Abstract
“Candidatus Chloracidobacterium thermophilum” is a recently discovered chlorophototroph from the bacterial phylum Acidobacteria, which synthesizes bacteriochlorophyll (BChl) c and chlorosomes like members of the green sulfur bacteria (GSB) and the green filamentous anoxygenic phototrophs (FAPs). The pigments (BChl c homologs and carotenoids), quinones, lipids, and hopanoids of cells and chlorosomes of this new chlorophototroph were characterized in this study. “Ca. Chloracidobacterium thermophilum” methylates its antenna BChls at the C-82 and C-121 positions like GSB, but these BChls were esterified with a variety of isoprenoid and straight-chain alkyl alcohols as in FAPs. Unlike the chlorosomes of other green bacteria, “Ca. Chloracidobacterium thermophilum” chlorosomes contained two major xanthophyll carotenoids, echinenone and canthaxanthin. These carotenoids may confer enhanced protection against reactive oxygen species and could represent a specific adaptation to the highly oxic natural environment in which “Ca. Chloracidobacterium thermophilum” occurs. Dihydrogenated menaquinone-8 [menaquinone-8(H2)], which probably acts as a quencher of energy transfer under oxic conditions, was an abundant component of both cells and chlorosomes of “Ca. Chloracidobacterium thermophilum.” The betaine lipid diacylglycerylhydroxymethyl-N,N,N-trimethyl-β-alanine, esterified with 13-methyl-tetradecanoic (isopentadecanoic) acid, was a prominent polar lipid in the membranes of both “Ca. Chloracidobacterium thermophilum” cells and chlorosomes. This lipid may represent a specific adaptive response to chronic phosphorus limitation in the mats. Finally, three hopanoids, diploptene, bacteriohopanetetrol, and bacteriohopanetetrol cyclitol ether, which may help to stabilize membranes during diel shifts in pH and other physicochemical conditions in the mats, were detected in the membranes of “Ca. Chloracidobacterium thermophilum.”
INTRODUCTION
Green bacteria are chlorophyll (Chl)-based phototrophs (i.e., chlorophototrophs) that occur in three phyla within the domain Bacteria. They include green sulfur bacteria (GSB) in the phylum Chlorobi, filamentous anoxygenic phototrophs (FAPs) belonging to the genera Chloroflexus, Chloronema, Chlorothrix, and Oscillochloris in the phylum Chloroflexi, and “Candidatus Chloracidobacterium thermophilum,” a recently discovered member of the phylum Acidobacteria (9–11). These microorganisms share two unique properties: they synthesize bacteriochlorophyll (BChl) c, d, or e and they utilize these BChls to assemble light-harvesting organelles called chlorosomes (9–11, 15, 54).
Chlorosomes are large sac-like organelles that characteristically contain large numbers of BChl c, d or e molecules (>200,000 per chlorosome) (15, 52). Because of the very large number of light-harvesting molecules and the resulting high antenna-to-reaction-center ratio, chlorosomes confer a remarkable ability to harvest light at low irradiance levels. Organisms that produce these organelles are found in environments that cannot support the growth of other chlorophototrophs (3, 15, 46). Chlorosomes are surrounded by a protein-stabilized (glyco)-lipid-containing monolayer membrane and, in addition to BChls, contain carotenoids, quinones, and wax esters. In the GSB Chlorobaculum tepidum, the chlorosome envelope contains 10 proteins that belong to four structure motif families (75). The chlorosome envelopes of “Ca. Chloracidobacterium thermophilum” contain seven different proteins, but only two of these have orthologs in C. tepidum (23). Both C. tepidum and the FAP Chloroflexus aurantiacus synthesize menaquinone species; C. aurantiacus synthesizes menaquinone-10, whereas C. tepidum synthesizes menaquinone-7 (13, 15, 19, 31). GSB strains also synthesize a derivative of menaquinone, chlorobiumquinone, which uniquely occurs in chlorosomes (19) and which probably functions in redox-dependent quenching of energy transfer (15, 19). Redox-dependent quenching of energy transfer also occurs in C. aurantiacus, but the magnitude of quenching under oxic conditions is smaller (6, 7, 19; A. M. Garcia Costas, unpublished observations).
A striking feature of BChls c, d, and e is their ability to self-aggregate and form supramolecular structures without the aid of proteins. This property is due to the absence of the 132-methylcarboxyl group, which would interfere with hydrogen bonding between neighboring BChls, and to the presence of a 31-hydroxyl group that coordinates the magnesium atom of an adjacent BChl molecule (21, 27, 43). In addition to these unique properties, BChls within chlorosomes also exhibit considerable heterogeneity in their molecular structures. In C. tepidum, a variety of alkyl groups can occur at the C-82 and C-121 positions of BChl c, and an individual chlorosome typically contains a complex mixture of BChl c molecules that arise from differences in methylation at these positions (28). Additional heterogeneity arises because of the occurrence of both R- and S-chirality at the C-31 carbon atom (27). This molecular heterogeneity increases the local structural disorder in the BChl aggregates in the suprastructure, and the resulting inhomogeneous line broadening increases the range of wavelengths that can be absorbed by chlorosomes (21, 28). In contrast, C. aurantiacus does not methylate its BChl c at the C-82 and C-121 positions, and as a result, its chlorosomes have spectroscopic properties that differ from those of C. tepidum. Another source of diversity in the BChls in chlorosomes is the esterifying alcohol of the C-17 propionic acid side chain. The majority (>95%) of the BChl c molecules in most GSB are esterified with the C-15 isoprenoid farnesol (12, 50, 70). However, BChl c molecules in FAPS are esterified with alcohols that include both isoprenoid and nonisoprenoid alkyl chains (5, 26, 27, 41, 43, 45).
Chlorosomes also contain large amounts of carotenoids, which have the ability to quench the triplet state of chlorophylls and the singlet state of oxygen and, thus, play a role in photoprotection (15, 17). They may additionally provide structural stability and expand the range of wavelengths that can be absorbed. The carotenoid contents of chlorosomes differ among green bacteria and can even vary within the members of one genus. Chlorobactene, an aromatic derivative of γ-carotene, is the major carotenoid found in chlorosomes of C. tepidum (15, 17). However, brown-colored, BChl e-containing GSB such as Chlorobium phaeobacteroides usually produce aromatic bicyclic carotenoids, isorenieratene and β-isorenieratene, instead of chlorobactene (32, 48, 49, 64). In addition to chlorobactene or isorenieratene, both green- and brown-colored GSB also synthesize small amounts of glycosylated carotenoids (47, 65). Under anoxic, phototrophic growth conditions, C. aurantiacus mainly synthesizes γ-carotene and β-carotene (15, 55, 59, 63).
We recently isolated chlorosomes from “Ca. Chloracidobacterium thermophilum,” described their ultrastructure and spectroscopic properties, and identified the proteins of the chlorosome envelope (23). However, little is known about other chlorosome properties of this unusual organism and what role, if any, these properties may play in its ability to perform chlorophototrophy under oxic conditions. Here, we describe the other components of the chlorosomes and membranes of “Ca. Chloracidobacterium thermophilum,” namely the BChl c homologs, carotenoids, quinones, and lipids. Additionally, we show that “Ca. Chloracidobacterium thermophilum” synthesizes hopanoids, which possibly contribute to the tolerance of this organism for alkaline pH and elevated temperature.
MATERIALS AND METHODS
Culture conditions.
“Ca. Chloracidobacterium thermophilum” was cultured in a HEPES-buffered medium supplemented with reduced organic carbon sources and ammonia as the nitrogen source, as previously described (9, 24). Cultures were incubated at 53°C with gentle shaking and harvested after 5 days, when BChl c absorption was maximal. “Ca. Chloracidobacterium thermophilum” cells were separated from other members of the enrichment by differential centrifugation as previously described (22, 24). Escherichia coli was grown anoxically at 37°C in Luria-Bertani medium in screw-cap test tubes with no air space.
Chlorosome isolation.
For the isolation of chlorosomes from “Ca. Chloracidobacterium thermophilum,” cells were harvested in chlorosome isolation buffer (CIB) containing 2.0 M sodium thiocyanate (NaSCN), 10 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1.0 mM phenylmethylsulfonyl fluoride (PMSF), and 2.0 mM dithiothreitol, incubated with lysozyme (3 mg ml−1) for 20 min, and subjected to 3 passes through a chilled (∼4°C) French pressure cell at 138 MPa. Unbroken cells and debris were pelleted by low-speed centrifugation (2,000 × g), and the resulting supernatant was collected and brought to 30% sucrose (wt/vol). A sucrose step gradient was generated with 45% (wt/vol) sucrose, followed by the low-speed supernatant fraction, 20% (wt/vol) sucrose (wt/vol), and 5% (wt/vol) sucrose, all in CIB. This gradient was centrifuged for 18 h at 250,000 × g. The chlorosome fraction appeared as a dark brown layer at the top of the gradient. This band was collected and diluted 3-fold in chlorosome buffer (10 mM potassium phosphate and 150 mM NaCl, pH 7.5), and chlorosomes were pelleted by high-speed centrifugation at 220,000 × g. This last step was repeated once more, and the resulting chlorosome pellet was resuspended in chlorosome buffer, aliquoted into cryovials, and stored at −80°C until required for other biochemical analyses.
Bacteriochlorophyll analyses.
