Abstract
Bacteria show remarkable adaptability under several stressful conditions by shifting themselves into a dormant state. Less is known, however, about the mechanism underlying the cell transition to dormancy. Here, we report that the transition to dormant states is mediated by one of the major toxin-antitoxin systems, RelEB, in a cell density-dependent manner in Escherichia coli K-12 MG1655. We constructed a strain, IKA121, which expresses the toxin RelE in the presence of rhamnose and lacks chromosomal relBE and rhaBAD. With this strain, we demonstrated that RelE-mediated dormancy is enhanced at high cell densities compared to that at low cell densities. The initiation of expression of the antitoxin RelB from a plasmid, pCA24N, reversed RelE-mediated dormancy in bacterial cultures. The activation of RelE increased the appearance of persister cells against β-lactams, quinolones, and aminoglycosides, and more persister cells appeared at high cell densities than at low cell densities. Further analysis indicated that amino acid starvation and an uncharacterized extracellular heat-labile substance promote RelE-mediated dormancy. This is a first report on the induction of RelE-mediated dormancy by high cell density. This work establishes a population-based dormancy mechanism to help explain E. coli survival in stressful environments.
INTRODUCTION
Dormancy is an adaptive response to environmental stress, and this strategy is observed with both eukaryotic organisms and bacteria. Bacteria can enter into dormant conditions as spores or persister cells and in viable but nonculturable (VBNC) states (30, 34). Dormant bacteria do not proliferate but are able to tolerate environmental stress and eventually recover under normal growing conditions.
Persister cells, which are a small subpopulation of apparently nongrowing multidrug-tolerant cells, are observed in bacterial biofilms (21, 24, 43). Biofilms are formed when bacterial cells attach to a surface and grow into a mass encapsulated by an exopolymer matrix (11). In biofilms, bacterial cells are very dense, and bacteria exhibit social behavior through the use of extracellular signals, a mechanism called quorum sensing (40). This mechanism enables bacteria to coordinate the activation and deactivation of multiple genes in a cell density-dependent manner via the secretion and recognition of several different types of signals. Thus, the biofilm is prominently involved in bacterial dormancy and quorum sensing.
It has been suggested that toxin-antitoxin (TA) modules are involved in the entry of Escherichia coli into dormant states (24, 31, 37). Several toxins, including HipA, RelE, YafQ, TisB, MqsR, CspD, and Hha, are associated with persister formation (13, 18, 24, 25). RelEB is among the most studied TA systems in E. coli and encodes a cytotoxin, RelE, which cleaves mRNA on translating ribosomes, and an antitoxin, RelB, which antagonizes RelE by direct protein-protein interaction (10). Transcription of relBE is autoregulated by the antitoxin RelB via binding to the relBE promoter region, and RelE enhances its repression by functioning as a corepressor (16). The transcriptional level of relBE is strongly induced by amino acid starvation (9, 10). While the expression of relBE is very low during exponential cell growth, its expression rapidly increases under amino acid starvation conditions (10). During starvation, the antitoxin RelB is degraded by the Lon protease, and RelE initiates the cleavage of mRNA (10).
Persister cells are frequently formed in biofilms, where bacteria can exist in high-cell-density states (24, 43). Several previous reports indicate an association between quorum-sensing signals, TA systems, and the appearance of persister cells. In E. coli, a TA system, YgiT-MqsR, is induced in biofilms and also by the quorum-sensing autoinducer AI-2 (39, 47), which is an interspecies-signaling molecule secreted by both Gram-negative and Gram-positive bacteria, including E. coli. Furthermore, extracellular death factor (EDF), Asn-Asn-Trp-Asn-Asn, functions as a quorum-sensing molecule and posttranscriptionally induces activity of the toxins MazF and ChpBK in E. coli (5, 28). In Pseudomonas aeruginosa, N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12 HSL), which is a major autoinducer, and pyocyanin, an extracellular virulence factor secreted during the stationary phase, significantly increase the number of persisters (32). These data indicate a potential link between quorum sensing, TA systems, and persister formation.
