyzed genomes were subdivided into individual taxonomic groups, each containing 3 to 19 genomes. Taxonomic groups with fewer than 3 genomes were excluded from the analysis because of the comparative genomics approach limitations. For large taxonomic groups (e.g., Clostridiaceae, Actinomycetales), reference genome sets were limited to a maximum of 19 genomes to adjust for computational limitations of the RegPredict Web server.
In each taxonomic group, we identified the lineage-specific Rex-binding motif and constructed the corresponding position weight matrix (PWM). For identification of conserved regulatory motifs, we utilized the iterative procedure “Discover profile” implemented in the RegPredict Web server (22) and the group-specific training sets of 5′ intergenic regions of candidate Rex target genes. The initial sets of Rex target genes have included the experimentally validated members of Rex regulons in S. coelicolor (4), S. aureus (25), and B. subtilis (11, 15, 32, 35) and their orthologs in related genomes. For those groups of genomes where there was no prior information on Rex-regulated genes, we used PWMs constructed for other groups to define sets of candidate Rex target genes. After construction of lineage-specific PWMs, we used the RegPredict tool to perform (i) genome-wide searches for additional Rex-binding sites in the analyzed groups of genomes and (ii) a cross-species comparison of the predicted sets of potentially coregulated genes to define regulon composition for each analyzed lineage (see Materials and Methods). As a result, we were able to reconstruct Rex regulons in all taxonomic groups selected for comparative analysis with only one exception of Bacteroidetes. The content of reconstructed Rex regulons for 11 taxonomic groups, including Actinomycetales, Chloroflexi, Desulfovibrionales, Deinococcus-Thermus, Thermotogales, and 6 groups from the Firmicutes phylum is summarized in Tables 1 and 2. Detailed information about the predicted DNA-binding sites and downstream regulated genes is provided in the RegPrecise database (regprecise.lbl.gov) (21).
Table 1.
Conserved core of Rex regulons in 11 taxonomic groups of bacteria
| Gene(s) and metabolism | No. of genomesa |
Functional role | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Thermotogales (11 genomes) | Clostridiaceae (19 genomes) | Thermoanaerobacterales (3 genomes) | Staphylococcaceae (7 genomes) | Bacillales (11 genomes) | Lactobacillaceae (15 genomes) | Streptococcaceae (15 genomes) | Chloroflexi (5 genomes) | Actinomycetales (18 genomes) | Deinococcus-Thermus (5 genomes) | Desulfovibrionales (10 genomes) | ||
| Energy metabolism | ||||||||||||
| atpBC | – | – | – | – | – | – | – | 3 | 5 | – | 10 | ATP synthase (EC 3.6.3.14) |
| cydABCD | # | – | # | – | 3b | – | – | – | 3b | – | – | Cytochrome d ubiquinol oxidase (EC 1.10.3.-) |
| hydABC | 11 | 8 | 3 | # | # | # | # | – | # | # | – | Fe-hydrogenase (EC 1.12.7.2) [NADH] |
| fnorAB | – | 4 | 2 | # | # | # | # | # | # | # | # | Ferredoxin-NADP+ reductase |
| ndh | – | – | 1 | – | 8b | – | – | 2 | 5 | 3 | – | NADH dehydrogenase (EC 1.6.99.3) |
| noxE | 8 | 6 | 2 | – | 3 | 7 | 6 | – | – | – | – | NADH oxidase (EC 1.6.99.3) |
| nuoA-nuoN | # | – | # | # | – | # | # | 4 | 13b | 5 | – | NADH ubiquinone oxidoreductase (EC 1.6.5.3) |
| Glycolysis | ||||||||||||
| gldA | 4 | – | # | – | – | 3 | – | # | – | # | # | Glycerol dehydrogenase (EC 1.1.1.6) [NAD] |
| gap | 10 | 2 | – | – | – | – | 8 | – | – | – | – | Glyceraldehyde-3P dehydrogenase (EC 1.2.1.12) [NAD] |
| pgk | 10 | 3 | – | – | – | – | 10 | – | – | – | – | Phosphoglycerate kinase (EC 2.7.2.3) |
| tpi | 8 | 2 | – | – | – | – | 7 | – | – | – | – | Triosephosphate isomerase (EC 5.3.1.1) |
| fba | – | 3 | – | – | – | – | 10 | # | – | – | – | Fructose-bisphosphate aldolase (EC 4.1.2.13) |
| Fermentation | ||||||||||||
| adhE | – | 11 | 1b | 2b | 2 | 13 | 14 | # | – | # | – | Alcohol/acetaldehyde dehydrogenase (EC 1.1.1.1/1.2.1.10) [NADH] |
| adhB | # | – | 2b | 4b | 1 | 6 | 9 | – | 2 | – | # | Alcohol dehydrogenase [Zn] (EC 1.1.1.1) [NADH] |
| adhA | 7 | 2 | 3b | # | # | # | # | # | # | # | – | Alcohol dehydrogenase [Fe] (EC 1.1.1.1) [NADH] |
| ldh | – | 5 | – | 3b | 9b | 4 | 12 | – | – | – | – | l-Lactate dehydrogenase (EC 1.1.1.27) [NADH] |
| pflBA | # | 7 | # | 4b | 3 | – | – | # | – | # | – | Pyruvate formate-lyase (EC 2.3.1.54) |
| dhaT | # | 4 | # | # | – | 3 | – | – | – | # | – | 1,3-Propanediol dehydrogenase (EC 1.1.1.202) [NADH] |
| bcd-hbd | – | 8 | 1 | # | # | # | # | # | # | # | # | Butanoate fermentation pathway [NADH] |
| butA | # | 4 | # | – | # | 3 | – | # | # | # | # | 2,3-Butanediol dehydrogenase (EC 1.1.1.4) [NADH] |
| NAD biosynthesis | ||||||||||||
| nadABC | – | 3 | – | # | 1 | # | # | – | – | 2 | – | De novo NAD biosynthesis enzymes |
| pncB-nadE | – | – | – | – | – | 1 | 8 | – | – | – | – | Nicotinate phosphoribosyltransferase, NAD synthase |
| Miscellaneous | ||||||||||||
| nirC | # | – | # | 3b | 2b | – | 9 | # | – | # | # | Formate/nitrite family of transporters |
| narK | # | 1 | # | 5 | – | – | – | # | – | # | – | Nitrate-nitrite antiporter |
| lctP | # | – | # | 6b | 4b | – | – | – | – | – | – | l-Lactate permease |
| rex | – | 5 | 3 | – | – | 5 | 13 | 5 | 14b | – | 6 | Transcriptional regulator Rex |
The numbers of genomes in the analyzed taxonomic groups are indicated in parentheses. The table shows the numbers of genomes in each taxonomic group where the target gene is preceded by a conserved Rex-binding site. –, genes without a Rex-binding site; #, the absence of orthologous gene(s) in the analyzed genomes.