Pigments were extracted from cell and chlorosome samples by sonication in acetone/methanol (7:2, vol/vol). Cell debris was removed by centrifugation and the resulting supernatant was filtered through a 0.2-μm filter. Reversed-phase high-performance liquid chromatography (RP-HPLC) separation of the extracted pigments was performed on a 25-cm by 4.6-mm Discovery 5-μm C18 column (Supelco, Bellefonte, PA) as previously described (19, 71). For studies on homolog composition as a function of light intensity, solvent A was changed to water/methanol/acetonitrile (62.5:21:16.5, vol/vol/vol). BChl c was monitored at 660 nm.
For BChl purification for mass spectrometric analyses, pigments were separated on a semipreparative 25-cm by 10-mm Discovery 5-μm C18 column (Supelco, Bellefonte, PA) with an elution program similar to that previously described (16, 19, 71). BChls were extracted in aceton/methanol (MeOH) (7:2, vol/vol), dried under a stream of N2, and resuspended in an equal volume of ether and methanol. Sterile deionized water was added dropwise until the ether phase separated. The ether phase containing the BChl was collected, dried under a stream of N2, resuspended in aceton/MeOH (7:2, vol/vol), filtered through a 0.2-μm filter, and loaded onto the semipreparative column. To identify the different homologs of BChl c, RP-HPLC was coupled with tandem mass spectrometry (MS-MS) as described previously (2). Formic acid, which demetalated the BChls and increased detection sensitivity in the MS-MS analyses, was added postcolumn (2). Mass spectrometric measurements were performed at the Mass Spectrometry Facility, Huck Institutes for the Life Sciences at The Pennsylvania State University, University Park, PA.
Quinone and carotenoid analyses.
Quinones and carotenoids were extracted from cells and chlorosome fractions by repeated sonication in acetone/MeOH (7:2, vol/vol). Cell debris was removed by centrifugation, and the resulting supernatant was dried under a stream of N2. For carotenoids, the dried extract was resuspended in acetone/MeOH (7:2, vol/vol) and filtered through a 0.2-μm filter. For quinones, the dried extract was resuspended in MeOH, and an equal volume of hexane was added. After mixing, an equal volume of 1.0 M NaCl was added, and the extract was shaken and centrifuged. BChl molecules precipitated between the aqueous and organic phases; the hexane phase containing quinones was collected, dried under a stream of N2, resuspended in acetone/MeOH (7:2, vol/vol), and filtered through a 0.2-μm filter. RP-HPLC analyses of the filtered extracts containing quinones and carotenoids were performed as described above. Quinones and carotenoids were detected by their absorption at 270 nm and 491 nm, respectively.
For mass spectrometry analyses, pigments and quinones were separated on a semipreparative 25-cm by 10-mm Discovery 5-μm C18 column (Supelco, Bellefonte, PA) with a similar elution program as previously described (9, 16). Mass spectrometric measurements were performed at the Mass Spectrometry Facility, Huck Institutes for the Life Sciences at The Pennsylvania State University, University Park, PA.
Lipid analyses.
For analysis of intact polar lipids (IPLs), chlorosomes were extracted using a modified Bligh and Dyer method (8). A known volume of single-phase solvent mixture of MeOH/dichloromethane (DCM)/phosphate buffer (2:1:0.8, vol/vol/vol) was added to the sample in a centrifuge tube and placed in an ultrasonic bath for 10 min. The extract and residue were separated by centrifugation at 1,000 × g for 5 min, and the solvent mixture was collected in a separate flask (3 times). The DCM and phosphate buffer were added to the single-phase extract to give a new ratio of MeOH/DCM/phosphate buffer (1:1:0.9, vol/vol/vol) and to induce phase separation. The extract was centrifuged at 1,000 × g for 5 min. The DCM phase was collected in a round-bottom flask, and the methanol/phosphate buffer phase was washed two additional times with DCM. The combined DCM phases were reduced under rotary vacuum and evaporated to dryness under a stream of N2.
The Bligh-Dyer extract was analyzed using HPLC-electron spray ionization (ESI)-ion trap MS. The extract was dissolved in hexane/2-propanol/water (72:27:1, vol/vol/vol) at a concentration of 2 mg ml−1 and filtered through a 0.45-μm regenerated cellulose filter (Alltech Associates, Inc., Deerfield, IL) prior to injection. Intact polar lipids (IPLs) were analyzed according to the method of Sturt et al. (62) with some modifications. An Agilent 1200 series LC (Agilent, San Jose, CA), equipped with auto-injector and column oven, which was coupled to a Thermo LTQ XL linear ion trap with Ion Max source with electrospray injection probe (Thermo Fisher Scientific), was used for the analyses. Separation was achieved on a Lichrosphere diol column (250 by 2.1 μm, 5-μm particles; Alltech Associates, Inc., Deerfield, IL) maintained at 30°C. The following elution program was used, with a flow rate of 0.2 ml min−1: 100% A for 1 min, followed by a linear gradient to 66% A; 34% B in 17 min, maintained for 12 min, followed by a linear gradient to 35% A; and 65% B in 15 min, where A is hexane/2-propanol/formic acid/14.8 M aqueous ammonia (NH3aq) (79:20:0.12:0.04, vol/vol/vol/vol) and B is 2-propanol/water/formic acid/14.8 M NH3aq (88:10:0.12:0.04, vol/vol/vol/vol). The total run time was 60 min, with a re-equilibration period of 20 min between runs. The ESI settings were as follows: capillary temperature, 275°C; sheath gas (N2) pressure, 25 (arbitrary units); auxiliary gas (N2) pressure, 15 (arbitrary units); sweep gas (N2) pressure, 20 (arbitrary units); and spray voltage, 4.5 kV (positive-ion ESI). The lipid extract was analyzed by an MS routine for which a positive ion scan (m/z 400 to 2,000) was followed by data-dependent MS-MS experiments, in which the four most abundant ions of the mass spectrum were fragmented (normalized collision energy, 25; isolation width, 5.0; activation Q, 0.175).
For identification of the fatty acids of the intact polar lipids, an aliquot of the Bligh-Dyer extract was hydrolyzed by refluxing with 5% HCl in methanol for 3 h. The acid hydrolysate was adjusted to pH 4 with 2 N KOH-MeOH (1:1, vol/vol) and, after the addition of water to a final 1:1 ratio of H2O-MeOH, extracted three times with DCM. The DCM fractions were collected and dried over sodium sulfate. The extract obtained was methylated with diazomethane and separated over an activated Al2O3 column into an apolar and a polar fraction using DCM and DCM-MeOH (1:1, vol/vol) as the eluent, respectively. The apolar fraction (containing the fatty acid methyl esters and hydrocarbons) was analyzed by gas chromatography (GC) and GC-MS.
To analyze the ester-bound compounds, particularly those of the BChls, the chlorosomes were extracted ultrasonically using a mixture of DCM and MeOH as described above. The extract was separated into an apolar and polar fraction using Al2O3 as stationary phase and DCM and DCM-MeOH (1:1, vol/vol) as eluents, respectively. The polar fraction was hydrolyzed with 1 N KOH-MeOH (96%) by reflux for 1 h. The hydrolyzate was neutralized with 2 N HCl-MeOH (1:1, vol/vol) to pH 4 and, after the addition of water, was extracted three times with DCM. The extract obtained was methylated with diazomethane and was silylated with N,O-bis(trimethylsilyl)-trifluoroacetamide in pyridine at 60°C for 20 min and analyzed by gas chromatography (GC) and GC-MS.
GC analyses were performed using a Hewlett-Packard (HP6890) instrument equipped with an on-column injector and a flame ionization detector. A fused-silica capillary column (25 m by 0.32 mm) coated with CP Sil-5 CB (film thickness, 0.12 mm) was used with helium as the carrier gas. The samples were injected at 70°C, and the oven temperature was programmed to increase to 130°C at a rate of 20°C min−1 and then to increase at 4°C min−1 to 320°C, at which temperature it was held for 10 min. GC-MS was performed on a Finnigan Trace ultra gas chromatograph interfaced with a Finnigan Trace DSQ mass spectrometer operated at 70 eV with a mass range of m/z 40 to 800 and a cycle time of 1.7 s. The gas chromatograph was equipped with a fused-silica capillary column as described for GC. The carrier gas was helium. The temperature program was the same as that for the GC analyses described above.
Detection and identification of hopanoids.
Lipids were extracted by sonicating cells in Teflon centrifuge tubes (VWR, Bridgeport, NJ) for 15 min at room temperature in 10 ml of 10:5:4 (vol/vol/vol) MeOH/DCM/water (8). Samples were centrifuged for 10 min at 3,000 × g, and the supernatant was transferred to a new tube. Cell pellets were sonicated a second time in 10 ml of MeOH/DCM/water (10:5:4, vol/vol/vol) and centrifuged, and the supernatant was combined with the first extraction. The samples were separated into two phases by adding 10 ml DCM and 5 ml water and centrifuged for 10 min at 3,000 × g, and the organic phase was transferred to a new vial. A second extraction with 10 ml DCM and 5 ml water was performed on the remaining aqueous phase. This sample was centrifuged, and the organic phase was combined with the previous extract. The organic solvents were evaporated under N2 and the total lipid extract (TLE) was incubated in 100 μl of acetic anhydride/pyridine (1:1, vol/vol) for 1 h at 70°C to derivatize alcohols into acetate esters.