Although a large amount of progress has been made in understanding persister cell formation, there are still many unsolved questions concerning the mechanism mediating bacterial entry into dormancy. In this study, we focused on one major TA system, RelEB, which is associated with the formation of persister cells. We constructed a strain to express relE from the E. coli chromosome and investigated the effects of cell density on RelE-mediated bacterial phenotype changes. We have shown that RelE-mediated dormancy occurs in a cell density-dependent manner.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli K-12 MG1655 and its isogenic mutants were used in this study. The strains and plasmids used in this study are listed in Table 1. Bacteria were grown in M9 liquid medium or Luria-Bertani (LB) medium at 37°C. When appropriate, the medium was supplemented with 0.2% Casamino Acids and 0.2% or 1% rhamnose. The PT5-lac promoter was induced by the addition of 100 μM isopropyl β-d-1-thiogalactopyranoside (IPTG). Antibiotics were used at the following concentrations: 100 μg/ml ampicillin, 5 μg/ml ofloxacin, 25 μg/ml kanamycin, and 25 μg/ml chloramphenicol.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| E. coli K-12 strains | ||
| MG1655 | F− λ−ilvG rfb-50 rph-1 | Laboratory collection |
| ΔrelBE strain | MG1655 ΔrelBE | This study |
| ΔrhaBAD strain | MG1655 ΔrhaBAD | This study |
| IKA119 | MG1655 ΔrhaBAD ΔrelBE | This study |
| IKA121 | MG1655 ΔrhaBAD ΔrelBE PrhaB-relE | This study |
| IKA121 ΔtnaA | IKA121 ΔtnaA::Kmr | This study |
| Plasmids | ||
| pKD13 | Kmr; template for mutant construction | 12 |
| pKD46 | Ampr; temperature-sensitive λ red recombinase expression vector | 12 |
| pCP20 | Ampr Cmr; temperature-sensitive replication and thermal induction of FLP recombinase | 7 |
| pBAD18 | Ampr; arabinose-inducible expression plasmid | 17 |
| pCA24N | Cmr; lacIq pCA24N | 26 |
| pCA24N-relB | Cmr; lacIq pCA24N PT5-lac::relB+ | 26 |
Construction of mutants.
MG1655 ΔrelBE, ΔrhaBAD, ΔtnaA, and IKA119 (MG1655 ΔrhaBAD ΔrelBE) strains were constructed using the one-step inactivation method previously described by Datsenko and Wanner (12). IKA121 (MG1655 ΔrhaBAD ΔrelBE PrhaB-relE) was constructed by starting with PCR amplification of chromosomal relE with the primers relEF-EcoRI (5′-CGGAATTCGGGAGTGAAACGATGGCGTATTTTCTGGATTTTGAC-3′) and relER-XbaI (5′-GCTCTAGATCAGAGAATGCGTTTGACCG-3′) from MG1655 genomic DNA. The DNA fragment was digested with EcoRI and XbaI before being ligated into a multicloning site of a pBAD18 plasmid digested with EcoRI and XbaI to generate pBAD18::relE. Then, a fragment containing aphA (a kanamycin resistance gene) was amplified using the primers P1-SpeI (5′-CGACTAGTGTGTAGGCTGGAGCTGCTTC-3′) and P4-HindIII (5′-GTAACAAGCTTATTCCGGGGATCCGTCGACC-3′) from pKD13, digested with SpeI and HindIII, and ligated downstream of relE in pBAD18::relE digested with XbaI and HindIII to generate pBAD18::relE-aphA. A DNA fragment containing relE and aphA was amplified with the primers relEF-rhaBADKO (5′-ATTCAGGCGCTTTTTAGACTGGTCGTAATGAAATTCAGCAGGATCACATTATGGCGTATTTTCTGGATTTTGAC-3′) and P4-rhaBADKO (5′-ATGCCTAAGTTAGCCGCAGGATCAAGCTGGACGTTACGGAAGAATTTGCCATTCCGGGGATCCGTCGACC-3′) from pBAD18::relE-aphA and transformed into IKA119/pKD46. Kanamycin-resistant colonies were retransformed with pCP20, and aphA was removed from the bacterial chromosome.
Determining the effects of RelE toxin.
E. coli IKA121 was grown until it reached exponential phase (optical density at 600 nm [OD600], 0.6) in M9 containing 0.2% Casamino Acids and was diluted to approximately 104 or 107 cells/ml in fresh medium in the presence or absence of 0.2% rhamnose. To examine the relationship between initial cell density and a decrease in CFU, the initial cell concentrations were diluted to 104, 105, 106, or 107 cells/ml. Glucose was not added to avoid the repression of the rhaB promoter. To determine CFU counts, samples were withdrawn at a particular incubation time, diluted with 0.85% NaCl, and spread on LB plates. After incubation for 16 h, CFU were counted. To examine the recovery from dormancy, E. coli IKA121/pCA24N and IKA121/pCA24N-relB were incubated in M9 containing 0.2% Casamino Acids and 25μg/ml chloramphenicol in the presence and absence of 1% rhamnose, and CFU were counted with LB agar containing 100 μM IPTG to induce expression of the plasmid-based antitoxin RelB in pCA24N-relB. To examine the effects of extracellular substances, E. coli IKA121 was grown to an OD600 of 0.3 (exponential phase) or an OD600 of 1.0 (stationary phase) in M9 medium containing 0.2% Casamino Acids, the cells and the supernatants were separated by centrifugation, and the supernatants were passed through 0.2-μm filters. When necessary, heat treatment of the supernatant was conducted at 60, 80, or 100°C for 20 min. Exponentially growing cells were washed and inoculated into the filtered supernatants containing 0.2% rhamnose.
