Abstract
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacterium best known as the predominant opportunistic pathogen infecting the lungs of cystic fibrosis patients. In this context, it is thought to form biofilms, within which locally reducing and acidic conditions can develop that favor the stability of ferrous iron [Fe(II)]. Because iron is a signal that stimulates biofilm formation, we performed a microarray study to determine whether P. aeruginosa strain PA14 exhibits a specific transcriptional response to extracellular Fe(II). Among the genes that were most upregulated in response to Fe(II) were those encoding the two-component system BqsR/BqsS, previously identified for its role in P. aeruginosa strain PAO1 biofilm decay (13); here, we demonstrate its role in extracellular Fe(II) sensing. bqsS and bqsR form an operon together with two small upstream genes, bqsP and bqsQ, and one downstream gene, bqsT. BqsR/BqsS sense extracellular Fe(II) at physiologically relevant concentrations (>10 μM) and elicit a specific transcriptional response, including its autoregulation. The sensor distinguishes between Fe(II), Fe(III), and other dipositive cations [Ca(II), Cu(II), Mg(II), Mn(II), Zn(II)] under aerobic or anaerobic conditions. The gene that is most upregulated by BqsR/BqsS, as measured by quantitative reverse transcription-PCR (qRT-PCR), is PA14_04180, which is predicted to encode a periplasmic oligonucleotide/oligosaccharide-binding domain (OB-fold) protein. Coincident with phenazine production during batch culture growth, Fe(II) becomes the majority of the total iron pool and bqsS is upregulated. The existence of a two-component system that senses Fe(II) indicates that extracellular Fe(II) is an important environmental signal for P. aeruginosa.
INTRODUCTION
Iron is an essential element for nearly all forms of life. Iron generally presents itself to biological systems in one of two oxidation states: ferric [Fe(III)] or ferrous [Fe(II)] iron. Given that Fe(II) is oxidized by atmospheric oxygen, it is commonly assumed that organisms most frequently acquire iron in the Fe(III) form. In oxic environments at a circumneutral pH, Fe(II) rapidly oxidizes to Fe(III), precipitating as sparingly soluble ferric (hydr)oxide minerals (35). Within living systems, host proteins, such as members of the transferrin family, bind Fe(III) (1, 19). Given Fe(III)'s low solubility, considerable attention has been paid to Fe(III) sensing and acquisition by bacteria, and much is understood about these processes (43, 53, 59, 67). Less is understood about Fe(II) sensing and uptake despite the facts that many environments contain Fe(II) (2, 29) and that increased solubility of Fe(II) renders it more bioavailable than Fe(III) (35). In particular, Fe(II) is known to be important in acidic and/or reducing environments (35, 38, 57), and such conditions can easily arise at the microscale in diverse habitats (28, 46), including biofilms (21).
Pseudomonas aeruginosa is a Gram-negative bacterium capable of aerobic and anaerobic respiration. It inhabits many environments, ranging from soils (20) to marine sediments (24). It is best known as an opportunistic pathogen that infects immunocompromised individuals and, in particular, the mucus that accumulates on the surface of lung epithelial cells in individuals with cystic fibrosis (CF). Within this environment, Pseudomonas is thought to form biofilms—sessile, multicellular communities (26, 49) that are more resistant to conventional antimicrobial therapies than are their free-living counterparts (9). P. aeruginosa requires iron to form cofactors of enzymes that play essential roles in electron transfer and other important cellular processes. Iron also signals biofilm formation (5, 48), but the level of iron necessary to promote biofilm formation exceeds that required for assimilatory purposes (4, 40). In addition, the production of several virulence determinants, such as exotoxin A, is regulated in response to iron (30).
P. aeruginosa can acquire iron in several ways. One strategy is to secrete high-affinity Fe(III)-binding molecules, called siderophores, which outcompete transferrin and other host proteins for Fe(III) (43). For example, pyoverdine is one of the dominant siderophores made by P. aeruginosa. When pyoverdine-bound Fe(III) enters the periplasm via TonB-dependent outer membrane transporters, iron is thought to be released from the pyoverdine complex via reduction facilitated by its binding to an inner membrane ABC transporter, allowing Fe(II) to enter the cytoplasm (59). In addition, P. aeruginosa can excrete citrate, which also chelates Fe(III), albeit with lower affinity than siderophores, causing the iron to enter the cytoplasm as Fe(II) through a different TonB-dependent porin and the G-protein-like transporter FeoB (31, 32). Pseudomonas also contains two heme uptake systems whose outer membrane receptors are HasR and PhuR, which are energized by an ABC transporter and TonB, respectively (39).