The acetylated TLEs were analyzed by high-temperature GC-MS as previously described (77) and by LC-MS. The LC-MS system comprises a 1200 series HPLC (Agilent Technologies, Santa Clara, CA) equipped with an autosampler and a binary pump linked to a quadrupole time of flight (Q-TOF) 6520 mass spectrometer (Agilent Technologies) via an atmospheric pressure chemical ionization (APCI) interface (Agilent Technologies) operated in positive mode. The analytical procedure was adapted from Talbot et al. (67). A Poroshell 120 EC-C18 column (2.1 by 150 mm, 2.7 μm; Agilent Technologies), set at 30°C, was eluted isocratically with MeOH/water (95:5, vol/vol) for 2 min at a flow rate of 0.15 ml min−1 and then using a linear gradient up to 20% (vol) of isopropyl alcohol (IPA) over 18 min at a flow rate of 0.19 ml min−1 and isocratically for 10 min. The linear gradient was then set to 30% (vol) of IPA at 0.19 ml min−1 over 10 min and maintained for 5 min. The column was subsequently eluted using a linear gradient up to 80% IPA (vol) over 1 min at a flow rate of 0.15 ml min−1 and isocratically for 14 min. Finally, the column was eluted with MeOH/water (95:5, vol/vol) at 0.15 ml min−1 for 5 min. The APCI parameters were as follows: gas temperature, 325°C; vaporizer temperature, 350°C; drying gas (N2) flow, 6 liters min−1; nebulizer (N2) flow, 30 liter min−1; capillary voltage, 1,200 V; corona needle, 4 μA; and fragmentor, 150 V. Data were recorded by scanning from m/z 100 to 1,600. Identification of the hopanoids was based on their exact mass and by comparison of retention times and mass spectra with published data (68, 69).
RESULTS
Bacteriochlorophyll c homologs.
Previous RP-HPLC analyses had suggested that “Ca. Chloracidobacterium thermophilum” synthesizes a unique set of BChl c homologs that are different and significantly more complex than the BChl c homologs produced by C. tepidum and C. aurantiacus (9, 27). The genome of “Ca. Chloracidobacterium thermophilum” encodes homologs of bchQ and bchR (24); the products of these genes in C. tepidum are radical S-adenosylmethionine (SAM)-dependent methyltransferases that methylate the C-82 and C-121 positions, respectively, of BChl c (28). Thus, it was anticipated that some adjacent BChl c homologs would differ by 14 Da, i.e., the mass of a methylene unit (Table 1). To confirm the initial pigment analyses and to identify the BChl c species synthesized by “Ca. Chloracidobacterium thermophilum,” BChls were analyzed by RP-HPLC and RP-HPLC combined with MS or MS-MS (Fig. 1 and Table 1; also see Fig. S1 in the supplemental material). Alcohols obtained by alkaline hydrolysis of chlorosomes were also analyzed by GC-MS (see Fig. 3).
Table 1.
Masses and identities of BChl c species found in “Candidatus Chloracidobacterium thermophilum”
| Peak | (M+H) | Macrocycle | [C8, C12 substituents]b | Mass of alkyl fragment (Da) | Assignment |
|---|---|---|---|---|---|
| 1 | 799 | 595 | [n-Pr, Et] | 204 | Farnesyl |
| 2 | 813 | 609 | [i-Bu, Et] | 204 | Farnesyl |
| 3 | 827 | NDc | [n-Pe, Et]a | ND | Farnesyla |
| 4 | 853 | ND | [Et, Et]a | ND | Geranylgeraniola |
| 5 | 867 | 595 | [n-Pr, Et] | 272 | Geranylgeraniol |
| 6 | 881 | 609 | [i-Bu, Et] | 272 | Geranylgeraniol |
| 7 | 805 | ND | [n-Pr, Et] | 210 | Pentadecanol |
| 8 | 819 | ND | [i-Bu, Et]/[Et, Me]a | 224/238 | Hexadecanol/heptadecanol |
| 9 | 817 | 581 | [Et, Et] | 236 | Heptadecanol (1) |
| 10 | 833 | 595 | [n-Pr, Et] | 238 | Heptadecanol |
| 11 | 847 | 609 | [i-Bu, Et] | 238 | Heptadecanol |
| 12 | ND | ND | [Et, Et]a | ND | ND |
| 13 | 847 | 595 | [n-Pr, Et] | 252 | Octadecanol |
| 14 | 861 | 609 | [i-Bu, Et] | 252 | Octadecanol |
| 15 | 875 | ND | [n-Pe, Et]a | ND | Octadecanol |
Assignments made based on full mass spectrum and/or adjacent homologs.
Et, ethyl; M, methyl; nPr, normal propyl; ibu, iso-butyl; nPe, neo-pentyl.
ND, not determined.
Fig 1.
RP-HPLC elution profile of BChls extracted from “Ca. Chloracidobacterium thermophilum” cells. Profile was obtained by monitoring at 667 nm. See text for further details and Table 1 for peak assignments.
Fig 3.
Gas chromatogram of base-hydrolyzed polar fraction of total extract of chlorosomes of “Ca. Chloracidobacterium thermophilum.” Indicated are the major alcohols present in this fraction and which may be ester bound to BChls. Other peaks represent nonalcohol compounds, such as fatty acids.
RP-HPLC analyses of BChls extracted from whole cells and isolated chlorosomes revealed a complex mixture of BChl c homologs with the most prominent homolog (peak 14) eluting at about 46.5 min (Fig. 1). The pattern of BChl c homologs eluting between 20 and 27 min resembled that for other green sulfur bacteria, including C. tepidum, in which the majority of the BChl c homologs are esterified with farnesol (15, 18, 27, 28). MS and MS-MS analyses of three of these homologs (corresponding to peaks 1, 2, and 3 in Fig. 1) showed that these compounds had increasing methylation and corresponded to [8-n-Pr, 12-Et]-BChl cF, [8-i-Bu, 12-Et]-BChl cF, and [8-n-Pe, 12-Et]-BChl cF. Based upon its elution time, it can further be assumed that the peak labeled “0” at about 22.5 min corresponded to [8-Et, 12-Et]-BChl cF. As can be seen in Fig. 1, no compound eluting at ∼21 min was observed, which would be the expected elution time for the parental compound, [8-Et, 12-Me]-BChl cF. This indicates that nearly all BChl c homologs are methylated at the C-12 position in “Ca. Chloracidobacterium thermophilum.” Furthermore, homologs carrying an isobutyl side chain at C-8 were more abundant than those carrying an n-propyl or neo-pentyl side chains at that position. This information was helpful in assigning the methylation status of other BChl c homologs.
The BChl c homologs eluting between 30 and 36 min had very similar patterns of abundance to those of the farnesyl derivatives, and the MS and MS-MS data identified a similar pattern of homologs differentiated by methylation. The esterifying alcohol for these homologs was identified as geranylgeraniol from the MS fragmentation data (Table 1). The most abundant homologs were found in peaks 5 and 6, which were identified as [8-n-Pr, 12-Et]-BChl cGG and [8-i-Bu, 12-Et]-BChl cGG, respectively.
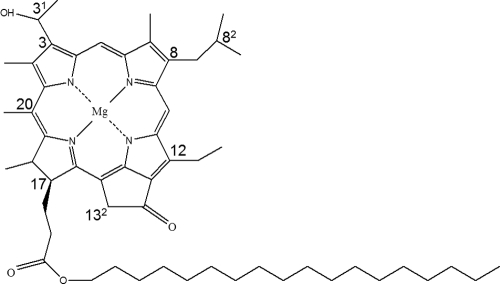
The pattern of BChl c homologs eluting between 35 and 47 min was very complex, and it was anticipated that these homologs would have several different esterifying alcohols. The compounds in peaks 13 to 15 corresponded to the most hydrophobic BChl c homologs and included the most abundant BChl c homolog (peak 14). The parental ion of demetalated compound 14 had an m/z of 861, and after loss of the esterifying alcohol from the C-17 propionate, the macrocycle had an m/z of 609.4. The difference of 252 Da indicated that the esterifying alcohol group was a C18 alcohol, which was assigned as octadecanol (stearol) (Table 1). The macrocycle fragment had an m/z value that was 42 Da greater than that expected for [8-Et, 12-Me]-BChl c (i.e., m/z = 567), the parent compound of the BChl c homolog family. This indicated that the macrocycle of this compound carried three additional methyl groups, and thus, this molecule was identified as [8-i-Bu, 12-Et]-BChl cS (2, 50) (Table 1 and Fig. 2). Supporting this assignment, MS-MS analyses of this peak resulted in a fragmentation pattern that was consistent with the pattern for BChl c as described previously (2) (see Fig. S1 in the supplemental material). It should be noted that the peak intensity pattern of the BChl c homologs esterified with octadecanol was similar to that observed for the farnesyl and geranylgeranyl derivatives discussed above, i.e., the most abundant homolog corresponded to the [8-i-Bu, 12-Et] homolog. Peaks 13 and 15 were correspondingly identified as the [n-Pr, 12-Et] and [n-Pe, 12-Et] BChl cS homologs and peak 12 was inferred to be the [8-Et, 12-Et] homolog. The compounds in peaks 10 and 11 had heptadecanol as the major esterifying alcohol, but peak 8 in particular contained BChl c homologs with both hexadecanol and heptadecanol esterifying alcohols (Table 1). Compound 7 had an m/z of 805, and if the esterifying alcohol is assumed to be pentadecanol, this would correspond to [8-n-Pr, 12-Et]-BChl c. In summary, the HPLC, MS, and MS-MS data in Fig. 1 and Table 1 indicate that BChl c in “Ca. Chloracidobacterium thermophilum” can be esterified with at least seven different alcohols.