Persistence assay.
Persistence was measured by evaluating survival following exposure to antibiotics. E. coli IKA121/pCA24N-relB was preincubated in M9 containing 0.2% Casamino Acids and 25 μg/ml chloramphenicol before being diluted to 104 or 107 cells/ml in fresh medium in the presence or absence of 1% rhamnose and was then incubated for 3 or 10 h. Antibiotic (ampicillin, ofloxacin, or kanamycin) was added to the culture and incubated for 3, 6, or 12 h at 37°C. The cells were collected with a 0.2-μm-pore polyvinylidene difluoride (PVDF) membrane filtration unit (Millipore). The membrane-collected cells were resuspended in 0.85% NaCl, sonicated for 5 min, and vortexed for 1 min. CFU were counted using LB agar plates containing 100 μM IPTG.
Quantitative real-time RT analysis.
The expression of rhaA was quantified using a real-time reverse transcription (RT) assay. E. coli MG1655 was preincubated in M9 medium containing 0.2% Casamino Acids with or without 0.2% rhamnose to adjust the initial cell density to approximately 104 or 107 cells/ml. After 6 h of incubation, the total RNA was extracted from approximately 108 cells by using an RNeasy minikit (Qiagen, Valencia, CA) using the manufacturer's protocol. The presence of extracted RNA was confirmed using a Cosmo-i microchip SV1210 electrophoretic instrument (Hitachi, Tokyo, Japan). The RNA was converted into cDNA by using an ExScript RT reagent kit (Takara, Otsu, Japan). The remaining RNA was digested with RNase H, and the first-strand cDNA was used directly as a template for PCRs with SYBR premix Ex Taq (Takara) using an ABI7000 sequence detection system (Applied Biosystems, United States). The rhaA-specific primers (rhaA-RTF, 5′-CGCCGTTGAGAGCAAATTGT-3′, and rhaA-R, 5′-GGTGGCATACCCCATGTAAAACT-3′) were used. The expression values were calculated with the following formula: (cDNA of rhaA)/(ng of total RNA). The experiments were repeated three times.
Bacterial viability assay.
The viability of cells in planktonic shaking culture was determined using a BacLight Live/Dead bacterial viability staining kit (Molecular Probes, Inc., Eugene, OR) as described previously (44). Two stock solutions of stain (SYTO9 and propidium iodide) were diluted to a concentration of 3 μl/ml in medium. The live SYTO9-stained cells and dead propidium-stained cells were excited with a cool argon laser and a helium neon laser, respectively, and detected with 560-nm and 505-nm long-pass filters, respectively, using a scanning confocal laser microscope (LSM 5 PASCAL; Carl Zeiss, Oberkochen, Germany). The results are expressed as the means and standard deviations (SD) from five representative images.
RESULTS
RelE-mediated CFU decrease is enhanced at a high cell density.
In order to investigate the effects of cell density on the activity of the toxin RelE, we constructed the strain E. coli IKA121, which lacks rhaBAD and relBE transcriptional units but has the relE gene incorporated downstream of the rhaB promoter in the chromosome of E. coli K-12 MG1655 (Fig. 1). This strain possesses a rhamnose-inducible RelE toxin system. Exponentially growing cells were diluted to approximately 104 cells/ml (low cell densities) or 107 cells/ml (high cell densities) in M9 minimal medium containing 0.2% Casamino Acids with or without 0.2% rhamnose and incubated for 10 h. CFU were then determined at several time points (Fig. 2A). In the absence of rhamnose, the CFU count increased when the initial cell concentrations were 104 and 107 cells/ml. In the presence of rhamnose, the CFU count decreased when the initial cell concentration was 107 but not when the initial cell concentration was 104 cells/ml. To determine the critical cell concentration, exponentially growing cells were inoculated into M9 medium in the presence of rhamnose with various initial cell concentrations, ranging from 104 cells/ml to 109 cells/ml. The results showed that CFU decreased remarkably at an initial cell concentration of 108 cells/ml (Fig. 2B). A CFU decrease was observed in IKA121 at an initial cell concentration of 107 cells/ml but not in IKA119 (ΔrhaBAD ΔrelBE), MG1655 wild type, and the relBE mutant (Fig. 2C), suggesting that other toxins do not cause CFU decrease under these experimental conditions and that RelE-mediated CFU decrease occurs when RelE is overexpressed. A CFU decrease at high cell density was also observed when LB medium was used (Fig. 2D). A quantitative analysis of rhaA expression using real-time reverse transcription-PCR confirmed that the difference in initial cell densities (104 and 107 cells/ml) did not alter the activation of the rhaB promoter in MG1655 in the presence of rhamnose (Fig. 2E). These data indicate that a RelE-mediated CFU decrease is more evident at high cell densities.
Fig 1.