Rex regulation was previously shown for the model organism from this group.
Table 2.
Taxon-specific members of the Rex regulons in bacteria
| Taxonomic groupa | Target gene(s) | No. of genomesb | Functional role | FCc |
|---|---|---|---|---|
| Thermotogales (11 genomes) | sat-hpr | 5 | Serine aminotransferase; hydroxypyruvate reductase [NADH] | C |
| gckA | 4 | d-Glycerate 2-kinase | C | |
| Clostridiaceae (20 genomes) | phf | 9 | Fe-hydrogenase (EC 1.12.7.2) | E |
| bdh | 9 | Butyraldehyde and butanol dehydrogenases [NADH] | F | |
| thlA | 4 | Acetyl coenzyme A (acetyl-CoA) acetyltransferase (EC 2.3.1.9) | F | |
| glcD-glcA | 4 | Glycolate oxidase (EC 1.1.99.14), glycolate permease | C | |
| pgm | 4 | Phosphoglycerate mutase (EC 5.4.2.1) | C | |
| fucO | 3 | Lactaldehyde reductase (EC 1.1.1.77) [NADH] | C | |
| cat-sucD-abfDH | 3 | Anaerobic succinate degradation [NADH] | C | |
| maeA | 3 | NAD-dependent malic enzyme (EC 1.1.1.38) | C | |
| narAB | 2 | Assimilatory nitrate reductase | R | |
| fld-por | 2 | Flavodoxin, pyruvate-flavodoxin oxidoreductase (EC 1.2.7.-) | F | |
| fprA | 2 | H2O-forming NADH oxidase (EC 1.6.3.-) | E | |
| asrTABC | 2 | Anaerobic sulfite reductase and transporter | R | |
| ctfAB | 2 | Butyrate-acetoacetate CoA-transferase (EC 2.8.3.9) | F | |
| frdA | 2 | Fumarate reductase flavoprotein subunit (EC 1.3.99.1) | C | |
| nadO | 1 | NADH oxidase | E | |
| Thermoanaerobacterales (3 genomes) | hydG | 3 | Fe-hydrogenase maturation protein | E |
| echABCDEF | 2 | Energy-conserving Ni-Fe hydrogenase (EC 1.12.7.2) [NADH] | E | |
| hypABFCDE | 2 | Ni-Fe hydrogenase maturation factors | E | |
| Staphylococcaceae (7 genomes) | nirR-sirA-nasDEFd | 6 | Assimilatory nitrite reductase [NAD(P)H] | R |
| ddhd | 5 | d-Lactate dehydrogenase (EC 1.1.1.28) [NADH] | F | |
| narGHJI-nreABCd | 5 | Respiratory nitrate reductase | R | |
| frpd | 4 | NAD(P)H-flavin oxidoreductase | E | |
| arcABDCRd | 5 | Arginine catabolism | M | |
| srrABd | 4 | Respiration regulation two-component system | M | |
| hmpd | 3 | Nitric oxide dioxygenase (EC 1.14.12.17) [NADPH] | R | |
| ald1d | 1 | Alanine dehydrogenase (EC 1.4.1.1) [NADPH] | C | |
| Bacillales (11 genomes) | qoxABCD | 2 | Cytochrome aa 3-600 quinol oxidase (EC 1.9.3.-) | E |
| nirS | 1 | Copper-containing nitrite reductase (EC 1.7.2.1) | R | |
| coxAB | 1 | Cytochrome c oxidase [B(O/a)3-type] (EC 1.9.3.1) | E | |
| Lactobacillaceae (13 genomes) | ackA | 2 | Acetate kinase (EC 2.7.2.1) | F |
| PF02525 | 1 | Putative NAD(P)H-quinone dehydrogenase | E | |
| Streptococcaceae (15 genomes) | eno | 10 | Enolase (EC 4.2.1.11) | C |
| ahpCF | 3 | Alkyl hydroperoxide reductase (EC 1.11.1.15) | M | |
| gapN | 2 | Glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.9) [NADP] | C | |
| hemH | 2 | Ferrochelatase, protoheme ferro-lyase (EC 4.99.1.1) | M | |
| Chloroflexi (5 genomes) | hepT | 5 | Heptaprenyl-PP synthase (EC 2.5.1.30), menaquinone biosynthesis | M |
| fpr | 4 | NADPH-ferredoxin reductase (EC 1.18.1.2) | E | |
| asc | 2 | Acetyl-CoA synthetase (EC 6.2.1.1) | C | |
| nuoM3 | 2 | NADH ubiquinone oxidoreductase subunit paralog | E | |
| Actinomycetales (18 genomes) | hemACDd | 14 | Heme biosynthesis | M |
| whiB | 15 | Transcriptional regulator involved in differentiation and sporulation | M | |
| COG0644 | 6 | Putative NAD-dependent reductase | M | |
| resA-ccd-resBC | 3 | Cytochrome c biosynthesis | E | |
| fadD | 3 | Long-chain-fatty-acid–CoA ligase (EC 6.2.13) | M | |
| choD | 3 | Cholesterol oxidase (EC 1.1.3.6) | M | |
| hppA | 3 | Pyrophosphate-energized proton pump (EC 3.6.1.1) | E | |
| pntAB | 2 | NAD(P) transhydrogenase (EC 1.6.1.2) | N | |
| pdhABC | 2 | Pyruvate dehydrogenase (EC 1.2.4.1) | F | |
| cooMSLFDGE | 2 | Carbon monoxide dehydrogenase (EC 1.2.99.2) | E | |
| Desulfovibrionales (10 genomes) | apsBA | 9 | Adenylyl-sulfate reductase (EC 1.8.99.2) | R |
| sat | 9 | Sulfate adenylyltransferase, dissimilatory type (EC 2.7.7.4) | R | |
| adk | 9 | Adenylate kinase (EC 2.7.4.3) | R | |
| ppaC | 9 | Manganese-dependent inorganic pyrophosphatase (EC 3.6.1.1) | R | |
| dsrABD | 8 | Sulfite reductase (EC 1.8.99.1) | R | |
| qrcABCD | 7 | Cytochrome c3, menaquinone oxidoreductase | E | |
| qmoABC | 7 | Quinone membrane-bound oxidoreductase | R | |
| dsrMKJOP | 5 | Sulfite reduction-associated complex | R | |
| dhcA-rnfCDGEAB | 3 | Decaheme cytochrome c, electron transport complex | R | |
| phsAB | 2 | Thiosulfate reductase | M |
Numbers of genomes in the analyzed taxonomic group are indicated in parentheses.
The numbers of genomes in a taxonomic group where the target gene has a conserved Rex-binding site.