Fig 2.
Structure of the major species of BChl c found in the chlorosomes of “Ca. Chloracidobacterium thermophilum.” [8-i-Bu, 12-Et]-BChl c esterified with octadecanol. The numbering of selected carbon atoms is indicated.
Figure 3 shows the GC elution profile for alcohols released from chlorosomes by alkaline hydrolysis and identified by mass spectrometry. Eleven different alcohols were identified by this method. Because wax esters were not analyzed separately, and wax esters can occur in significant amounts in chlorosomes (15, 60), it is possible that some of the alcohols identified might be derived from these components. There is no precedent to suggest that the 14-methyl-pentadecanol (iso-C16 alcohol) and 15-methyl-hexadecanol (iso-C17 alcohol) could be esterifying alcohols for BChls, while they can be prominent components in wax esters of some FAPs (74) and, thus, may be derived from wax esters. Four isoprenoid alcohols were identified, including farnesol (i-C15:3-OH), geranylgeraniol (i-C20:4-OH), phytol (i-C20:1-OH), and phytadienol (i-C20:2-OH). Phytol was previously shown to be the esterifying alcohol for BChl a in “Ca. Chloracidobacterium thermophilum” (9), and because chlorosomes contain some BChl a, phytol was expected to be present in this fraction. As described above, MS and MS-MS analyses were employed to identify BChl c homologs esterified with farnesol and geranylgeraniol. It is likely that some of the minor peaks eluting between 35 to 47 min, as shown in Fig. 1, are BChl c homologs esterified with phytadienol (iC20:2-OH), but this was not directly confirmed by MS and MS-MS analysis. Straight-chain alcohols, ranging in length from 15 to 19 carbons, were also detected (Fig. 3). Octadecanol (C18-OH) was the most abundant straight-chain alcohol; lesser and roughly equal amounts of hexadecanol (C16-OH) and heptadecanol (C17-OH) were also present. The alcohol analysis (Fig. 3) generally supported the inferences made from the HPLC and MS-MS analyses (Fig. 1 and Table 1).
The proportions and distribution of BChl species seemed to change as a function of the light conditions for growth (see Fig. S2 in the supplemental material). RP-HPLC analyses of BChls extracted from cells grown at irradiance values ranging from 30 to 120 μmol photons m−2 s−1 showed that the proportion of homologs esterified with isoprenoid tails (i.e., farnesol and geranylgeraniol) was highest at the lowest irradiance value. Conversely, the proportion of BChl c homologs esterified with unbranched alkyl chains was highest at the highest irradiance value and decreased with decreasing irradiance (see Fig. S2 in the supplemental material).
Carotenoids.
Figure 4 shows the elution profile from the RP-HPLC analysis of chlorosome pigments with monitoring at 491 nm for the detection of carotenoids. The most abundant carotenoid in both whole cells (data not shown) and chlorosomes eluted at 45.3 min. Based upon its absorption spectrum with a single, broad maximum at 465 nm (Fig. 4C), its m/z of 551, and its coelution with authentic echinenone from Synechococcus sp. strain PCC 7002 (29, 78), this compound was identified as echinenone. Other minor carotenoids were also identified using similar criteria. These included canthaxanthin (30 min, m/z = 564) (Fig. 4B), lycopene (52.2 min), γ-carotene (53.7 min), and β-carotene (55.3 min). The minor carotenoids of peaks 2, 3, and 4, shown in Fig. 4A, were not identified in this study; compounds 3 and 4 will be further described elsewhere (72).
Fig 4.
Carotenoids in “Ca. Chloracidobacterium thermophilum” chlorosomes. (A) HPLC elution profile monitored at 491 nm of carotenoids extracted from “Ca. Chloracidobacterium thermophilum.” Identified peaks are canthaxanthin (1), echinenone (5), lycopene (6), γ-carotene (7), and β-carotene (8). Peaks 2, 3, and 4 are unidentified carotenoids. (B) Absorption spectrum of peak 1 (canthaxanthin). (C) Absorption spectrum of peak 5 (echinenone). AU, absorbance units.
Quinones.
The fluorescence emission of “Ca. Chloracidobacterium thermophilum” cells and isolated chlorosomes is strongly quenched (up to 17-fold) under oxic conditions (23). This behavior suggested that a quenching agent, which was likely to be an isoprenoid quinone (6, 7, 18–20, 71), was present in the chlorosomes. Furthermore, the genome of “Ca. Chloracidobacterium thermophilum” predicts that the organism should be capable of synthesizing menaquinone (24). RP-HPLC analyses of quinones extracted from both whole cells and isolated chlorosomes revealed a hydrophobic quinone, eluting at ∼60 min, which had the absorption spectrum of menaquinone (Fig. 5A and B). This hydrophobic, menaquinone-like compound eluted slightly later than an authentic menaquinone-8 standard, which had been isolated from E. coli cells grown under anoxic conditions (data not shown) (51). This indicated that the quinone in “Ca. Chloracidobacterium thermophilum” was slightly more hydrophobic than menaquinone-8, and mass spectrometry showed that the mass differed by two mass units (m/z = 718) from the menaquinone-8 standard (m/z = 716) prepared from E. coli. This difference probably arises from the reduction of a single double bond in one of the eight isoprenoid units comprising the hydrophobic tail of the menaquinone. Although the role of such reductions is not clear, menaquinone-8(H2) derivatives with a reduced double bond in one of the isoprenoid units of quinones have been observed in many organisms, most notably in soil organisms (33).
Fig 5.
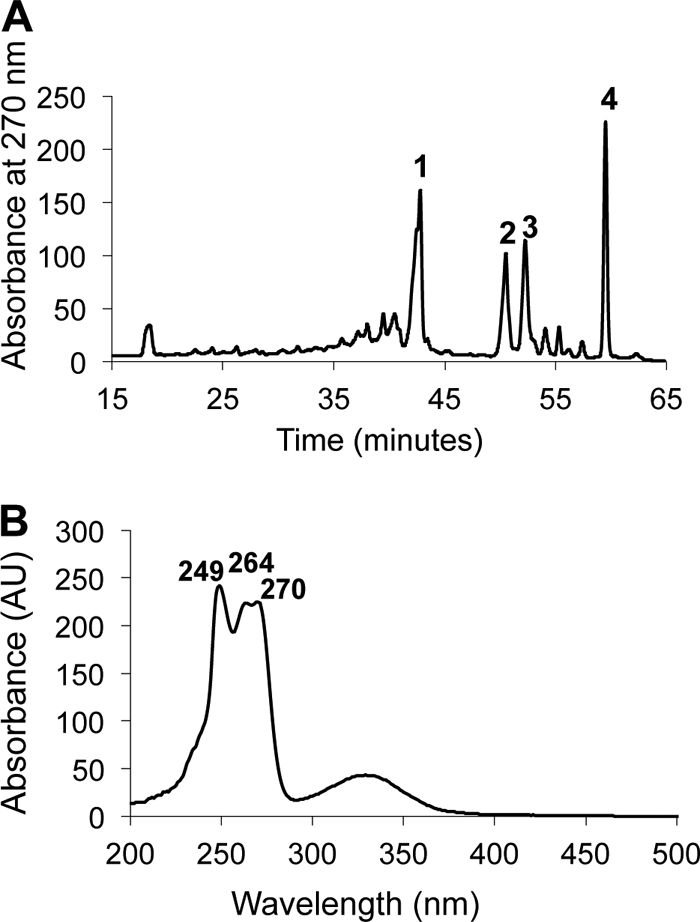
Quinones in “Ca. Chloracidobacterium thermophilum” cells. (A) HPLC elution profile of quinones extracted from “Ca. Chloracidobacterium thermophilum” monitored at 297 nm. Peak 4 represents menaquinone-8(H2). Peaks 1, 2, and 3 were identified as carotenoids by their absorption spectra. Other minor peaks represent BChls or bacteriopheophytins. (B) Absorption spectrum of peak 4 [menaquinone-8(H2)]. The absorption spectrum of menaquinone-8 extracted from E. coli was identical (data not shown).
Lipids.
Figure 6 shows the elution profile of fatty acids identified by GC-MS analysis of fatty acid methyl esters derived from the acid-hydrolyzed lipid extract of chlorosomes from “Ca. Chloracidobacterium thermophilum.” The most abundant fatty acid was 13-methyl tetradecanoic acid (isoC15), and smaller amounts of 14-methyl-pentadecanoic (isohexadecanoic) acid and n-hexadecanoic acid were also observed. Interestingly, the chlorosome fraction also contained a C18 n-alkane. This pattern was very similar to the fatty acid composition of whole cells (see Fig. S3 in the supplemental material), except that 13-methyl-tetradecanoic acid was proportionally an even greater percentage of the total fatty acids derived from the cells. The fatty acid composition of the heterotrophic members of this enrichment community had a distinctive and quite different fatty acid composition (see Fig. S4 in the supplemental material). Although 13-methyl-tetradecanoic acid was also present in the lipids of these two organisms, the proportion of 14-methyl-pentadecanoic acid was much greater and 12-methyl-tridecanoic (isotetradecanoic) acid, 12-methyl-tetradecanoic (anteisopentadecanoic) acid, and 14-methyl-hexadecanoic (anteisoheptadecanoic) acid were also present. These data showed that the procedure for removing the heterotrophic bacterial cells from the “Ca. Chloracidobacterium thermophilum” cells was highly effective and that the cell preparations analyzed were almost completely depleted of contaminating cells from Anoxybacillus and Meiothermus spp. (compare Fig. 7 and Fig. S6 in the supplemental material).