Overview of the rhaBAD operon and relBE operon in E. coli K-12 MG1655 and its derivative mutant IKA121. The expression of relE is controlled by PrhaB in IKA121.
Fig 2.
RelE-mediated CFU decrease is enhanced at a high cell density. (A) The effect of RelE on CFU decrease at low and high cell densities. E. coli IKA121 was inoculated at an initial concentration of 104 (open symbols) or 107 (closed symbols) cells/ml and cultured in the absence (dashed lines) and presence of 0.2% rhamnose (solid lines) in M9 medium containing 0.2% Casamino Acids, and the CFU were determined at each time point. (B) Exponentially growing cells of IKA121 were washed and inoculated at an initial concentration of 104, 105, 106, 107, 108, or 109 cells/ml and cultured for 6 h in M9 medium containing 0.2% Casamino Acids and 0.2% rhamnose. The percentage of culturable cells was calculated by comparing the CFU count of the induced culture at the final time point to the CFU count at the zero time point. (C) Exponentially growing cells of the IKA121, IKA119, MG1655, or ΔrelBE strain were inoculated at an initial concentration of 104 (black bars) or 107 (white bars) cells/ml and cultured for 6 h in M9 medium containing 0.2% Casamino Acids, and the CFU were determined. (D) IKA121 was inoculated at an initial concentration of 104 or 107 cells/ml and cultured for 6 h in LB containing 0.2% rhamnose. The percentage of culturable cells was calculated by comparing the CFU of the induced culture at the final time point to the CFU at the zero time point. (E) Quantitative real-time RT-PCR analysis of the rhaA transcript levels in MG1655 during the exponential phase in M9 medium containing 0.2% Casamino Acids with or without 0.2% rhamnose. The cells were grown to the exponential growth phase, diluted to 104 or 107 cells/ml, and incubated for 4 h. The data points represent the average results from triplicates, and the error bars indicate standard deviations.
RelE promotes a shift to dormancy at high cell densities.
We examined whether induction of RelE transcription induces cell death or induces a shift to the dormant state in which the cells are still viable but unable to proliferate. A Live/Dead assay indicated that more than 99% of the cells were still alive despite the activation of the RelE toxin (data not shown). These results suggest that the RelE toxin does not kill the cells but instead promotes a shift to dormancy. To further corroborate this RelE-mediated bacterial dormancy, the CFU counts of IKA121/pCA24N and IKA121/pCA24N-relB grown on LB agar plates with or without 100 μM IPTG were examined. Dormant IKA121/pCA24N-relB cells recovered growth capability when the antitoxin RelB was expressed on an LB agar plate containing IPTG. As the presence of 1% but not 0.2% rhamnose decreased the CFU of IKA121/pCA24N after 10 h of incubation (data not shown), RelE was activated with 1% rhamnose in the later experiments when using strains harboring pCA24N. The CFU of IKA121/pCA24N-relB decreased on an LB plate but not on an LB plate containing IPTG (Fig. 3), suggesting that RelE-mediated dormancy could be reversed by the expression of the antitoxin RelB. These data indicate that the production of the RelE toxin does not kill cells but induces cell dormancy under high-cell-density conditions. This dormancy condition could be reversed by the expression of the RelB antitoxin.
Fig 3.
RelE causes dormancy, and the antitoxin RelB can restore culturability. IKA121/pCA24N and IKA121/pCA24N-relB were inoculated at an initial concentration of 104 or 107 cells/ml and cultured in M9 medium containing 0.2% Casamino Acids and 1% rhamnose for 10 h. The CFU were determined using LB agar plates with or without 100 μM IPTG. The percentage of culturable cells was calculated by comparing the CFU of the induced culture at the final time point to the CFU at the zero time point. The data points represent the average results from triplicate experiments, and the error bars indicate the standard deviations.
RelE-mediated persister cells appear at a high cell density.
RelE is known to elicit the appearance of persister cells (24). To investigate whether cell density affects the RelE-induced rate of appearance of persister cells, the persistence of IKA121/pCA24N-relB cells against the β-lactam ampicillin, the quinolone ofloxacin, and the aminoglycoside kanamycin was measured. Exponentially growing cells were diluted to low or high cell densities (104 or 107 cells/ml, respectively), were incubated with or without rhamnose for 3 h, and were then exposed to each antibiotic. When rhamnose was not added, the persister cells appeared at a rate of less than 1/1,000. These tendencies were not altered by differences in cell densities (Fig. 4A). On the other hand, when RelE was induced by 1% rhamnose, the rate of persister cell appearance varied depending on the cell densities (Fig. 4B). As shown in Fig. 4C, a higher number of persister cells appeared at high cell densities than at low cell densities. These results indicate that cell density is a critical factor in the RelE-mediated appearance of persister cells.
Fig 4.