Abbreviations for the functional categories: E, energy metabolism (including electron transport chains, hydrogenases, ATP synthase); F, fermentation (includes pyruvate utilization pathways); C, central carbohydrate metabolism (includes glycolytic catabolic pathways); N, NAD(P) biogenesis; R, nitrogen or sulfur reduction; M, miscellaneous pathways.
Rex-dependent regulation of this target was previously known in model organisms.
(iv) Coevolution of Rex regulators and their DNA recognition motifs.
The obtained species-specific DNA recognition motifs of Rex regulators demonstrated significant conservation across the analyzed taxonomic groups of bacteria (Fig. 1). Eight lineage-specific Rex-binding motifs have a common con-sensus (TTGTGAANNNNTTCACAA), the Rex motif in the Deinococcus-Thermus group has a slightly different consensus (TCGTGANNNNNNTCACGA), whereas in two Clostridia groups (Clostridiaceae and Thermoanaerobacterales), the modified Rex-binding motif has consensus TTGTTAANNNNTTAACAA. The observed overall conservation of the Rex recognition DNA motifs in distant phylogenetic groups is in good correlation with high evolutional conservancy of their DNA-binding domains. Using a multiple sequence alignment of 119 Rex regulators and the set of ∼1,000 candidate Rex operators, we computed correlations between amino acid sequences of Rex DNA-binding domains and nucleotide sequences of their DNA-binding sites. As a result, we identified 26 significantly correlated pairs of residues/nucleotides that correspond to 16 positions in Rex proteins and 4 positions in DNA operators (see Fig. S3 in the supplemental material). Only two symmetrical positions in the palindromic Rex-binding motif, T/G5 and A/C14, appear to have the largest number of correlations with the sequences of Rex DNA-binding domains. All correlated residues were mapped on the tertiary structure of the T. aquaticus Rex regulator in complex with its DNA operator (see Fig. S4 in the supplemental material). The most correlated residue, Lys47, is inserted into the major groove of DNA and forms a hydrogen bond with G5 of the DNA operator. Lys47 and Arg46 are the only residues in the DNA recognition helix that form direct hydrogen bonds with bases of the DNA operator, and their functional importance has been validated by Rex protein mutagenesis (17). In contrast to Lys47, which is substituted to Arg/Gln in multiple Rex proteins, Arg46 is an absolutely conserved residue in the Rex family. Further analysis of correlations revealed that the largest contribution to correlation scores comes from two groups of Clostridia that have the most diverged DNA-binding domains of regulators and their operators (Fig. 1). Glu47 was found exclusively in the Rex proteins from Clostridia, and only Rex recognition motifs from Clostridia possess the T5 and A14 nucleotides. In summary, we report a correlation of the specificity-determining position Lys47 (which can be substituted by Arg47 in bacilli) and two contacting bases in a DNA recognition motif, G5 and C14.
Fig 1.
Sequence logos of DNA recognition motifs for Rex regulators and amino acid distribution of correlated residues in their DNA-binding domains. The numbers of analyzed Rex regulators in each lineage are shown in parenthesis. DNA operator sequence logos were constructed based on alignment of all Rex sites identified in each group of genomes (available in the RegPrecise database). Positions of 16 correlated residues in amino acid logos correspond to the Rex protein from Thermus aquaticus, for which the structure of its complex with DNA is available. Amino acid residues contacting with DNA are shown in black.
(v) Conserved and lineage-specific members of Rex regulons.
The reconstructed Rex regulons control mostly genes involved in energy metabolism, central glycolytic pathways, and fermentation. However, the specific content of Rex regulons is highly variable not only between different lineages but also between different species in the same taxonomic group. Below, we provide functional descriptions of genes identified within the conserved and variable parts of Rex regulons.
We defined the core conserved part of the Rex regulons as a set of 22 genes and operons that are preceded by candidate Rex-binding sites in more than one taxonomic group (Table 1). Most of the core regulated genes encode NAD-depending dehydrogenases involved in energy metabolism and electron transport chains (various NADH dehydrogenases), fermentation (alcohol, lactate, propanediol, butanediol dehydrogenases), glycerol metabolism (GldA), and glycolysis (Gap). In addition, the core Rex regulon includes some other pyridine cofactor-independent enzymes involved in respiration (ATP synthase, cytochrome d uniquinol oxidase), glycolysis (Pgk, Tpi, Fba), and fermentation (Pfl). Rex-dependent regulation of the fermentation genes is a characteristic of all five studied lineages from the Firmicutes phylum, whereas the glycolysis genes were found under regulation by Rex in Thermotogales, Clostridiaceae, and Streptococcaceae. The residual part of the Rex regulon core embraces genes with miscellaneous functions, such as transporters (lctP, narK, nirC), NAD biosynthesis genes (nadABC, pncB-nadE), and the rex gene itself. Previously, autoregulation of the rex gene was shown only in S. coelicolor, where rex is cotranscribed with the heme synthesis genes hemACD (4). Here, we demonstrated that Rex regulates expression of its own gene in 51 out of 119 analyzed genomes from seven different lineages (Table 1).
More than 75 predicted target genes and operons form a group of taxon-specific regulon members that are preceded by candidate Rex-binding sites only in the genomes from a single taxonomic group. The taxon-specific members of Rex regulons were found in all studied taxonomic groups, with the only exception being the Deinococcus-Thermus phylum. The largest sets of taxon-specific Rex targets were detected in the groups of Clostridiaceae, Staphylococcaceae, Actinomycetales, and Desulfovibrionales (12 to 15 target operons per group). Other analyzed taxonomic groups have 2 to 6 taxon-specific members of Rex regulons per group. Table 2 summarizes information on 62 taxon-specific Rex regulon members with certain functional annotations. Most of the taxon-specific members of Rex regulons are involved in energy metabolism and electron transport chains, hydrogen production, fermentation, and central carbohydrate metabolism, as well as nitrogen and sulfur reduction pathways. In addition, some taxonomic groups also contain Rex-regulated genes involved in the biosynthetic pathways for menaquinone, heme, and NAD cofactors, the anaerobic arginine catabolism pathway, and the nitric oxide and hyperoxide stress protection.
Reconstruction and experimental validation of the Rex regulon in Thermotogales. (i) Overview of the Thermotoga maritima Rex regulon.