Limiting cells for Fe(III) has received considerable attention from a therapeutic perspective (3, 34, 48). This is because Fe(III) limitation has been shown to significantly reduce biofilm formation in various in vitro systems (48). However, these systems are commonly operated under aerobic conditions and may not reflect in vivo conditions. Some evidence suggests anaerobic niches may exist in CF lung mucus where bacteria form dense biofilms (66), and biofilms themselves are known to harbor microdomains that are more acidic (21) or more reducing (54) than the bulk environment. Because steep chemical gradients can develop within biofilms over a small spatial scale (28), it is reasonable to expect that Fe(II) could exist in appreciable amounts in these microhabitats. Although the dominant iron oxidation state in the CF lung is unclear (44), it likely changes as infections progress and the local environment becomes increasingly reducing. On a related note, one reason to postulate that Fe(II) may be important for P. aeruginosa is that phenazines are known to be produced at micromolar concentrations in the CF lung (65). Phenazines can reduce host protein-bound Fe(III) (10), as well as Fe(III) bound up in mineral phases (60), to Fe(II). Fe(II) crosses the outer membrane via generic outer membrane porins and is actively transported across the cytoplasmic membrane via FeoB (31). Recently, it was shown that phenazine carboxylic acid can promote P. aeruginosa biofilm formation by stimulating Fe(II) acquisition in the presence of conalbumin (61). In addition, the presence of Fe(II) has been detected within a P. aeruginosa biofilm producing a gradient of the reduced phenazine pyocyanin (25).
Given these observations, we hypothesized that P. aeruginosa might respond differently to Fe(II) than to Fe(III). To test this, we performed a microarray experiment to measure the transcriptional response to Fe(II) or Fe(III) shock. Not only did we observe different responses depending on the iron oxidation state, we identified a two-component system that specifically senses extracellular Fe(II).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. Stationary-phase cultures of P. aeruginosa strains were grown aerobically at 37°C in 3 g/liter Bacto tryptic soy broth containing 100 mM KNO3. The overnight cultures were diluted to an optical density at 500 nm (OD500) of 0.01 in minimal metal medium (MMM). MMM consists of 0.3 g/liter Bacto tryptic soy broth, 50 mM glutamate, 1% glycerol, 100 mM KNO3, and 50 mM MOPS (morpholinepropanesulfonic acid); the pH was adjusted to 7.0 with NaOH, and the medium was treated with 1% (wt/vol) Amberlite chelating resin (Sigma) for 1 h and filtered into acid-washed medium bottles prior to autoclaving. After autoclaving, MMM was immediately sparged to maintain anaerobic conditions and brought into a vinyl anaerobic chamber containing a 5% hydrogen, 15% carbon dioxide, and 80% nitrogen gas mixture (Coy Laboratory Products). Ten milliliters of medium was added to 25-ml anaerobic Balch tubes, which were acid-washed in 1 M HCl to remove residual metals. Unless otherwise specified, inoculations were performed anaerobically in the anaerobic chamber with the previously stated gas mixture and incubated and shaken at 37°C. Spectrophotometric measurements were performed on a Beckman Spec20 unless otherwise specified.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa PA14 | ||
| WT | Clinical isolate UCBPP-PA14 | 38 |
| ΔbqsR mutant | Deletion in bqsR | This study |
| ΔbqsR mutant and pMQ64 | Deletion in bqsR; contains plasmid pMQ64 | This study |
| ΔbqsR mutant and pMQ64-bqsR | Deletion in bqsR; contains plasmid pMQ64-bqsR | This study |