Fig 6.
Total ion chromatogram of acid-hydrolyzed extract of chlorosomes of “Ca. Chloracidobacterium thermophilum.” isoC14, 12-methyl-tridecanoic acid; isoC15, 13-methyl-tetradecanoic acid; isoC16, 15-methyl-hexadecanoic acid; nC16, hexadecanoic acid; nC18, octadecanoic acid.
Fig 7.
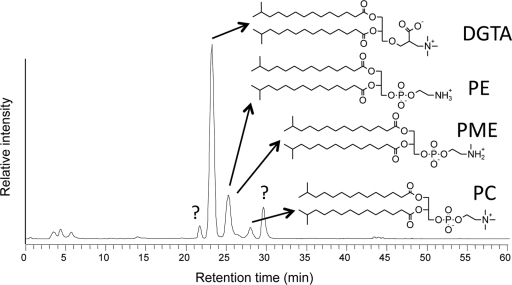
HPLC-ESI-MS ion chromatogram of intact polar lipids in chlorosomes of “Ca. Chloracidobacterium thermophilum.” DGTA, diacylglycerylhydroxymethyl-(N,N,N)-trimethylalanine; PE, phosphatidylethanolamine; PME, phosphatidyl-(N)-methylethanolamine; PC, phosphocholine. Example structures with two isopentadecanoic acid (13-methyl-tetradecanoic acid) moieties are illustrated. Other fatty acids present in the polar lipids are isotetradecanoic acid (12-methyl-tridecanoic acid), isoheptadecanoic (15-methyl-hexadecanoic acid) acid, and hexadecanoic acid.
Figure 7 shows an HPLC–MS-MS analysis of the intact polar lipids extracted from the chlorosome envelope membrane. The most prominent lipid observed was the betaine lipid, diacylglycerylhydroxymethyl-N,N,N-trimethyl-β-alanine (DGTA). The fatty acids associated with DGTA were 13-methyl-tetradecanoic acid (isoC15) but also included molecules esterified with both 13-methyl-tetradecanoic acid (isoC15) and 14-methyl-pentadecanoic acid (isoC16). Three other lipids were identified, phosphatidylethanolamine (PE), phosphatidylcholine (PC), and phosphatidylmonomethylethanolamine (PME), all mostly esterified with 13-methyl-tetradecanoic acid (isoC15). Two additional polar lipids were detected but could not be identified. The intact polar lipids of whole cells were similar and mostly contained DGTA, PME, and PE (see Fig. S5 in the supplemental material). As with the fatty acid analyses, a parallel experiment with the heterotrophic contaminants revealed a distinctive and different polar lipid composition of these organisms with respect to that of “Ca. Chloracidobacterium thermophilum” (see Fig. S6 in the supplemental material), providing further confirmation that our technique is effective in separating “Ca. Chloracidobacterium thermophilum” cells from cells of Anoxybacillus and Meiothermus spp. that are found in the enrichment culture.
Hopanoids.
The presence of putative genes for enzymes of bacteriohopanepolyol biosynthesis in the “Ca. Chloracidobacterium thermophilum” genome indicated that this organism probably had the potential to produce these compounds (24). To determine if this was the case, total lipids were extracted from “Ca. Chloracidobacterium thermophilum” cells and were analyzed by GC-MS and LC-MS. Through these analyses, it was determined that “Ca. Chloracidobacterium thermophilum” produces the C30 hydrocarbon diploptene (data not shown), as well as two functionalized hopanoids: bacteriohopanetetrol and bacteriohopanetetrol cyclitol ether (Fig. 8). We did not attempt to resolve whether some of the diploptene originated from dehydration of diplopterol during the isolation and analysis, although the absence of other hopene isomers, such as hop-17, 21-ene, suggested that this was not the case. Previous studies have shown that acetylation of bacteriohopanetetrol cyclitol ether can result in two species, which are hepta-acetylated (1,002-Da) and octa-acetylated (1,044-Da) compounds (66). As shown in Fig. 8, both of these compounds were produced under the acetylation conditions employed. The 1,002-Da molecular ion is also indicative of the bacteriohopanetetrol glycoside, another functionalized hopanoid that is produced by a variety of bacteria, including Zymomonas mobilis (53). However, the presence of the 984-Da peak, as well as MS-MS analysis of the 1,002-Da molecular ion (data not shown), indicated that bacteriohopanetetrol glycoside is not produced by “Ca. Chloracidobacterium thermophilum.”
Fig 8.
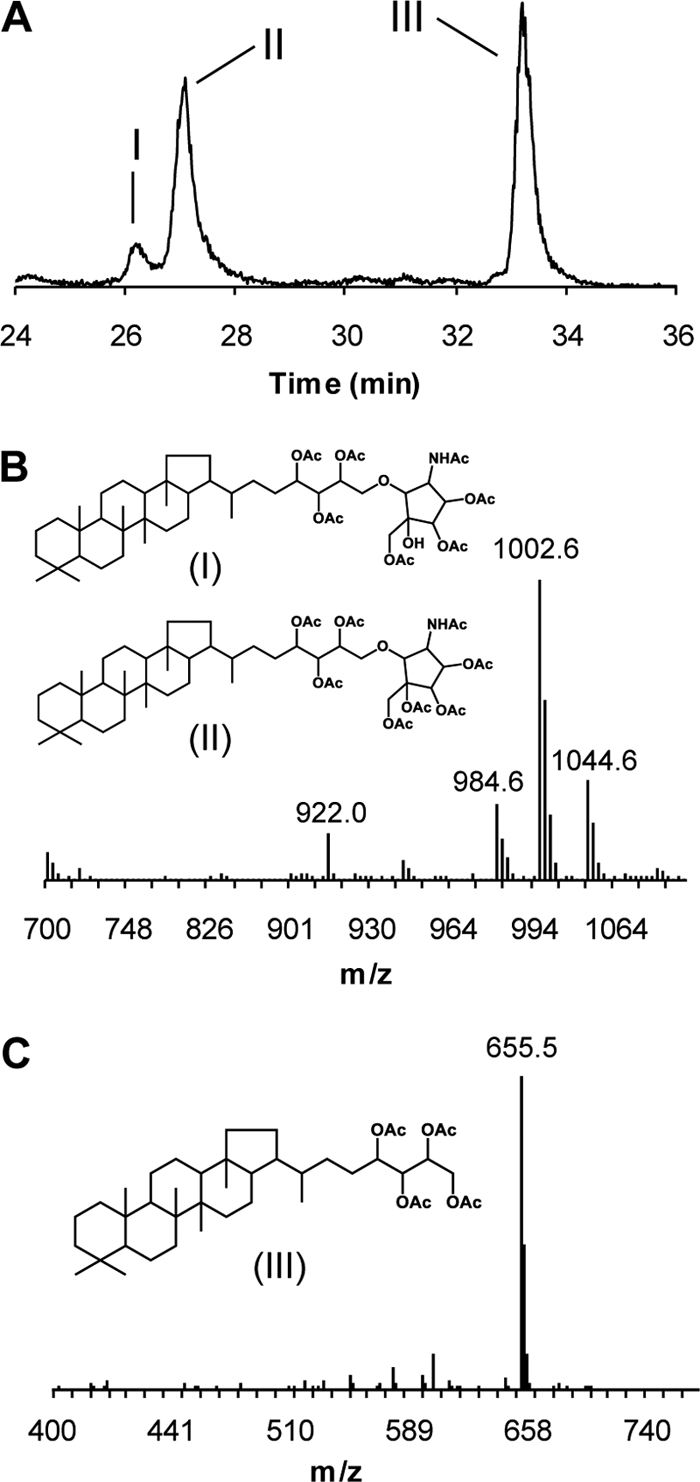
Bacteriohopanepolyols produced by “Ca. Chloracidobacterium thermophilum.” (A) LC-MS extracted ion chromatogram (m/z 655 and 1,002) of acetylated total lipid extract demonstrating the production of bacteriohopanecyclitol ether (I and II) and bacteriohopanetetrol (III). (B) APCI-mass spectrum of acetylated bacteriohopanecyclitol ether showing the pseudomolecular ions at 1,002 Da and 1,044 Da. The 1,002 ion corresponds to the heptacetylated compound and the 1,044 ion indicates the octa-acetylated compound. (C) APCI-mass spectrum of acetylated bacteriohopanetetrol showing the pseudomolecular ion at 655 Da. OAc, acetylated hydroxyl group; NHAc, acetylated amino group.