The reconstructed Rex regulon in Thermotoga maritima contains 40 genes organized in 12 putative operons (Table 3). The dnaX-TM0687-gap-pgk-tpi operon encodes three glycolytic enzymes, including the NAD-dependent glyceraldehyde-3-phosphate dehydrogenase Gap, a DNA polymerase III subunit, and a hypothetical protein. The TM1586-gckA and sat-hpr operons encode glycerate-2-kinase, serine-pyruvate aminotransferase, and NADH-dependent hydroxypyruvate reductase that form the serine utilization pathway previously characterized by us in T. maritima (38). The gldA gene encodes NAD-dependent glycerol dehydrogenase. The noxE gene and the TM1420-1-2-3-hydCAB-rex1 operon encode NADH oxidase and NADH-reducing [FeFe]hydrogenase, respectively. The hycABC-TM0009, hycD, and hycE-hycC2 operons encode components of putative quinine-dependent NADH dehydrogenase. The TM1814-TM1807 operon encodes a hypothetical CRISPR-like system possibly implicated in prokaryotic immunity. Finally, the predicted Rex regulon in T. maritima includes the hypothetical genes TM0179 and the putative operon TM0983-TM0979. An unusual feature of T. maritima and other Thermotogales is the presence of two paralogs of Rex encoded in the genome, TM0169 and TM1427. Although by a combination of criteria such as phylogenetic distribution and genomic context of orthologs, TM0169 appeared to be a stringer candidate for the master regulator, the participation of TM1427 in interactions with at least some of the predicted sites could not be ruled out without experimental testing (see below).
Table 3.
Rex-binding sites in Thermotoga spp. validated by fluorescence polarization binding assay
| First gene locus taga | Rex-regulated operon | Rex site positionb | TSS positionc | Rex site sequence | Rex site scored | Kd (nM)e |
|---|---|---|---|---|---|---|
| TM0012 | hycABC-TM0009 | −45 | −74 | TTGTGAAAAAATTCCCGA | 4.91 | 9.4 |
| TM0179 | TM0179 | −30 | −14 | TTGTTATAATAGTAACAA | 4.62 | 12.4 |
| TM0201 | hycD | −54 | −32 | TTGAGAAATTTATCACAA | 5.15 | 9.5 |
| TM0227 | hycE-hycC2 | −33 | −36 | TTGTGAAATTTTTGACGA | 5.08 | 18.4 |
| TM0379 | noxE | −32 | −38 | TCGTGAAAAATTTCTCTT | 4.87 | 10.1 |
| TM0423 | gldA | −52 | −14 | TCGTGAAAAATTTAACAA | 5.38 | 17.9 |
| TM0686 | dnaX-TM0687-gap-pgk-tpi | −104 | −32 | TAGTTATTTTTTTCACGA | 5.02 | 17.7 |
| TM0983 | TM0983-979 | −41 | −46 | TTGAGATTTTTTTCACAA | 5.03 | 20.8 |
| TM1400 | sat-hpr | −47 | −142 | TAGTTAAAAAAATAACAA | 5.01 | 23.4 |
| TM1420 | TM1420-1423–hydCAB-rex1 | −45 | −69 | TAGTGAAAAATATAACGT | 4.91 | 26.8 |
| TM1586 | TM1586-gckA | −177 | −161 | GAGTAAAATATTTCACAA | 4.78 | 5.6 |
| TM1814 | TM1814-1807 | −40 | −38 | ATGATAAAATTATCACAA | 4.85 | 8.9 |
| TRQ2_0578 | adhE | −85 | GTGAGAAAATTTTCACAA | 4.95 | 16.6 | |
| −61 | TTGTTATGAAATTCACAA | 4.70 | 11.8 |
Genes from the genomes of T. maritima MSB8 and Thermotoga sp. RQ-2 are shown with the prefixes TM and TRQ2, respectively.
Positions of transcription start sites determined by 5′ RACE are given relative to the translational start.
Positions of the 5′ end of Rex sites are given relative to the translational start.
Scores of Rex sites were calculated by the RegPredict program as the sum of positional nucleotide weights using the Thermotoga Rex-binding site recognition profile.
Apparent Kd values for Rex protein interacting with the tested DNA fragments were measured by fluorescence polarization assay.
(ii) Transcription start sites of Rex-regulated operons in T. maritima.
To determine the position of predicted Rex-binding sites in relation to the promoter regions of the candidate Rex regulon genes, the transcription start sites (TSSs) were detected by the 5′ RACE method in T. maritima. The determined TSSs for 12 Rex target operons were located between 14 and 161 bp upstream of the translational gene starts of the proximal target genes (Table 3). To cross-map the identified TSSs, promoter elements, and Rex operators for each predicted Rex-regulated gene in T. maritima, we performed multiple alignment of orthologous upstream gene regions from five related genomes of Thermotoga spp. (see Fig. S5 in the supplemental material). For most target genes, the identified Rex operators either overlap with or are located downstream of the putative conserved promoter motifs (−35 and −10 boxes), suggesting that Rex is a repressor of these genes. The only exception from this rule was the TM0686 gene, a first gene in the candidate dnaX-TM0687-gap-pgk-tpi operon, which has a Rex operator located 72 bp upstream of its determined TSS. The functional significance of this alternative localization (often detected for transcriptional activators), which is conserved in all Thermotoga spp., is unknown. Interestingly, other Thermotogales (Thermosipho, Fervidobacterium, Petrotoga spp.) have orthologs of the dnaX gene without Rex regulation; however, the gap-pgk-tpi operons in these genomes are preceded by candidate Rex-binding sites located closely to the translational start site.
(iii) T. maritima Rex binds to its cognate binding sites in vitro.
We used in vitro binding assays to address three main goals: (i) experimentally test all predicted Rex regulatory sites in T. maritima; (ii) compare the abilities of two Rex paralogs, TM0169 and TM1427, to interact with predicted DNA motifs and, thus, establish which of the two proteins constitutes a master Rex regulator in Thermotogales; and (iii) test whether the NADH/NAD+ balance retains its role as an effector in the evolutionary remote branch of Thermotogales. For this purpose, we cloned and overexpressed in E. coli both genes, TM0169 and TM1427, as fusions with Smt3-His6 tags, purified both recombinant proteins, and explored their ability to bind to the predicted DNA operators by fluorescence polarization (FP) assay and electrophoretic mobility shift assay (EMSA) at 22°C.