| ΔbqsS mutant | Deletion in bqsS | This study |
| feoB mutant | feoB::MAR2xT7; transposon mutant containing Gmr cassette | 26 |
| E. coli | ||
| UQ950 | E. coli DH5α λ(pir) host for cloning; F-Δ(argF-lac)169 θ80 dlacZ58(ΔM15) glnV44(AS) rfbD1 gyrA96(Nalr) recA1 endA1 spoT1 thi-1 hsdR17 deoR λpir+ | D. Lies, Caltech |
| BW29427 | Donor strain for conjugation; thrB1004 pro thi rpsL hsdS lacZ ΔM15RP4-1360 Δ(araBAD)567 ΔdapA1341::[erm pir(wt)] | W. Metcalf, University of Illinois |
| S. cerevisiae | ||
| InvSc1 | MATa/MATα leu2/leu2 trp1-289/trp1-289 ura3-52/ura3-52 his3-Δ1/his3-Δ1 | Invitrogen |
| Plasmids | ||
| pMQ30 | pEX18-Gmr, URA3, CEN6, ARSH4; allelic replacement vector | 42 |
| pΔbqsR | 2-kb fusion PCR fragment containing the ΔbqsR mutant cloned into the BamHI/EcoRI site of pMQ30; used to make the ΔbqsR mutant | This study |
| pΔbqsS | 2-kb fusion PCR fragment containing the ΔbqsS mutant cloned into the BamHI/EcoRI site of pMQ30; used to make the ΔbqsS mutant | This study |
| pMQ64 | pMQ30 with a deletion of SacB | 42 |
| pMQ64-bqsR | bqsR cloned into pMQ64 | This study |
Gmr, gentamicin resistant; Nalr, nalidixic acid resistant.
Construction of bqsR and bqsS unmarked deletions.
The bqsR unmarked deletion was created using a pMQ30-based construct, which was made by applying the Saccharomyces cerevisiae-based molecular tool kit (47) (Table 1). This procedure takes advantage of yeast homologous recombination to combine the vector with PCR-amplified regions upstream and downstream from the gene of interest. The 5′ region (∼1 kb in length) of the sequence flanking bqsR was amplified using primers JW1_F and JW1_R, and the 3′ region (∼1 kb in length) of the sequence flanking bqsR was amplified using primers JW2_F and JW2_R (see Table S1 in the supplemental material). Both primer pairs contained regions that were homologous to each other, and the 5′ ends of the JW1_F and the JW2_R primers contained 40-bp regions of homology to pMQ30.
S. cerevisiae strain InvSc1 (Table 1) was cotransformed with plasmid linearized with EcoRI and BamHI as well as with the upstream and downstream PCR fragments; recombinant plasmids were generated by yeast homologous recombination (47). Recombinants were selected on synthetic defined medium lacking uracil (SD−Ura). Plasmids were liberated from uracil-auxotrophic yeast using the QIAprep Spin miniprep kit (Qiagen) yeast protocol and electroporated into Escherichia coli DH5α. Plasmids obtained from DH5α were electroporated into E. coli BW29427. The deletion plasmid was mobilized from BW29427 into PA14 using biparental conjugation (64). PA14 single recombinants (merodiploid containing the intact bqsR gene and a deleted gene) were selected on LB agar containing 100 μg/ml gentamicin. These colonies were then grown in the absence of selection to resolve merodiploids. Potential bqsR deletion mutants (resolved merodiploids) were identified by selecting for colonies that grew in the presence of 10% sucrose. Strains with properties of double recombination were further analyzed by PCR and sequenced to verify bqsR deletion (ΔbqsR). The above protocol was performed for the bqsS deletion (ΔbqsS), substituting primers dbqsS 1 and dbqsS 2 for primers JW1_F and JW1_R and dbqsS 3 and dbqsS 4 for primers JW2_F and JW2_R (see Table S1 in the supplemental material for primer sequences). Because the reading frames in the bqs operon overlap, we were careful to construct our deletions in this region so as to both render them nonpolar and preserve the integrity of the remaining genes in the operon.
Analysis of global gene expression using P. aeruginosa Affymetrix GeneChips.