DISCUSSION
The data presented here describe the pigments, lipids, and quinones of the membranes and chlorosomes of the only known chlorophototrophic member of the phylum Acidobacteria, “Ca. Chloracidobacterium thermophilum.” Although unique features that seem to correlate with the oxic lifestyle of this organism were identified, the chlorosomes closely resemble those found in the chlorophototrophic Chlorobi and the green Chloroflexi. Like all known GSB, “Ca. Chloracidobacterium thermophilum” methylates its BChls at the C-82 and C-121 positions. These methylations have been shown to improve the light-harvesting capacity of C. tepidum, making it more competitive in light-limiting environments (28), and we presume that these methylations play a similar role in the chlorosomes of “Ca. Chloracidobacterium thermophilum.” Interestingly, all of the BChl c homologs seem to be methylated at the C-121 position, and highly methylated BChl c species were favored at the C-82 position. Increased methylation of BChls shifts the absorbance maximum of chlorosomes toward longer wavelengths (28). In “Ca. Chloracidobacterium thermophilum,” this red shift could be advantageous for two reasons. Recent in situ metatranscriptomic studies of samples collected throughout the day from the phototrophic mats of Mushroom Spring, where “Ca. Chloracidobacterium thermophilum” is found in nature, have revealed that “Ca. Chloracidobacterium thermophilum” is transcriptionally highly active at dusk, when the available light is naturally enriched in longer wavelengths (44). Moreover, “Ca. Chloracidobacterium thermophilum” must compete for light with Chloroflexus spp. and “Candidatus Thermochlorobacter aerophilum,” which are also present in the phototrophic mats of Mushroom and Octopus Springs (38). The latter organism has chlorosomes containing BChl d and seems to occur closer to the mat surface than “Ca. Chloracidobacterium thermophilum” (Z. Liu, C. G. Klatt, M. Ludwig, D. B. Rusch, S. I. Jensen, M. Kühl, D. M. Ward, and D. A. Bryant, submitted). Most members of the Chloroflexi do not methylate BChl c, and therefore, the Qy absorbance maximum of the BChl c aggregates in the chlorosomes of these organisms is typically slightly blue shifted with respect to those of C. tepidum and “Ca. Chloracidobacterium thermophilum” (23).
Unlike C. tepidum, farnesol, an isoprenoid alcohol, is not the major esterifying alcohol of BChl c in “Ca. Chloracidobacterium thermophilum.” Instead, the data indicate that the major esterifying alcohol for BChl c is the straight-chain, saturated alcohol octadecanol (stearol). In addition to this dominant BChl c species, “Ca. Chloracidobacterium thermophilum” synthesizes amounts of BChl c esterified to other alcohols, including pentadecanol, hexadecanol, heptadecanol, geranylgeraniol, and under light-limiting conditions, the isoprenoid farnesol. In this respect, “Ca. Chloracidobacterium thermophilum” resembles the GSB C. phaeobacteroides or green FAPs, such as Chloronema sp., which similarly display a diversity of methylation patterns and esterifying alcohols, including unbranched saturated alcohols (25, 42, 50). In both “Ca. Chloracidobacterium thermophilum” and C. phaeobacteroides, the proportion of isoprenoid esterifying alcohols increases under light-limiting conditions (see Fig. S2 in the supplemental material). It has been suggested that the availability of reducing power, rather than light intensity, might be responsible for the increased production of isoprenoid chains under light-limiting conditions. The biosynthesis of saturated alcohols requires more reducing power than the biosynthesis of isoprenoid alcohols (50). For “Ca. Chloracidobacterium thermophilum” in its ecological niche in the mats, higher light intensities would correspond to higher oxygen partial pressures (58). The synthesis of saturated, nonisoprenoid alcohols is also probably an adaptation to avoid peroxidation of the double bonds in esterifying isoprenoid alcohols (73).
The chlorosomes of “Ca. Chloracidobacterium thermophilum” have a unique carotenoid composition. Whereas the chlorosomes of other members of the Chlorobiales mostly contain chlorobactene or isorenieratene and green FAPs synthesize γ-carotene as the major carotenoid, the dominant carotenoid in “Ca. Chloracidobacterium thermophilum” is echinenone. Lesser amounts of lycopene, γ-carotene, β-carotene, and canthaxanthin were also found in the chlorosomes. This carotenoid profile differs from that of the reaction center cores (predominantly lycopene) and that of the reaction center-associated, carotenoid-binding protein (echinenone and glycosylated derivatives of deoxyflexixanthin) (72). Both echinenone and canthaxanthin are xanthophylls and possess 4-keto group(s) on their rings. Xanthophyll carotenoids have been shown to play roles in photoprotection in other microorganisms (34, 40, 61, 78). In particular, echinenone might be involved in protection against peroxide radicals (36, 37, 57). Because of the high irradiances and the oxic environment that “Ca. Chloracidobacterium thermophilum” encounters in its natural habitat (58), these ketocarotenoids probably also play an important photoprotective role in this microorganism. When cells were grown in the presence of N,N-diethyl-N-[2-(4-methylphenoxy)ethyl]amine (MPTA) to inhibit lycopene cyclase and the synthesis of cyclic carotenoids, the resulting cells exhibited much higher rates of photobleaching than untreated control cells (22).
Although “Ca. Chloracidobacterium thermophilum” is an aerobe, excitation energy transfer from BChl c to BChl a associated with the baseplate protein CsmA was redox dependent and was very strongly quenched under oxic conditions (23). In other organisms that exhibit this behavior, the quencher is known to be a quinone (6, 7, 18–20, 71), and HPLC and mass spectrometric analyses revealed the presence of menaquinone-8 in the chlorosomes of “Ca. Chloracidobacterium thermophilum.” Chlorobiumquinone, which is present in chlorosomes of C. tepidum (20) and other GSB strains, was not present in the chlorosomes of “Ca. Chloracidobacterium thermophilum.” Other hydroxylated quinones, which have been postulated to be responsible for fluorescence quenching in C. aurantiacus when added exogenously (71), were likewise not present.
The lipid composition in “Ca. Chloracidobacterium thermophilum” chlorosomes seems to reflect an adaptation to the alkaline, hot spring environment from which this organism was isolated, and the composition differs from that of other chlorosome-containing chlorophototrophs (15). The major lipid, DGTA, in “Ca. Chloracidobacterium thermophilum” is a betaine lipid, which contains nitrogen instead of phosphorus. DGTA is synthesized instead of phospholipids in purple sulfur bacteria at low phosphate levels (4). The phosphate levels in Octopus and Mushroom Springs are naturally low, and other chlorophototrophs in the mat communities of these springs have adaptations to the low phosphate concentrations associated with these thermal features (1). Although the phospholipid fraction is quite similar to that of chlorosomes from other green bacteria, the glycolipids typically found in the membranes of other chlorosomes, such as monogalactosyldiacylglycerol or rhamnolipids (39, 60), were not present in the chlorosomes of “Ca. Chloracidobacterium thermophilum.” The fatty acid fraction was dominated by branched fatty acids, mostly 13-methyl-tetradecanoic acid, but 12-methyl-tridecanoic acid and 14-methyl-pentadecanoic acid were also present. This finding is consistent with lipid analyses conducted on Octopus Spring mats that found branched fatty acids ranging from 14 to 18 carbons (76). Interestingly, the lipid and fatty acid analyses of the isolated chlorosomes revealed the presence of a C18 n-alkane in addition to the branched fatty acids. The biosynthetic pathway for this alkane is currently unknown, but the genome of “Ca. Chloracidobacterium thermophilum” exhibits redundancy in dehydrogenases that are homologous to 3-oxoacyl reductase (data not shown). The biosynthesis and significance of this alkane will be the subject of future studies.
“Ca. Chloracidobacterium thermophilum” synthesizes the common bacteriohopanepolyol precursor diploptene, as well as bacteriohopanetetrol and bacteriohopanetetrol cyclitol. To our knowledge, this is the first demonstration of hopanoid synthesis in a member of the microbial mats of Mushroom and Octopus Springs. Hopanoids have been postulated to play a role in strengthening the cell membrane of the organisms that synthesize these compounds, in a manner analogous to the way sterols strengthen eukaryotic membranes (77). There have been few studies, however, that have provided any information on how hopanoids specifically affect the membranes. Recent findings in Rhodopseudomonas palustris TIE-1 suggest that hopanoids increase pH tolerance by preventing proton leakage from cells (30, 77). However, it is not yet known whether this is a general property of all hopanoids or a property of specific types (77). The phototrophic mats of Mushroom and Octopus Spring undergo large diel variations of both oxygen partial pressure and pH (58), and it seems reasonable to hypothesize that the hopanoids found in “Ca. Chloracidobacterium thermophilum” might enhance membrane stability to withstand these conditions.
In conclusion, combined ultrastructural, spectroscopic, and biochemical analyses of isolated chlorosomes from “Ca. Chloracidobacterium thermophilum” presented here and elsewhere (23) show that, although “Ca. Chloracidobacterium thermophilum” is an aerobe, its chlorosomes are remarkably similar to those of other green bacteria, all of which produce and utilize these organelles under anoxic conditions. The chlorosomes from this aerobe share many common properties with those of the members of the Chlorobiales and the green FAPs Chloroflexales, including side chain methylation of BChl c, spectroscopic properties, and the occurrence of menaquinone as a redox-dependent quencher of energy transfer (23; this study). However, the most obvious adaptations to oxic conditions appear to be the presence of large amounts of ketocarotenoids, specifically echinenone and canthaxanthin, and the reduced usage of polyunsaturated isoprenoid compounds as esterifying alcohols for BChl c. This finding is in agreement with other recent studies of chlorosomes, which have shown that these antenna complexes have specific adaptations to protect against excessive illumination and oxidation while retaining the capacity for highly efficient energy transfer at exceptionally low irradiances under anoxic conditions (3, 25, 28, 35, 56). The lipid and hopanoid composition of “Ca. Chloracidobacterium thermophilum” cells also seem to reflect the environmental conditions that this organism encounters in situ, namely, a phosphorus-deficient, thermophilic habitat that is probably subject to large diel shifts in physical and chemical parameters.