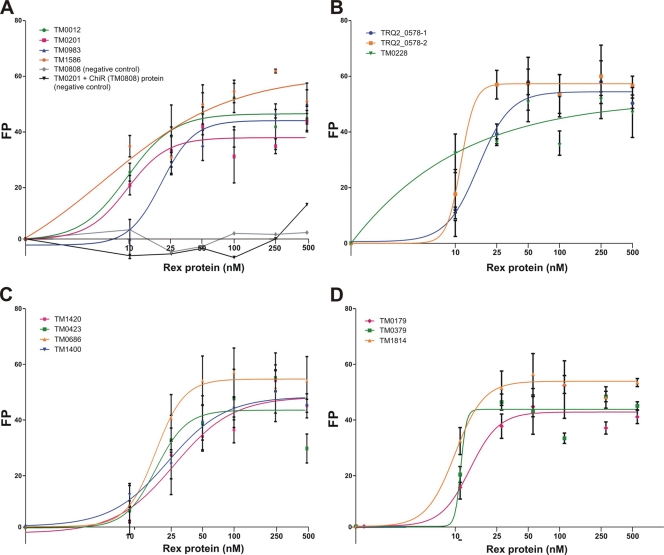
First, we used the FP assay to assess the specific binding of TM0169 and TM1427 proteins to all 12 predicted Rex operators in T. maritima (Table 3). In addition, we tested two Rex sites predicted in the upstream region of the adhE gene in the closely related strain Thermotoga sp. RQ-2, since this gene is missing from the T. maritima genome. The results show that the recombinant TM0169 (Rex) protein can specifically bind to the synthetic 28-bp DNA fragments containing Rex operators. All tested DNA fragments demonstrated a Rex concentration-dependent increase of fluorescence polarization, confirming specific interaction between the Rex protein and DNA fragments (Fig. 2). As negative controls, we assessed the binding of another T. maritima regulator (TM0808) with the Rex target DNA fragments, as well as the Rex protein with the TM0808 target DNA fragment. The apparent dissociation constant (Kd) values for the Rex protein interacting with the tested DNA fragments were in the range of 5 to 27 nM (Table 3).
Fig 2.
Fluorescence polarization binding assay of Rex (TM0169) with target DNA operators in T. maritima MSB8 and Thermotoga sp. RQ-2. Increasing Rex protein concentrations (10 to 500 nM) were mixed with 28-bp fluorescence-labeled DNA fragments (1 nM) of gene regions containing Rex-binding sites: TM0012, TM0201, TM0983, TM1586 (A); TRQ2-0578 (1), TRQ2-0578 (2), TM0228 (B); TM1420, TM0423, TM0686, TM1400 (C); TM0179, TM0379, TM1814 (D). As a negative control, the T. maritima regulator ChiR (TM0808) and DNA fragment of the TM0808 gene containing its cognate DNA-binding operator were used. The values are means from three measurements, and the standard deviations are shown. The linear regression with variable slope method from the GraphPad Prism software was used to plot the curves.
Second, we performed EMSA experiments to test the influence of the NADH/NAD+ effectors on the interaction between the recombinant TM0169 (Rex) protein and its cognate operators. We chose EMSA instead of the FP method because NADH may quench fluorescence signals. Binding to a 28-nucleotide DNA fragment containing a high-scored Rex-binding site at the TM0201 gene was assessed. A substantial shift of the DNA band was observed with the T. maritima Rex protein at concentrations of 50 nM or higher (Fig. 3A). In the second EMSA experiment, interaction between the same DNA fragment (0.1 nM) and the Rex protein (50 nM) was assessed in the presence of variable concentrations of NAD+ and NADH (Fig. 3B). The presence of 2 μM NADH significantly decreased Rex binding to DNA (Fig. 3B, lane 3); however, the increasing concentrations of NAD+ (up to 2 mM) did not interfere with the Rex-DNA complex formation (Fig. 3B, lanes 8 to 11). To test the effect of the NADH/NAD+ ratio on the Rex-DNA complex, the increasing NAD+ concentrations were used on the background of 2 μM NADH (Fig. 3B, lanes 4 to 7). As a result, Rex binding to DNA was restored at NAD+ concentrations of 100 μM or higher. This corresponded to ∼2% NADH as a proportion of the total NAD(H) cofactor pool. We concluded that (i) NADH, but not NAD+, can actively dissociate Rex that is bound to its DNA target and (ii) Rex responds to the redox poise of the NAD(H) cofactor pool rather than NADH itself. These results confirm that the T. maritima Rex protein has regulatory characteristics similar to those of the previously described Rex proteins from B. subtilis and S. coelicolor (4, 35).
Fig 3.
EMSA with T. maritima Rex (TM0169) protein and TM0201 DNA fragment. (A) Titration of the protein in the absence of an effector. EMSA was performed in the absence (lane 1) and in the presence (lanes 2 to 6) of increasing Rex protein concentrations (5, 12, 25, 50, and 100 nM, respectively). (B) Assessment of effector influence. EMSA was performed with the same DNA fragment in the absence (lane1) or the presence (lanes 2 to 11) of 50 nM Rex protein. To assign the influence of the NADH/NAD+ ratio, EMSA was performed with different concentrations of NADH/NAD+: lane 2, no NADH/NAD+; lane 3, 2 μM NADH; lanes 4 to 7, 2 μM NADH plus the increasing NAD+ concentrations, respectively, 0.05, 0.1, 0.5, and 2 mM (correspond to 4%, 2%, 0.4%, and 0.1% NADH/NAD+ ratio); lanes 8 to 11, the increasing NAD+ concentrations, respectively, 0.05, 0.1, 0.5, and 2 mM.
(iv) Rex-binding motif corresponds to the previously described GK box.
Our previous analysis of the T. maritima genome revealed an 18-bp palindromic motif (termed GK box, for glycerate kinase regulation) located upstream of the serine, glycerol, and glucose utilization operons sat-hpr, TM1586-gckA, gldA, and dnaX-TM0687-gap-pgk-tpiA (38). In this study, the combined comparative genomics and experimental techniques allow us to identify these and other T. maritima operons as members of the redox-controlled Rex regulon and to assign the predicted GK box motif as a binding motif of the Rex transcription factor.
(v) Potential functional role of the Rex1 paralog in Thermotogales.
Multiple sequence alignment and phylogenetic analysis of the Rex protein family demonstrated high divergence of Rex1 paralogs in Thermotogales, suggesting their possible functional diversification. In contrast to the main branch of Rex proteins that are present in all studied Thermotogales genomes, Rex1 paralogs were not found in two Thermotogales genomes: Petrotoga mobilis and Kosmotoga olearia. Since Rex1 is encoded by the last gene in the hyd operon in T. maritima (TM1420-TM1427) and this genomic context is conserved in multiple genomes, we hypothesized that Rex1 possibly operates as a local regulator of this operon. Comparison of upstream regions of TM1420 and its orthologs allowed us to predict a conserved DNA motif with consensus TTGTGAANNNNGTAACAA that is similar to the previously validated Rex-binding motif (see Fig. S5 in the supplemental material). The putative Rex1 site is located upstream of the validated Rex-binding site and overlaps the −10 promoter element of the TM1420 gene. We decided to test the possibility that this novel upstream motif can serve as a binding site of Rex1. We cloned, expressed, and purified the Rex1 (TM1427) protein to test its binding ability to the predicted upstream motif. However, the FP experiments conducted with the recombinant Rex1 protein did not show any specific binding with the predicted DNA fragment. Then, we performed EMSA experiments to test specific binding of Rex1 and Rex with the 200-bp DNA fragment of the TM1420 upstream region containing both the confirmed Rex-binding site and the predicted Rex1 site. Under the conditions tested (room temperature), Rex demonstrated a specific electromobility shift of the tested 200-bp DNA fragment at 100 nM, whereas Rex1 did not show a shift at concentrations of up to 1 μM (see Fig. S6 in the supplemental material). These results confirm that the TM0169 protein is a master Rex regulator in Thermotoga; however, further experiments will be needed for assessing the potential role of TM1427/Rex1 in the control of the hyd operon in Thermotoga.