RelE-mediated appearance of persisters at high cell densities. Exponentially growing IKA121/pCA24N-relB was inoculated at an initial concentration of 104 (open symbols) or 107 (closed symbols) cells/ml and cultured in the absence (A) and presence (B) of 1% rhamnose in M9 medium containing 0.2% Casamino Acids and 25 μg/ml chloramphenicol to maintain plasmid. After 3 h of incubation, either ampicillin (squares), ofloxacin (diamonds), or kanamycin (circles) was added, and the culture was incubated for another 3 h and 6 h. The CFU were determined using LB agar containing 100 μM IPTG. The data represent the average results from triplicate experiments, and the error bars indicate the standard deviations. (C) Percentage of surviving cells after exposure to antibiotics for 6 h. The data points represent the average results from triplicate experiments, and the error bars indicate the standard deviations. Amp, ampicillin; Ofl, ofloxacin; Km, kanamycin.
To further investigate the effect of the RelE toxin on resistance to antibiotics at high cell densities, RelE was induced for different periods (3 h and 10 h), and the effects of longer exposure (12 h) to each antibiotic were examined (Fig. 5). When RelE was induced for 3 h, the number of bacterial cells tolerant to ampicillin was much higher than the number of cells resistant to ofloxacin or kanamycin. Conversely, when RelE was induced for 10 h, the number of cells tolerant to ofloxacin was the same as the number of cells resistant to ampicillin. These data suggest that a long-term induction of RelE enables bacterial cells to survive despite being exposed to ofloxacin.
Fig 5.
RelE induction time changes resistance to antibiotics. Exponentially growing IKA121/pCA24N-relB was inoculated at an initial concentration of 107 cells/ml and cultured in the presence of 1% rhamnose in M9 medium containing 0.2% Casamino Acids and 25 μg/ml chloramphenicol to maintain plasmid. After a 3-h (short-term) or 10-h (long-term) incubation, either ampicillin, ofloxacin, or kanamycin was added, and the culture was incubated for another 12 h. The CFU were determined using LB agar containing 100 μM IPTG. The data points represent the average results from triplicate experiments, and the error bars indicate the standard deviations.
Amino acid starvation and an extracellular heat-labile substance promote RelE-mediated dormancy.
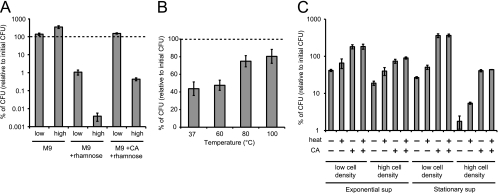
The question is why RelE-mediated dormancy is enhanced at high cell densities. It is hypothesized that nutrition exhaustion and accompanying starvation occurs more easily under high-cell-density conditions than low-cell-density conditions. To corroborate whether such starvation induces dormancy, cells were diluted in M9 medium with or without Casamino Acids. The CFU count decreased when Casamino Acids was absent (Fig. 6A). Previously, it has been reported that mazEF and hicAB, as well as relBE, are induced by amino acid starvation (9, 10, 22). A CFU decrease following Casamino Acids starvation only occurred when rhamnose was added (relE is expressed) (Fig. 6A), suggesting that RelE, not other toxins, affected dormancy under these experimental conditions. RelE-mediated dormancy was promoted by initial cell densities regardless of the presence of Casamino Acids (Fig. 6A), suggesting that other factors also influence cell density-dependent RelE-mediated dormancy.
Fig 6.
Amino acid starvation and a heat-labile extracellular substance promote RelE-mediated dormancy. (A) The effects of amino acids on RelE-mediated dormancy. IKA121 was inoculated into M9 with or without rhamnose and Casamino Acids (CA) at an initial concentration of 104 (low) or 107 (high) cells/ml and cultured for 6 h. (B) The effects of the supernatant on RelE-mediated dormancy. The culture in the M9 medium with Casamino Acids was collected during the exponential phase, and the cells and the supernatant were separated. The supernatant was incubated at 37, 60, 80, or 100°C for 20 min, and rhamnose was added to obtain a concentration of 0.2%. Washed cells were then inoculated into the supernatants at an initial concentration of 104 cells/ml and cultured for 6 h. (C) The effects of the combination of amino acid starvation and the supernatant (sup) on RelE-mediated dormancy. The cells and the supernatant from the M9 culture with Casamino Acids during the exponential phase or the stationary phase were separated, and rhamnose was added to the supernatant. Washed cells were inoculated into the supernatant containing rhamnose at an initial concentration of 104 cells/ml and cultured for 6 h. In all experiments, the percentage of culturable cells was calculated by comparing the CFU count of the induced culture at the final time point to that at the zero time point. The data points represent the average results from triplicate experiments, and the error bars indicate the standard deviations.