The GeneChip P. aeruginosa genome array (Affymetrix) contains probe sets for more than 5,500 genes from P. aeruginosa PAO1 and 117 additional genes from strains other than PAO1. Due to the strong similarity between strains PAO1 and PA14, the Affymetrix array has been used for gene expression analysis of both strains (12, 33, 63). P. aeruginosa PA14 was grown in anaerobic MMM as described in “Bacterial strains and growth conditions.” The cultures were grown anaerobically at 37°C until they reached exponential phase (OD500 of 0.3), at which time they were removed from the incubator and were taken into the anaerobic chamber containing a gas mixture of 5% H2 and 95% N2 (Coy Laboratory Products). Water, 100 μM FeCl3, or 100 μM FeCl2 was added to the cultures. Iron concentrations were calculated by weight, assuming 100% purity of Fe(III) and Fe(II) stocks. The cultures were vortexed every 5 min over a 30-min period, after which 5 ml of culture was removed and mixed with 10 ml of bacterial RNAprotect (Qiagen). The cultures were incubated for 5 min at room temperature before cells were pelleted (10 min, 5,000 × g). DNA-free RNA was isolated from the cell pellet using the RNeasy minikit (Qiagen) that includes on-column DNase treatment. The preparation of the cDNA, as well as the processing of the P. aeruginosa GeneChip arrays, was performed by the BioMicro Center at the Massachusetts Institute of Technology (MIT) using an Affymetrix fluidics station. GeneChip arrays were performed in biological triplicate. Signal intensities were background corrected, normalized, and summarized using the robust multiarray averaging (RMA) method in the Bioconductor package affy (8, 14, 22). Linear models for each gene were fit using the program limma, and moderated t statistics were calculated with limma's empirical Bayes approach (50). P values were corrected for multiple testing using the Benjamini-Hochberg method to control the false-discovery rate (6); these adjusted values are referred to as q values. A threshold q value of 0.01 was used to identify differentially expressed genes, resulting in a false-discovery rate of 1% among genes called significant. Verification of GeneChip data was performed with quantitative reverse transcriptase PCR (qRT-PCR).
Validating iron concentrations.
Fe(II) readily oxidizes in the presence of trace levels of oxygen. Different Fe(II) salts have different stabilities in water. For most experiments, the Fe(II) source was ferrous chloride; it was used because the anion, chloride, was expected to have little effect on the metabolism of PA14. However, this form is less stable in water than ferrous ammonium sulfate. To be certain of the total iron concentration and to distinguish between Fe(II) and Fe(III), the ferrozine assay (56) was used. Ferrozine binds to Fe(II) and provides a colorimetric readout of Fe(II) concentration compared to that of a standard. Ferrous ammonium sulfate in acid was used as a standard because it is the most stable form of Fe(II) and, under acidic conditions, all the iron is expected to remain in the Fe(II) state. Iron-bound ferrozine was measured at 562 nm. Total iron concentration was determined by reducing the sample with hydroxylamine hydrochloride to convert all Fe(III) to Fe(II).
Fe(II) shock experiments.
Genes found to be strongly induced in the array experiment were examined using qRT-PCR (primers are listed in Table S1 in the supplemental material). P. aeruginosa strains listed in Table 1 were grown anaerobically in 10-ml cultures in triplicate as described in “Bacterial strains and growth conditions.” The Fe(II) shock was performed in the anaerobic chamber after the cultures grown in MMM reached early exponential phase. Five-milliliter samples of each culture were removed immediately before the Fe(II) shock and mixed with 2 volumes of bacterial RNAprotect (Qiagen), incubated for 5 min at room temperature, and centrifuged for 10 min at 5,000 × g. These served as the Fe-free control. A 50 μM concentration of Fe(NH4)2(SO4)2 was added to the cultures, mixed, and incubated for 30 min. After the 30-min shock, the remaining 5-ml culture was immediately added to 10 ml RNAprotect, incubated for 5 min at room temperature, and centrifuged for 10 min at 5,000 × g. The pellets were stored at −80°C until RNA isolation, and qRT-PCR analysis was performed.
Anaerobic metal shock experiments.
PA14 was grown in MMM to early exponential phase and treated as described in “Fe(II) shock experiments,” substituting Fe(III) and various dipositive cations for Fe(II). The metals tested were 100 μM Fe(NH4)2(SO4)2 [containing 100 μM Fe(II), as determined by weight and the ferrozine assay], 100 μM FeCl3, 5 mM CaCl2, 100 μM MnCl2, 10 mM MgCl2, 100 μM ZnCl2, and 100 μM CuCl2. The concentrations of calcium and magnesium were selected to approximate physiologically relevant concentrations (27).
Aerobic metal shock experiments.