ACKNOWLEDGMENTS
This research was supported by grant DE-FG02-94ER20137 from the U.S. Department of Energy to D.A.B. Y.T. was supported by postdoctoral fellowships from the Japan Society for the Promotion of Science (no. 181481) and The Uehara Memorial Foundation. E. C. Hopmans (NIOZ) provided analytical support for the lipid analysis. S.S. acknowledges funding from the Netherlands Organization of Scientific Research through a VICI grant. Research at MIT was supported by an NSF Minority Postdoctoral Fellowship to P.V.W. and a grant from the NSF Chemical Oceanography Program (OCE-0849940) to R.E.S.
Footnotes
Published ahead of print 30 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Adams M, Gomez-Garcia MR, Grossman AR, Bhaya D. 2008. Phosphorus deprivation responses and phosphonate utilization in a thermophilic Synechococcus sp. from microbial mats. J. Bacteriol. 190:8171–8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Airs RL, Keely BJ. 2002. Atmospheric pressure chemical ionization liquid chromatography/mass spectrometry of Bchls from Chlorobiaceae: characteristic fragmentations. Rapid Commun. Mass Spectrom. 16:453–461 [DOI] [PubMed] [Google Scholar]
- 3. Beatty JT, et al. 2005. An obligately photosynthetic bacterial anaerobe from a deep-sea hydrothermal vent. Proc. Natl. Acad. Sci. U. S. A. 102:9306–9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benning C, Huang Z-H, Gage DA. 1995. Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch. Biochem. Biophys. 317:103–111 [DOI] [PubMed] [Google Scholar]
- 5. Betti JA, Blankenship RE, Natarajan LV, Dickinson LC, Fuller RC. 1982. Antenna organization and evidence for the function of a new antenna pigment species in the green photosynthetic bacterium Chloroflexus aurantiacus. Biochim. Biophys. Acta 680:194–201 [Google Scholar]
- 6. Blankenship RE, Olson JM, Miller M. 1995. Antenna complexes from green photosynthetic bacteria, p 399–435 In Blankenship RE, Madigan MT, Bauer CE. (ed), Advances in photosynthesis and respiration, vol 2 Anoxygenic photosynthetic bacteria Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 7. Blankenship RE, et al. 1993. Redox regulation of energy transfer efficiency in antennas of green photosynthetic bacteria. Photochem. Photobiol. 57:103–107 [DOI] [PubMed] [Google Scholar]
- 8. Bligh EG, Dyer WJ. 1959. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 9. Bryant DA, et al. 2007. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 317:523–526 [DOI] [PubMed] [Google Scholar]
- 10. Bryant DA, Frigaard N-U. 2006. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 14:488–496 [DOI] [PubMed] [Google Scholar]
- 11. Bryant DA, et al. 2011. Comparative and functional genomics of anoxygenic green bacteria from the taxa Chlorobi, Chloroflexi, and Acidobacteria, p 47–102 In Burnap RL, Vermaas W. (ed). Advances in photosynthesis and respiration, vol. 33. Functional genomics and evolution of photosynthetic systems. Springer, Dordrecht, The Netherlands [Google Scholar]
- 12. Caple MB, Chow H, Strouse CE. 1978. Photosynthetic pigments of green sulfur bacteria: the esterifying alcohols of bacteriochlorophylls c from Chlorobium limicola. J. Biol. Chem. 253:6730–6737 [PubMed] [Google Scholar]
- 13. Eisen JA, et al. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. U. S. A. 99:9509–9514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reference deleted.
- 15. Frigaard N-U, Bryant DA. 2006. Chlorosomes: antenna organelles in photosynthetic green bacteria, p 79–114 In Shively JM. (ed), Complex structures in prokaryotes, vol. 2 Springer, Berlin, Germany [Google Scholar]
- 16. Frigaard N-U, Voigt GD, Bryant DA. 2002. Chlorobium tepidum mutant lacking bacteriochlorophyll c made by inactivation of the bchK gene, encoding bacteriochlorophyll c synthase. J. Bacteriol. 184:3368–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frigaard N-U, Maresca JA, Yunker CE, Jones AD, Bryant DA. 2004. Genetic manipulation of carotenoid biosynthesis in the green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 186:5210–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frigaard N-U, Matsuura K, Hirota M, Miller M, Cox RP. 1998. Studies of the location and function of isoprenoid quinones in chlorosomes from green sulfur bacteria. Photosynth. Res. 58:181–190 [Google Scholar]
- 19. Frigaard N-U, Takaichi S, Hirota M, Shimada K, Matsuura K. 1997. Quinones in chlorosomes of green sulfur bacteria and their role in the redox-dependent fluorescence studied in chlorosome-like bacteriochlorophyll c aggregates. Arch. Microbiol. 167:343–349 [Google Scholar]
- 20. Frigaard N-U, Tokita S, Matsuura K. 1999. Exogenous quinones inhibit photosynthetic electron transfer in Chloroflexus aurantiacus by specific quenching of the excited bacteriochlorophyll c antenna. Biochim. Biophys. Acta 1413:108–116 [DOI] [PubMed] [Google Scholar]
- 21. Ganapathy S, et al. 2009. Alternating syn-anti bacteriochlorophylls form concentric helical nanotubes in chlorosomes. Proc. Natl. Acad. Sci. U. S. A. 106:8525–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garcia Costas AM. 2010. Isolation and characterization of Candidatus Chloracidobacterium thermophilum. Ph.D. Thesis The Pennsylvania State University, University Park, PA [Google Scholar]
- 23. Garcia Costas AM, et al. 2011. Ultrastructural analysis and identification of envelope proteins of “Candidatus Chloracidobacterium thermophilum” chlorosomes. J. Bacteriol. 193:6701–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garcia Costas, et al. 2012. Complete genome of Candidatus Chloracidobacterium thermophilum, a chlorophyll-based photoheterotroph belonging to the phylum Acidobacteria. Environ. Microbiol. 14:177–190 [DOI] [PubMed] [Google Scholar]
- 25. Gich F, et al. 2003. Characterization of the chlorosome antenna of the filamentous anoxygenic phototrophic bacterium Chloronema sp. strain UdG9001. Arch. Microbiol. 180:417–426 [DOI] [PubMed] [Google Scholar]
- 26. Gloe A, Risch N. 1978. Bacteriochlorophyll cs, a new bacteriochlorophyll from Chloroflexus aurantiacus. Arch. Microbiol. 118:153–156 [DOI] [PubMed] [Google Scholar]
- 27. Gomez Maqueo Chew A, Bryant DA. 2007. Chlorophyll biosynthesis in bacteria: the origins of structural and functional diversity. Annu. Rev. Microbiol. 61:113–129 [DOI] [PubMed] [Google Scholar]
- 28. Gomez Maqueo Chew AGM, Frigaard N-U, Bryant DA. 2007. Bacteriochlorophyllide c C-8(2) and C-12(1) methyltransferases are essential for adaptation to low light in Chlorobaculum tepidum. J. Bacteriol. 189:6176–6184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Graham JE, Bryant DA. 2009. The biosynthetic pathway for myxol-2′ fucoside (myxoxanthophyll) in the cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 191:3292–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haines TH. 2001. Do sterols reduce proton and sodium leaks through lipid bilayers? Prog. Lipid Res. 40:299–324 [DOI] [PubMed] [Google Scholar]
- 31. Hale MB, Blankenship RE, Fuller RC. 1983. Menaquinone is the sole quinone in the facultatively aerobic green photosynthetic bacterium Chloroflexus aurantiacus. Biochim. Biophys. Acta 723:376–382 [Google Scholar]
- 32. Hirabayashi H, Ishii T, Takaichi S, Inoue K, Uehara K. 2004. The role of carotenoids in the photoadaptation of the brown-colored sulfur bacterium Chlorobium phaeobacteroides. Photochem. Photobiol. 79:280–285 [DOI] [PubMed] [Google Scholar]
- 33. Iizuka T, et al. 2003. Plesiocystis pacifica gen. nov., sp. nov., a marine myxobacterium that contains dehydrogenated menaquinone, isolated from the Pacific coasts of Japan. Int. J. Syst. Evol. Microbiol. 53:189–195 [DOI] [PubMed] [Google Scholar]
- 34. Khaneja R, et al. 2010. Carotenoids found in Bacillus. J. Appl. Microbiol. 108:1889–1902 [DOI] [PubMed] [Google Scholar]
- 35. Kim H, Li H, Maresca JA, Bryant DA, Savikhin S. 2007. Triplet exciton formation as a novel photoprotection mechanism in chlorosomes of Chlorobium tepidum. Biophys. J. 93:192–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kirilovsky D. 2010. The photoactive orange carotenoid protein and photoprotection in cyanobacteria. Adv. Exp. Med. Biol. 675:139–159 [DOI] [PubMed] [Google Scholar]
- 37. Kirilovsky D, Kerfeld CA. 2012. The orange carotenoid protein in photoprotection of photosystem II in cyanobacteria. Biochim. Biophys. Acta 1817:158–166 [DOI] [PubMed] [Google Scholar]
- 38. Klatt CG, et al. 2011. Metagenomic analyses of phototrophic hot spring microbial mat communities. ISME J. 5:1262–127821697961 [Google Scholar]
- 39. Knudsen E, Jantzen E, Bryn K, Ormerod JG, Sirevåg R. 1982. Quantitative and structural characteristics of lipids in Chlorobium and Chloroflexus Arch. Microbiol. 132:149–154 [Google Scholar]