Another possibility is that an extracellular substance secreted by bacterial cells may affect RelE-mediated dormancy. To investigate the effects of these extracellular substances, an exponentially growing culture was separated into cells and supernatant. The cells were then washed, diluted to 104 cells/ml in the filtered supernatant, and incubated with rhamnose. The CFU count decreased after 6 h of incubation, and the effective factors in the supernatants were partially inactivated after incubation at over 80°C (Fig. 6B). When exponentially growing culture was inoculated to a fresh medium that was heated or not heated, the heat treatment did not alter the CFU count (data not shown), suggesting that heating M9 itself did not affect RelE-mediated dormancy. Hence, it is suggested that an extracellular heat-labile substance may promote RelE-mediated dormancy.
In E. coli, at least two quorum-sensing signals are known: indole, which regulates biofilm formation and antibiotic resistance (20, 29), and the pentapeptide EDF, which can signal induce cell death via another major TA system, MazEF (27, 28). We confirmed that the addition of EDF does not affect RelE-mediated dormancy in this experiment (data not shown). Furthermore, we constructed an IKA121 ΔtnaA mutant, which does not synthesize indole, and confirmed that this strain presents a tendency toward RelE-mediated CFU decrease similar to that of a wild-type strain (data not shown). From these data, we suggest that a novel uncharacterized extracellular heat-labile signal may enhance RelE-mediated dormancy.
To further investigate the contribution of these factors to RelE-mediated dormancy, the combination of amino acid starvation and extracellular heat-labile substance was examined. Under conditions of both low and high cell densities, the heat treatment of supernatants of the exponential phase decreased the CFU count when Casamino Acids was not supplemented (Fig. 6C). Moreover, the decrease in CFU was remarkable when bacterial cells were exposed to the supernatant of stationary-phase cells (Fig. 6C), suggesting that an extracellular heat-labile substance may accumulate in the supernatant during the stationary phase and promote RelE-mediated dormancy under conditions of amino acid starvation.
DISCUSSION
The mechanism mediating RelE toxin inhibition of translation has been studied extensively (8–10, 33, 35, 36, 38, 42), and the relationship between RelE and the appearance of persister cells has also been shown (24, 31). On the other hand, the relationship between bacterial populations and the toxic actions of RelE is still unclear, although the appearance of persister cells in biofilms has frequently been reported (21, 24, 43). In this study, we report on several important pieces of experimental evidence that aid in understanding RelE-mediated dormancy, including the importance of cell density on the action of the RelE toxin and the changes of cellular phenotypes induced by the RelE toxin.
Several studies have indicated the importance of cell density in the appearance of persister cells (21, 23, 32, 43). Spoering and Lewis have shown that persister cells appear in a stationary planktonic population or inside a biofilm but not in exponential planktonic conditions in P. aeruginosa cultures (43). In a study of E. coli, Keren et al. have indicated that the number of persister cells increases dramatically in the mid-exponential phase (24). There seem to be at least two reasons for the high persister appearance rates under such conditions: environmental stresses and extracellular signals. In regard to environmental stresses, oxygen limitation is generally cited as a major factor inducing the appearance of persister cells inside biofilms. The repression of metabolic activity has been confirmed inside mature biofilms of P. aeruginosa, Klebsiella pneumoniae, Staphylococcus epidermidis, and Shewanella oneidensis (1, 3, 45, 46). These metabolically inactivated cells have resistance to antibiotics (3, 46). Another environmental stress presumed to be associated with dormancy is nutrient starvation. Transcription of TA-coding genes, including relBE, mazEF, and hicAB, is strongly induced by amino acid starvation (9, 10, 22). In the case of relBE, the activity of the Lon protease, which degrades RelB, increases during amino acid starvation. The transcription of relBE increases dramatically, and RelE initiates the cleavage of mRNA (10, 36). In our experiments, amino acid starvation increased RelE-mediated dormancy in our experimental strain (Fig. 6A and C), in which relB is deleted and relE expression is controlled by the rha promoter. These results suggest that amino acid starvation may increase RelE's toxic activity by means other than a simple increase in relBE transcription. Amino acid limitations increase the intercellular concentration of guanosine tetraphosphate (ppGpp) and cause pleiotropic physiological changes; this phenomenon is defined as the stringent response (6). Because ppGpp inhibits the transcription of stable RNA operons (6), we believe that this shutdown of stable RNA synthesis may increase RelE-mediated dormancy.
Our results also imply the existence of an extracellular signal that promotes RelE-mediated dormancy (Fig. 6B and C). This substance was heat labile and could therefore be a peptide. Before now, the mechanism of population toxin activation of MazEF has been explored by Engelberg-Kulka and her colleagues (5, 27, 28). MazF-mediated cell death occurs under various stressful conditions, including bacteriostatic antibiotic exposure and UV irradiation (19). In addition, MazF-mediated cell death is promoted by the presence of the quorum-sensing pentapeptide EDF (28). Recently, it has been demonstrated that EDF specifically binds to the toxin MazF and amplifies the endonucleotic activity of MazF in vitro (5). Our results indicate that an undefined extracellular substance affects RelE activity in the strain lacking the antitoxin RelB. The detailed mechanism mediating this heat-labile substance in the activation of RelE remains unclear, but it may be that RelE toxin sensitivity is affected, for instance, by inhibition of mRNA cleavage by RelE. Further chemical identification of the heat-labile substance will be required to understand the mechanism of RelE activation in bacterial populations.