PA14 was grown in MMM aerobically to early exponential phase and shocked with the metal concentrations given above. The Fe(II) concentration in the Fe(NH4)2(SO4)2 was determined by the ferrozine assay before and after the aerobic shock to determine if any iron remained in the Fe(II) state after the 30-min exposure to oxygen. The initial Fe(II) concentration was 100 μM, and the concentration after the 30-min shock was 4.9 μM.
Fe(II) titration.
Triplicate samples of 50 ml MMM in 125-ml acid-washed serum vials were inoculated from cultures grown as described in “Bacterial stains and growth conditions” and sealed. When the cells reached early exponential phase, the Fe(II) shock was performed. Five milliliters of culture was treated with RNAprotect before the iron shock and used as the no-iron control. Subsequently, 5 ml of culture was added to tubes containing iron and incubated for 30 min. The Fe(II) concentrations assayed were 6 μM, 19 μM, 31.6 μM, 38 μM, 51 μM, and 63 μM (10 μM, 30 μM, 50 μM, 60 μM, 80 μM, and 100 μM FeCl2 by weight, respectively). After the shock, the cultures were added to 10 ml RNAprotect for RNA stabilization.
RNA isolation and qRT-PCR analysis.
Total RNA was extracted from the cell pellet using the RNeasy minikit (Qiagen), including the optional DNase treatment step, according to the manufacturer's instructions. cDNA was generated using the extracted RNA as a template for an iScript (Bio-Rad) random-primed reverse transcriptase reaction by following the manufacturer's protocol. The cDNA was used as a template for quantitative PCR (Real Time 7500 PCR machine; Applied Biosystems) using SYBR green with the ROX detection system (Bio-Rad). Samples were assayed in biological triplicate. The threshold cycle (CT) values of recA and clpX were used as endogenous controls (12). Fold changes were calculated using the ΔΔCT method. Briefly, all samples were normalized to each other by subtracting the CT value for the control gene recA in the shock condition from the value in the control condition (ΔCT recA). This value was then converted from the log2 using the following formula, which accounts for machine error: a = 2ΔCT recA. The change in the genes of interest was calculated by subtracting the CT value of the gene in the shock condition from the CT value for the gene in the control condition (ΔCT gene). This value was then converted from the log2 using the following formula: b = 2ΔCT gene. The final relative fold change is calculated by the following formula: fold change = b/a. To ensure recA was constant under all conditions tested, the relative fold change for the internal control clpX, whose expression was also expected to remain constant across all our treatments, was calculated as described above. Only those samples with a clpX fold change between 0.5 and 2 were used. Primers (Integrated DNA Technologies) were designed using Primer3, with settings modified for qRT-PCR primers.
Growth, Fe(III) reduction, and bqsR expression experiment.
PA14 was grown aerobically on LB agar plates at 37°C. Two single colonies were picked, and each was transferred to a fresh tube containing 3 ml of sterilized MOPS minimal medium (50 mM MOPS, 2.2 mM KH2PO4, 43 mM NaCl, 93 mM NH4Cl, 40 mM succinate, 1 mM MgSO4, and 3.6 μM FeSO4 · 7H2O; pH 7.1) and grown overnight at 37°C. One milliliter of turbid inoculum was then added to 50 ml of sterilized MOPS medium with 500 μM FeCl3 · 6H2O and incubated at 37°C in triplicate, and a low-iron control culture was inoculated in MOPS minimal medium without additional Fe(III). At given time points, aliquots were removed from the culture for OD measurements, pyocyanin (a phenazine) concentration measurements, tests to distinguish the iron oxidation state, and qRT-PCR measurements of the bqs operon. A sample of 500 μl was taken for OD measurements, 1 ml was frozen for subsequent high-performance liquid chromatography (HPLC) analysis, 1.6 ml was taken to distinguish between iron oxidation states, and 2 to 5 ml was taken for RNA isolation. The OD500 was measured in triplicate using the Beckman DU 800 spectrophotometer. Pyocyanin HPLC peaks were identified from experimental samples and quantified with known concentrations of a pyocyanin reference on a Beckman System Gold instrument with an acetonitrile gradient, a Waters Symmetry C18 reverse-phase column (5-μm particle size; 4.6 by 250 mm), and a 168 diode array detector. The ferrozine assay was used to determine the iron concentrations and the oxidation states of the samples. All iron oxidation state distinctions were conducted in an anaerobic chamber to avoid reoxidation of ferrous ions, and measurements were made with a Biotek Synergy 4 plate reader. Relative fold changes for qRT-PCR were calculated by comparing samples from MOPS minimal medium plus Fe to samples with the same OD500 from MOPS minimal medium without Fe.