DISCUSSION
NAD(H) is an indispensable metabolic cofactor in hundreds of redox reactions. It plays the central role in electron transfer from the oxidized substrates to the electron transport chain. Transcriptional regulation of gene expression in response to changes in cellular NADH/NAD+ is an important mechanism to control redox metabolism in bacteria. The Rex repressor plays a key role in NADH/NAD+ sensing and transcriptional control in several model microorganisms, including S. coelicolor (4), B. subtilis (11, 15, 32, 35), and S. aureus (25). However, apart from these few model species, where the repressor controls expression of 3 to 18 operons, Rex regulons in other bacteria were mostly uncharacterized. For instance, despite the knowledge of tertiary structures of Rex proteins from T. aquaticus (33, 37) and T. thermophilus (20), their cellular targets remained unknown. The availability of complete genomes of hundreds of bacterial species opens an opportunity to perform bioinformatics analyses of the genomic sequences with the aim of identifying transcriptional regulatory sites and inferring new regulatory interactions. Here, we applied the comparative genomics approach to reconstruct the Rex regulons in the sequenced genomes encoding this transcription factor. We demonstrated high conservation of Rex proteins and their DNA operator motifs that are opposed to high flexibility in the repertoires of Rex-regulated genes between genomes. Furthermore, we used the in vitro DNA-binding assays to validate the novel Rex regulon identified in the hyperthermophilic bacterium Thermotoga maritima.
The Rex family of transcriptional regulators is highly conserved in many distantly related taxonomic groups of bacteria that include aerobic, anaerobic, and facultative-anaerobic chemoheterotrophs but exclude chemoautotrophs (see Table S2 in the supplemental material). Comparative genomics reconstruction of regulatory interactions in 114 microbial genomes demonstrated that Rex regulates key enzymes involved in energy metabolism and redox status balance. Conservation of key residues involved in protein-DNA interaction in Rex proteins correlates with the observed strong similarity of DNA-binding regulatory motifs across bacterial genomes. On the other hand, the content of Rex regulons is rapidly changing during the evolution of taxonomic groups, lineages, or even within a single genus, being most likely under the influence of different lifestyles and ecological niches occupied by the individual species. Among 11 analyzed taxonomic groups, there are no genes with absolutely conserved Rex regulation. The core Rex regulon was defined as a set of 22 genes/operons that are regulated by Rex in several genomes from at least two different taxonomic groups (Table 1). The rest of the predicted Rex regulon members constitute the group of lineage-specific targets, which is at least three times larger than the core (Table 2). The average size of Rex regulons constitutes 2 to 4 operons per genome in the Thermus-Deinococcus, Lactobacillaceae, Bacillales, and Actinomycetales lineages, whereas in other studied lineages the average regulon size is 7 to 10 operons (Fig. 4A). The size of reconstructed Rex regulons often varies between genomes within the same taxonomic group. For instance, the Rex regulon in Thermus includes only one target operon, nuo, which was determined in this study for the first time, whereas the related Deinococcus genus has two additional Rex-regulated operons, ndh and nadCA. Another example of the high variability of Rex regulons is represented by the Actinomycetales lineage. The largest Rex regulons in this group (10 target operons per genome) are present in Streptomyces spp., whereas the smallest Rex regulons (2 operons per genome) were found in Frankia and Nocardia spp. Among Clostridium spp., the reconstructed Rex regulons in C. butyricum and C. scindens contain 16 and 2 targets, respectively, whereas a single Clostridia genome, Clostridium sp. L2-50, lacks both the Rex regulator ortholog and candidate Rex-binding sites. These observations confirm the high flexibility of the Rex transcriptional networks.
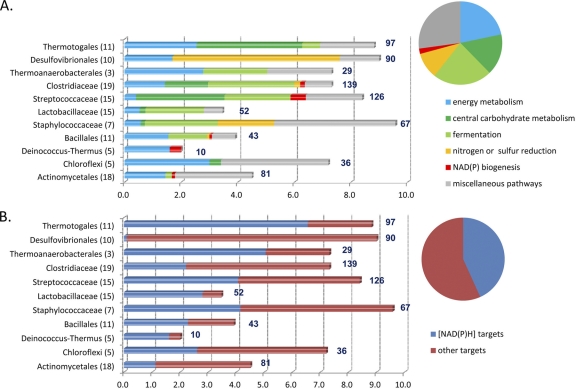
Fig 4.
Breakdown of the average numbers of Rex-regulated operons per genome by functional categories (A) and NAD(P)H dependence (B) of enzymes encoded in the target operons. The numbers of genomes in the analyzed taxonomic group are indicated in parentheses. The total numbers of identified Rex-regulated operons in each taxonomic group are indicated at the right of each bar. Pie charts show the breakdown by functional categories calculated for the total number of Rex-regulated operons in all taxonomic groups.
To understand the role of Rex-dependent regulation in microbial cell physiology, we analyzed functional categories and the cofactor dependency of the predicted Rex-regulated enzymes (Fig. 4). Overall, three-quarters of Rex targets can be attributed to one of five functional categories: (i) energy metabolism, including numerous dehydrogenases from electron transport chains, hydrogenases, and the ATP synthase complex; (ii) carbohydrate metabolism, including the glycolysis and the peripheric sugar catabolic pathways; (iii) fermentation pathways producing ethanol, lactate, formate, acetate, and butanol/butyrate; (iv) nitrate/nitrite and sulfate/sulfite reduction pathways; and (v) NAD(P)H biogenesis pathways, including the de novo cofactor biosynthesis and salvage pathways and the NAD(P) transhydrogenase enzyme. At that, 42% of Rex targets encode NAD(P)H-dependent enzymes. However, the distribution of functional gene categories controlled by Rex varies substantially between 11 analyzed taxonomic groups (Fig. 4). The observed functional variability of Rex regulons can be attributed to the existence of a large variety of NADH/NAD+-utilizing redox enzymes that can potentially influence the cellular NADH/NAD+ balance.