The timing of persister appearance has been studied, using a microfluidic device, by Gefen et al. (15). When bacterial cells are transferred from a stationary culture to fresh medium, protein is still synthesized for a certain period, and a small population of bacterial cells will develop persistence against ampicillin after the lag phase. Although the molecular mechanism responsible for the generation of subpopulations of persister cells after the lag phase remains unknown, it may be that some environmental signals induce the appearance of persister cells. The results of our study support the possibility that the existence of extracellular signals induces the appearance of persister cells.
We found that persister cells induced by RelE present different tolerances to antibiotics (Fig. 4 and 5). The variation in antibiotic sensitivity of persister cells has been noted in several reports (2, 21, 48). We propose a model in which dormant states are classified into at least three different phases based on metabolic conditions (Fig. 7). In phase 1, bacterial growth is repressed, but DNA synthesis and protein synthesis still occur, and the cells at this stage are able to resist β-lactams. In phase 2, bacterial DNA synthesis is arrested, but protein synthesis is not completely shut down. The cells at this stage are able to resist β-lactams and quinolones. In phase 3, bacterial protein synthesis is finally and completely stopped. This stage enables cells to be resistant to β-lactams, quinolones, and aminoglycosides. Previously, Balaban et al. showed two types of persister cells: type I persisters, which constitute a preexisting population of nongrowing cells, and type II persisters, which constitute a subpopulation of slowly growing cells (4, 14). Because both types of persister cells are resistant to ampicillin (4), growth does not seem to have to be stopped to obtain persistence against β-lactams in phase 1. It has been previously shown that cells inside mature E. coli biofilms formed in a flow cell are resistant to ampicillin but not to ofloxacin and kanamycin (21). This result suggests that environmental stresses, such as nutrient depletion or low concentrations of oxygen, cause cells to enter into phase 1 dormancy. In our study, the short-term induction of RelE enabled high-density cells to resist short-term exposure to three kinds of antibiotics (Fig. 4). This result is consistent with a previous report by Keren et al. (24). However, experiments with long-term exposure to antibiotics revealed that the duration of RelE induction modulates the dormancy stages. Short-term RelE induction brings the population to phase 1, while long-term RelE induction leads to phase 2 (Fig. 5). In this way, RelE toxin induction length and cell density significantly influence dormancy levels. Recently, Allison et al. reported that aminoglycosides but not β-lactams and quinolones eradicate persister cells in the presence of a carbon source (2). As they obtained persister cells by treatment with ofloxacin, the bacterial cells may have been situated in phase 2, and it is likely that only aminoglycosides could kill the persister cells. Mycobacterium tuberculosis has three homologues of RelE, and they have different effects on survival rates in the presence of antibiotics (41). While RelE2 specifically affects bacterial survival in the presence of rifampin, RelE3 increases those in the presence of gentamicin, levofloxacin, and isoniazid. In this way, each toxin may affect bacterial persistence in media with antibiotics through different mechanisms of action. It will be interesting to understand the contribution of other toxins to antibiotic survival in E. coli in the future.
Fig 7.
Schematic representation of RelE-mediated dormancy. The E. coli relBE operon synthesizes the labile antitoxin RelB and toxin RelE. RelE cleaves mRNA on the ribosome and inhibits translation. RelB forms a tight complex with RelE and neutralizes the toxicity. The dashed lines indicate the effects proposed in this study. Amino acid starvation and an unidentified extracellular heat-labile substance promote entry into dormancy by RelE. It is proposed that dormant states can be classified into at least three different levels. In phase 1, bacterial growth is repressed, and cells at this stage can resist β-lactams. In phase 2, bacterial DNA synthesis is stopped, and cells at this stage can resist β-lactams and quinolones. In phase 3, bacterial protein synthesis is stopped, and cells at this stage can resist β-lactams, quinolones, and aminoglycosides.
Here, we have demonstrated that RelE-mediated dormancy is enhanced in high-density bacterial populations using an experimental strain in which single-copy relE is located on the chromosome and in which relE expression is controlled by the addition of rhamnose. Amino acid starvation and an uncharacterized extracellular heat-labile substance enhance RelE-mediated dormancy. The cell density leads to an increased number of persister cells, and the length of the RelE induction period leads to changes in dormancy level. Our findings add to the growing understanding of the mechanism of RelE toxic activity and persister cell physiology.
ACKNOWLEDGMENTS
This research was financially supported by CREST, Japan Science and Technology Agency. Yosuke Tashiro is supported by a Scientific Research Fellowship from the Japan Society for the Promotion of Sciences (JSPS).