- 40. Köcher S, Breitenbach J, Müller V, Sandmann G. 2009. Structure, function and biosynthesis of carotenoids in the moderately halophilic bacterium Halobacillus halophilus. Arch. Microbiol. 191:95–104 [DOI] [PubMed] [Google Scholar]
- 41. Larsen KL, Miller M, Cox RP. 1995. Incorporation of endogenous long chain alcohols into bacteriochlorophyll c homologs by Chloroflexus aurantiacus. Arch. Microbiol. 163:119–123 [Google Scholar]
- 42. Larsen KL, Cox RP, Miller M. 1994. Effects of illumination intensity on bacteriochlorophyll c homolog distribution in Chloroflexus aurantiacus grown under controlled conditions. Photosynth. Res. 41:151–156 [DOI] [PubMed] [Google Scholar]
- 43. Liu Z, Bryant DA. Biosynthesis and assembly of bacteriochlorophyll c in green bacteria: theme and variations. In Kadish KM, Smith KM, Guilard R. (ed), Handbook of porphyrin science, set IV-20, in press World Scientific Publishing, Hackensack, NJ [Google Scholar]
- 44. Liu Z, et al. 2011. Metatranscriptomic analyses of chlorophototrophs of a hot-spring microbial mat. ISME J. 5:1279–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lopez JC, Ryan S, Blankenship RE. 1996. Sequence of the bchG gene from Chloroflexus aurantiacus: relationship between chlorophyll synthase and other polyprenyltransferases. J. Bacteriol. 178:3369–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Manske AK, Glaeser J, Kuypers MM, Overmann J. 2005. Physiology and phylogeny of green sulfur bacteria forming a monospecific phototrophic assemblage at a depth of 100 meters in the Black Sea. Appl. Environ. Microbiol. 71:8049–8060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maresca JA, Bryant DA. 2006. Two genes encoding new carotenoid-modifying enzymes in the green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 188:6217–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maresca JA, Graham JE, Bryant DA. 2008. The biochemical basis for structural diversity in the carotenoids of chlorophototrophic bacteria. Photosynth. Res. 97:121–140 [DOI] [PubMed] [Google Scholar]
- 49. Maresca JA, Romberger SP, Bryant DA. 2008. Isorenieratene biosynthesis in green sulfur bacteria requires the cooperative actions of two carotenoid cyclases. J. Bacteriol. 190:6384–6391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Massé AM, Airs RL, Keely BJ, de Wit R. 2004. The impact of different intensities of green light on the bacteriochlorophyll homologue composition of the Chlorobiaceae: Prosthecochloris aestuarii and Chlorobium phaeobacteroides. Microbiology 150:2555–2564 [DOI] [PubMed] [Google Scholar]
- 51. Meganathan R. 2001. Ubiquinone biosynthesis in microorganisms. FEMS Microbiol. Lett. 203:131–139 [DOI] [PubMed] [Google Scholar]
- 52. Montaño GA, et al. 2003. Characterization of Chlorobium tepidum chlorosomes: a calculation of bacteriochlorophyll c per chlorosome and oligomer modeling. Biophys. J. 85:2560–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moreau RA, et al. 1995. Analysis of intact hopanoids and other lipids from the bacterium Zymomonas mobilis by high-performance liquid chromatography. Anal. Biochem. 224:293–301 [DOI] [PubMed] [Google Scholar]
- 54. Oostergetel GT, van Amerongen H, Boekema EJ. 2010. The chlorosome: a prototype for efficient light harvesting in photosynthesis. Photosynth. Res. 104:245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pierson BK, Castenholz RW. 1974. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch. Microbiol. 100:5–24 [DOI] [PubMed] [Google Scholar]
- 56. Pšenčík J, et al. 2009. Structure of chlorosomes from the green filamentous bacterium Chloroflexus aurantiacus. J. Bacteriol. 191:6701–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rengel D, et al. 2000. Exogenously incorporated ketocarotenoids in large unilamellar vesicles. Protective activity against peroxidation. Biochim. Biophys. Acta 1463:179–187 [DOI] [PubMed] [Google Scholar]
- 58. Revsbech NP, Ward DM. 1984. Microelectrode studies of interstitial water chemistry and photosynthetic activity in a hot spring microbial mat. Appl. Environ. Microbiol. 48:270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schmidt K, Maarzahl M, Mayer F. 1980. Development and pigmentation of chlorosomes in Chloroflexus aurantiacus strain Ok-70-fl. Arch. Microbiol. 127:87–97 [Google Scholar]
- 60. Sørensen PG, Cox RP, Miller M. 2008. Chlorosome lipids from Chlorobium tepidum: characterization and quantification of polar lipids and wax esters. Photosynth. Res. 95:191–196 [DOI] [PubMed] [Google Scholar]
- 61. Steiger S, Schafer L, Sandmann G. 1999. High-light-dependent upregulation of carotenoids and their antioxidative properties in the cyanobacterium Synechocystis PCC6803. J. Photochem. Photobiol. 52:14–18 [Google Scholar]
- 62. Sturt HF, Summons RE, Smith K, Elvert M, Hinrichs KU. 2004. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 18:617–628 [DOI] [PubMed] [Google Scholar]
- 63. Takaichi S. 2001. Carotenoids and carotenogenesis in anoxygenic photosynthetic bacteria, p 39–69 In Frank HA, Young AJ, Britton G, Cogdell RJ. (ed), The photochemistry of carotenoids. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 64. Takaichi S, Oh-oka H. 1999. Pigment composition in the reaction center complex from the thermophilic green sulfur bacterium, Chlorobium tepidum: carotenoid glucoside esters, menaquinone and chlorophylls. Plant Cell Physiol. 40:691–694 [Google Scholar]
- 65. Takaichi S, et al. 1997. New carotenoids from the thermophilic green sulfur bacterium Chlorobium tepidum: 1′,2′-dihydro-gamma-carotene, 1′,2′-dihydrochlorobactene, and OH-chlorobactene glucoside ester, and the carotenoid composition of different strains. Arch. Microbiol. 168:270–276 [DOI] [PubMed] [Google Scholar]
- 66. Talbot HM, Squier AH, Keely BJ, Farrimond P. 2003. Atmospheric pressure chemical ionisation reversed-phase liquid chromatography/ion trap mass spectrometry of intact bacteriohopanepolyols. Rapid Commun. Mass Spectrom. 17:728–737 [DOI] [PubMed] [Google Scholar]
- 67. Talbot HM, Watson DF, Murrell JC, Carter JF, Farrimond P. 2001. Analysis of intact bacteriohopanepolyols from methanotrophic bacteria by reversed-phase high-performance liquid chromatography-atmospheric pressure chemical ionisation mass spectrometry. J. Chromatogr. 921:175–185 [DOI] [PubMed] [Google Scholar]
- 68. Talbot HM, Rohmer M, Farrimond P. 2007. Structural characterisation of unsaturated bacterial hopanoids by atmospheric pressure chemical ionisation liquid chromatography/ion trap mass spectrometry. Rapid Commun. Mass Spectrom. 21:1613–1622 [DOI] [PubMed] [Google Scholar]
- 69. Talbot HM, Summons R, Jahnke L, Farrimond P. 2003. Characteristic fragmentation of bacteriohopanepolyols during atmospheric pressure chemical ionisation liquid chromatography/ion trap mass spectrometry. Rapid Commun. Mass Spectrom. 17:2788–2796 [DOI] [PubMed] [Google Scholar]
- 70. Tamiaki H, Shibata R, Mizoguchi T. 2007. The 17-propionate function of (bacterio)chlorophylls: biological implication of their long esterifying chains in photosynthetic systems. Photochem. Photobiol. 83:152–162 [DOI] [PubMed] [Google Scholar]
- 71. Tokita S, Frigaard N-U, Hirola M, Shimada K, Matsuura K. 2000. Quenching of bacteriochlorophyll fluorescence in chlorosomes from Chloroflexus aurantiacus by exogenous quinones. Photochem. Photobiol. 72:345–350 [DOI] [PubMed] [Google Scholar]
- 72. Tsukatani Y, Romberger SP, Golbeck JH, Bryant DA. Isolation and characterization of a homodimeric type-1 reaction center complex from “Candidatus Chloracidobacterium thermophilum”, an aerobic chlorophototroph. J. Biol. Chem., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Valko M, Morris H, Cronin MT. 2005. Metals, toxicity and oxidative stress. Curr. Med. Chem. 12:1161–1208 [DOI] [PubMed] [Google Scholar]
- 74. van der Meer MT, et al. 2010. Cultivation and genomic, nutritional, and lipid biomarker characterization of Roseiflexus strains closely related to predominant in situ populations inhabiting Yellowstone hot spring microbial mats. J. Bacteriol. 192:3033–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vassilieva EV, et al. 2002. Subcellular localization of chlorosome proteins in Chlorobium tepidum and characterization of three new chlorosome proteins: CsmF, CsmH and CsmX. Biochemistry 41:4358–4370 [DOI] [PubMed] [Google Scholar]
- 76. Ward DM, Panke S, Kloppel KD, Christ R, Fredrickson H. 1994. Complex polar lipids of a hot spring cyanobacterial mat and its cultivated inhabitants. Appl. Environ. Microbiol. 60:3358–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Welander PW, et al. 2009. Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 191:6145–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhu Y, et al. 2010. Roles of xanthophyll carotenoids in protection against photoinhibition and oxidative stress in the cyanobacterium Synechococcus sp. strain PCC 7002. Arch. Biochem. Biophys. 504:86–99 [DOI] [PubMed] [Google Scholar]