The tendency of Rex to control NAD(P)H-dependent processes is most pronounced in the Thermus-Deinococcus and Lactobacillaceae groups. The core Rex regulon in the former group of aerobic heterotrophs includes membrane-bound NADH dehydrogenases (Nuo, Ndh), key components of the oxidative respiratory chain. In contrast, the core Rex regulon in the latter group of facultative anaerobes includes various fermentation enzymes (Adh, Ldh, Dha, But), in agreement with the fact that many lactobacilli are associated with fermentations of plants, meat, or milk. The core Rex regulons in facultative anaerobic bacteria from the Bacillales and Streptococcaceae groups share NADH-specific l-lactate dehydrogenase (Ldh) that plays a key role in their fermentative metabolism. Only in the Streptococcaceae group, the Rex regulon is expanded to control central enzymes from the glycolysis pathway (Pgk, Tpi, Fba, Eno, Gap). This lineage-specific regulon expansion can be explained by the loss of the central glycolytic gene repressor CggR in the Streptococcaceae genomes.
The hydrogen-producing Thermotogales, Thermoanaerobacterales, and, partially, Clostridiaceae groups have similar Rex regulon cores that include NADH-specific hydrogenase (Hyd), NADH oxidase (Nox), and various alcohol dehydrogenases (Adh). Rex regulons in the latter two groups of Firmicutes also share ferredoxin-NAD(P) oxidoreductase (Fnor), which is important for the pyruvate oxidation pathway through puruvate:ferredoxin oxidoreductase. Thus, Rex plays an important role in the regulation of end-product pathways of carbohydrate catabolism in these anaerobic organotrophs. In addition, Thermotogales and Clostridiaceae share several glycolytic enzymes (Gap, Pgk, Tpi) in their Rex regulons. The observed large overlap between Rex regulons in the phylogenetically distinct groups of Thermotogales and thermophilic Clostridia could be explained by the fact that these species share a large proportion of genes that were likely acquired by massive horizontal gene transfer in the past (40).
Surprisingly, the Rex regulon in the Desulfovibrionales lineage does not include NAD(P)H-dependent enzymes; instead, its core contains ATP synthase and multiple enzymes constituting the sulfate reduction pathway (Sat, PpaC, ApsAB, Adk, DsrABD). This dramatic shift in the Rex regulon content could be linked to a special lifestyle of the sulfate-reducing Deltaproteobacteria that couples the pathway of oxidation of di- and tricarboxylic acids (i.e., malate, lactate) with the sulfate reduction pathway. In this metabolic process, energy is conserved due to electron transfer from NAD(P)H to the adenylyl phosphosulfate reductase ApsAB (14). However, the NoxE-like NADH oxidase required for the complete reduction of ApsAB by NADH (DVU3212 in Desulfovibrio vulgaris) (5) does not belong to the predicted Rex regulon in Desulfovibrio. The effector molecule of the Rex regulator in Desulfovibrio is not yet experimentally determined, although our comparative analysis of Rex proteins suggests that their ability to sense NADH should be preserved in this lineage. Validation experiments for the predicted Rex-dependent regulation in D. vulgaris are under way.
For the first time, we identified novel functional links between Rex-dependent regulation and pyridine cofactor metabolism. The NAD biosynthetic genes nadABC and pncB-nadE belong to the candidate Rex regulons in several genomes from four studied taxonomic groups (Fig. 4A). Moreover, the Rex regulon in Rhodococcus spp. includes the pyridine nucleotide transhydrogenase PntAB. The most relevant explanation for these interesting observations is that the cellular NADH/NAD+ ratio can be balanced not only via NADH oxidation but also by an increase of NAD+ concentration due to activation of the de novo cofactor biosynthesis or by transhydrogenation between NADH and NADP cofactors. Another novel finding of this work is that Rex can control transcription of various glycolytic enzymes in the Thermotogales, Clostridiaceae, and Streptococcaceae lineages. Among these enzymes, there are two enzymes that use NAD or NADP as cofactors (Gap and GapN), whereas most other Rex-controlled glycolytic enzymes are independent of NAD(P) cofactors. The metabolic implication of Rex-dependent derepression of glycolytic enzymes under the condition of a high NADH/NAD+ ratio is not clear. Connections with NADH/NAD balance are not clear for several other novel members of reconstructed Rex regulons, including the menaquinone biosynthesis enzyme HepT in Chloroflexi and a WhiB family transcriptional regulator required for differentiation and sporulation in Actinomycetales. Future in vivo metabolic and expression experiments will allow the researchers to uncover this and other puzzles on Rex-dependent regulation of cellular metabolism that were emphasized in this study.
Experimental testing of selected bioinformatic predictions (binding sites) provides an additional validation strategy that was used by us in some of the previous studies on genomic reconstruction of regulatory networks (16, 29–31). In this study, we chose to test the novel Rex regulon in T. maritima because of its several unique features. First, this is a model hyperthermophilic species from the deep-branched Thermotogales phylum that was recognized by its ability to produce hydrogen by fermenting a wide range of carbohydrates and thus was intensively studied (6). Fermentative hydrogen production in Thermotoga is catalyzed by multiple iron-containing hydrogenases that oxidize NADH generated during glucose catabolism. Thus, understanding the NADH-dependent metabolic regulation of the hydrogen production genes by Rex is an important goal. Second, another unique feature of the Thermotogales genomes is that they contain two highly diverged rex paralogs. The amino acid sequence comparison and genome context analysis suggest that one of the two Rex paralogs (TM0169 in T. maritima) is a primary candidate for the role of a global regulator. The second paralog, termed Rex1 (e.g., TM1427 in T. maritima) has numerous mutations in putative DNA- and NAD-binding domains and is missing in two Thermotogales genomes. Since rex1 genes are genomically linked to the iron-hydrogenase hydCBA genes, we speculate that Rex1 could play the role of a local regulator for this operon. Experimental testing of both Rex family proteins in T. maritima confirmed that TM0169 (but not TM1427) is the global transcription factor that senses the intracellular NADH/NAD+ balance and binds the operator sites located in promoter regions of multiple genes. Under the conditions tested (room temperature), the Rex1 paralog interacted neither with the Rex binding sites nor with the 200-bp promoter region of the hydCBA-containing operon, and thus its regulatory role in T. maritima remains unclear.