We are grateful for the ASKA strains provided by the Genome Analysis Project in Japan.
Footnotes
Published ahead of print 30 December 2011
REFERENCES
- 1. Agladze K, Wang X, Romeo T. 2005. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J. Bacteriol. 187:8237–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderl JN, Zahller J, Roe F, Stewart PS. 2003. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 47:1251–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625 [DOI] [PubMed] [Google Scholar]
- 5. Belitsky M, et al. 2011. The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol. Cell 41:625–635 [DOI] [PubMed] [Google Scholar]
- 6. Chatterji D, Kumar Ojha A. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160–165 [DOI] [PubMed] [Google Scholar]
- 7. Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 8. Christensen SK, Gerdes K. 2004. Delayed-relaxed response explained by hyperactivation of RelE. Mol. Microbiol. 53:587–597 [DOI] [PubMed] [Google Scholar]
- 9. Christensen SK, Gerdes K. 2003. RelE toxins from Bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48:1389–1400 [DOI] [PubMed] [Google Scholar]
- 10. Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. U. S. A. 98:14328–14333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Costerton J, Stewart P, Greenberg E. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 12. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dorr T, Vulic M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8:e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gefen O, Balaban NQ. 2009. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol. Rev. 33:704–717 [DOI] [PubMed] [Google Scholar]
- 15. Gefen O, Gabay C, Mumcuoglu M, Engel G, Balaban NQ. 2008. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:6145–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gotfredsen M, Gerdes K. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065–1076 [DOI] [PubMed] [Google Scholar]
- 17. Guzman L, Belin D, Carson M, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrison JJ, et al. 2009. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 53:2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hazan R, Sat B, Engelberg-Kulka H. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186:3663–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. 2005. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 55:1113–1126 [DOI] [PubMed] [Google Scholar]
- 21. Ito A, Taniuchi A, May T, Kawata K, Okabe S. 2009. An increase in antibiotic resistance of Escherichia coli in mature biofilms. Appl. Environ. Microbiol. 75:4093–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jorgensen MG, Pandey DP, Jaskolska M, Gerdes K. 2009. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 191:1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13–18 [DOI] [PubMed] [Google Scholar]
- 24. Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim Y, Wood TK. 2010. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 391:209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kitagawa M, et al. 2005. Complete set of ORF clones of Escherichia coli ASKA library (A complete Set of E. coli K-12 ORF Archive): unique resources for biological research. DNA Res. 12:291–299 [DOI] [PubMed] [Google Scholar]
- 27. Kolodkin-Gal I, Engelberg-Kulka H. 2008. The extracellular death factor: physiological and genetic factors influencing its production and response in Escherichia coli. J. Bacteriol. 190:3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. 2007. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318:652–655 [DOI] [PubMed] [Google Scholar]
- 29. Lee J, Jayaraman A, Wood TK. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56 [DOI] [PubMed] [Google Scholar]
- 31. Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 108:13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Möker N, Dean CR, Tao J. 2010. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J. Bacteriol. 192:1946–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neubauer C, et al. 2009. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139:1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34:415–425 [DOI] [PubMed] [Google Scholar]
- 35. Overgaard M, Borch J, Gerdes K. 2009. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J. Mol. Biol. 394:183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Overgaard M, Borch J, Jørgensen MG, Gerdes K. 2008. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol. 69:841–857 [DOI] [PubMed] [Google Scholar]
- 37. Pedersen K, Christensen SK, Gerdes K. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501–510 [DOI] [PubMed] [Google Scholar]
- 38. Pedersen K, et al. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131–140 [DOI] [PubMed] [Google Scholar]
- 39. Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515–524 [DOI] [PubMed] [Google Scholar]
- 40. Singh P, et al. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764 [DOI] [PubMed] [Google Scholar]
- 41. Singh R, Barry CE, III, Boshoff HIM. 2010. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J. Bacteriol. 192:1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sørensen MA. 2001. Charging levels of four tRNA species in Escherichia coli Rel+ and Rel− strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J. Mol. Biol. 307:785–798 [DOI] [PubMed] [Google Scholar]
- 43. Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tashiro Y, et al. 2008. Opr86 is essential for viability and is a potential candidate for a protective antigen against biofilm formation by Pseudomonas aeruginosa. J. Bacteriol. 190:3969–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Teal TK, Lies DP, Wold BJ, Newman DK. 2006. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl. Environ. Microbiol. 72:7324–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walters MC, III, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamaguchi Y, Park J-H, Inouye M. 2009. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 284:28746–28753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zuroff TR, et al. 2010. Robustness analysis of culturing perturbations on Escherichia coli colony biofilm beta-lactam and aminoglycoside antibiotic tolerance. BMC Microbiol. 10:185. [DOI] [PMC free article] [PubMed] [Google Scholar]