The Rex-dependent regulation was previously studied experimentally in several model species from various taxonomic groups, including S. aureus from Staphylococcaceae, B. subtilis from Bacillales, S. coelicolor from Actinomycetales, and T. ethanolicus from Thermoanaerobacterales. Using the comparative genomics approach, we identified a large number of novel members of Rex regulons in each of these lineages, e.g., one gene in Staphylococcaceae, seven operons in Bacillales, nine operons in Thermoanaerobacterales, and 12 operons in Actinomycetales. Although the Rex regulator in Thermus was intensively analyzed in previous studies, for the first time in this work we were able to determine its target operon, nuo, encoding NADH ubiquinone dehydrogenase. Finally, we identified a large number of Rex-regulated genes in the species from six other taxonomic groups, where regulation by Rex has not been analyzed experimentally (see Tables 1 and 2). In conclusion, the genomic reconstruction of Rex regulons over 119 genomes from 11 taxonomic groups of bacteria provides a framework for further studies of the Rex-dependent redox regulation in bacteria. Specifically in Thermotogales, it opens opportunity to exploit a biotechnological potential of these species for biohydrogen production.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the U.S. Department of Energy, Office of Science (Biological and Environmental Research), as part of Genomic Science Program contracts DE-FG02-08ER64686 with SBMRI and UCSD and DE-SC0004999 with SBMRI and LBNL. Additional funding was provided by National Science Foundation grant DBI-0850546, the Russian Foundation for Basic Research grant 10-04-01768, and the Russian Academy of Sciences under the Molecular and Cellular Biology program.
Footnotes
Published ahead of print 30 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alkema WB, Lenhard B, Wasserman WW. 2004. Regulog analysis: detection of conserved regulatory networks across bacteria: application to Staphylococcus aureus. Genome Res. 14:1362–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bitoun JP, Nguyen AH, Fan Y, Burne RA, Wen ZT. 2011. Transcriptional repressor Rex is involved in regulation of oxidative stress response and biofilm formation by Streptococcus mutans. FEMS Microbiol. Lett. 320:110–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brekasis D, Paget MS. 2003. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). EMBO J. 22:4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen L, Le Gall J, Xavier AV. 1994. Purification, characterization and properties of an NADH oxidase from Desulfovibrio vulgaris (Hildenborough) and its coupling to adenylyl phosphosulfate reductase. Biochem. Biophys. Res. Commun. 203:839–844 [DOI] [PubMed] [Google Scholar]
- 6. Conners SB, et al. 2006. Microbial biochemistry, physiology, and biotechnology of hyperthermophilic Thermotoga species. FEMS Microbiol. Rev. 30:872–905 [DOI] [PubMed] [Google Scholar]
- 7. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dehal PS, et al. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38:D396–D400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 11. Gyan S, Shiohira Y, Sato I, Takeuchi M, Sato T. 2006. Regulatory loop between redox sensing of the NADH/NAD(+) ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J. Bacteriol. 188:7062–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hung YP, Albeck JG, Tantama M, Yellen G. 2011. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 14:545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huson DH, et al. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinform. 8:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kremer DR, Nienhuis-Kuiper HE, Timmer CJ, Hansen TA. 1989. Catabolism of malate and related dicarboxylic acids in various Desulfovibrio strains and the involvement of an oxygen-labile NADPH dehydrogenase. Arch. Microbiol. 151:34–39 [Google Scholar]
- 15. Larsson JT, Rogstam A, von Wachenfeldt C. 2005. Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis. Microbiology 151:3323–3335 [DOI] [PubMed] [Google Scholar]
- 16. Leyn SA, et al. 2011. Control of proteobacterial central carbon metabolism by the HexR transcriptional regulator: a case study in Shewanella oneidensis. J. Biol. Chem. 286:35782–35794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McLaughlin KJ, et al. 2010. Structural basis for NADH/NAD+ redox sensing by a Rex family repressor. Mol. Cell 38:563–575 [DOI] [PubMed] [Google Scholar]
- 18. Mironov AA, Vinokurova NP, Gel'fand MS. 2000. Software for analyzing bacterial genomes. Mol. Biol. 34:253–262 [PubMed] [Google Scholar]
- 19. Mossessova E, Lima CD. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5:865–876 [DOI] [PubMed] [Google Scholar]
- 20. Nakamura A, Sosa A, Komori H, Kita A, Miki K. 2007. Crystal structure of TTHA1657 (AT-rich DNA-binding protein; p25) from Thermus thermophilus HB8 at 2.16 A resolution. Proteins 66:755–759 [DOI] [PubMed] [Google Scholar]
- 21. Novichkov PS, et al. 2010. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38:D111–D118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Novichkov PS, et al. 2010. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 38:W299–W307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Osterman AL, et al. 1995. Domain organization and a protease-sensitive loop in eukaryotic ornithine decarboxylase. Biochemistry 34:13431–13436 [DOI] [PubMed] [Google Scholar]
- 24. Overbeek R, et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1,000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pagels M, et al. 2010. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 76:1142–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pei J, et al. 2011. The mechanism for regulating ethanol fermentation by redox levels in Thermoanaerobacter ethanolicus. Metab. Eng. 13:186–193 [DOI] [PubMed] [Google Scholar]
- 27. Qiu Y, et al. 2010. Structural and operational complexity of the Geobacter sulfurreducens genome. Genome Res. 20:1304–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodionov DA. 2007. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem.Rev. 107:3467–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodionov DA, et al. 2008. Transcriptional regulation of NAD metabolism in bacteria: NrtR family of Nudix-related regulators. Nucleic Acids Res. 36:2047–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodionov DA, et al. 2008. Transcriptional regulation of NAD metabolism in bacteria: genomic reconstruction of NiaR (YrxA) regulon. Nucleic Acids Res. 36:2032–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodionov DA, et al. 2011. Comparative genomic reconstruction of transcriptional networks controlling central metabolism in the Shewanella genus. BMC Genomics 12(Suppl. 1):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schau M, Chen Y, Hulett FM. 2004. Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon. J. Bacteriol. 186:4585–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sickmier EA, et al. 2005. X-ray structure of a Rex-family repressor/NADH complex insights into the mechanism of redox sensing. Structure 13:43–45 [DOI] [PubMed] [Google Scholar]
- 34. Titolo S, Brault K, Majewski J, White PW, Archambault J. 2003. Characterization of the minimal DNA binding domain of the human papillomavirus e1 helicase: fluorescence anisotropy studies and characterization of a dimerization-defective mutant protein. J. Virol. 77:5178–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang E, et al. 2008. Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol. Microbiol. 69:466–478 [DOI] [PubMed] [Google Scholar]
- 36. Wang E, et al. 2011. Small-angle X-ray scattering study of a Rex family repressor: conformational response to NADH and NAD+ binding in solution. J. Mol. Biol. 408:670–683 [DOI] [PubMed] [Google Scholar]
- 37. Wood MJ, Storz G. 2005. Oxygen, metabolism, and gene expression: the T-Rex connection. Structure 13:2–4 [DOI] [PubMed] [Google Scholar]
- 38. Yang C, Rodionov DA, Rodionova IA, Li X, Osterman AL. 2008. Glycerate 2-kinase of Thermotoga maritima and genomic reconstruction of related metabolic pathways. J. Bacteriol. 190:1773–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao Y, et al. 2011. Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab. 14:555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhaxybayeva O, et al. 2009. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc. Natl. Acad. Sci. U. S. A. 106:5865–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.