Microarray data accession number.
Our microarray data have been deposited in EBI Array Express (accession number E-MEXP-3459).
RESULTS
P. aeruginosa exhibits a transcriptional response that depends on the iron oxidation state.
To determine if exogenous Fe(II) and Fe(III) are perceived differently at the cellular level by P. aeruginosa strain PA14, we used Affymetrix P. aeruginosa GeneChips to identify transcriptional changes in response to Fe(II) or Fe(III). To ensure that the iron oxidation state remained stable, strain PA14 was cultured anaerobically as described in Materials and Methods. Upon reaching exponential phase, triplicate cultures were exposed to approximately 100 μM Fe(II), 100 μM Fe(III), or water for 30 min, and total RNA was extracted, labeled, and hybridized to P. aeruginosa Affymetrix GeneChips.
In both the Fe(II) and Fe(III) shock conditions, relative to the no-iron control, iron-scavenging genes (e.g., pyochelin biosynthesis genes) were downregulated, as expected (see the supplemental material). To determine if any genes specifically responded to Fe(II), we compared the Fe(II) and Fe(III) shock treatments. Cultures exposed to extracellular Fe(II) displayed an expression profile that was distinct from that of Fe(III)-exposed cultures (Table 2). We found 3 genes that were downregulated and 18 genes that were upregulated at least 2-fold by Fe(II) with q values of ≤0.01. Of the upregulated genes, a putative operon containing two short putative membrane proteins with PepSY domains and a putative two-component system showed the highest upregulation (Table 2).
Table 2.
Exogenous Fe(II) upregulates a specific subset of genes in PA14
| Gene | PA14 IDa | Locus | Fold change | q value | Description |
|---|---|---|---|---|---|
| bqsP | PA14_29710* | PA2659 | 98.4 | 0 | Probable PepSY type peptidase |
| bqsQ | PA14_29720* | PA2658 | 41.1 | 0 | Probable PepSY type peptidase |
| bqsR | PA14_29730* | PA2657 | 34.1 | 0 | Probable two-component response regulator |
| PA14_04180* | PA0320 | 27.5 | 0 | Conserved hypothetical protein; periplasmic localization | |
| bqsS | PA14_29740* | PA2656 | 10.4 | 3.00E−5 | Probable two-component sensor |
| PA14_07070* | PA0545 | 6.5 | 4.00E−3 | Probable ferric reductase | |
| PA14_01240* | PA0102 | 6.5 | 2.00E−5 | Probable carbonic anhydrase | |
| PA14_01250* | PA0103 | 5.8 | 0 | Probable sulfate transporter | |
| PA14_04270* | PA0327 | 5.2 | 1.00E−5 | Probable transcription factor | |
| PA14_18800 | PA3520 | 4.4 | 5.14E−3 | Heavy metal transport protein/chaperone | |
| inaA | PA14_56930 | PA4378 | 3.9 | 5.60E−4 | InaA protein |
| PA14_52340 | PA0921 | 3.4 | 2.39E−3 | Hypothetical protein | |
| PA14_56930 | PA4379 | 3.2 | 5.60E−4 | Probable methyltransferase | |
| PA14_29750 | PA2655 | 3.1 | 2.33E−3 | Hypothetical protein | |
| PA14_34170 | PA2358 | 3.0 | 5.52E−3 | Hypothetical protein | |
| dsbB | PA14_07000 | PA0538 | 2.8 | 3.58E−3 | Disulfide bond formation protein |
| iscA | PA14_14750 | PA3812 | 2.7 | 2.75E−3 | Probable iron-binding protein IscA |
| PA14_72370 | PA5482 | 2.3 | 1.59E−3 | Hypothetical protein | |
| PA14_30410 | PA2604 | 1.8 | 2.62E−3 | Hypothetical protein | |
| pdxH | PA14_50800 | PA1049 | −1.6 | 5.61E−3 | Pyridoxamine 5′-phosphate oxidase |
| narH | PA14_13800 | PA3874 | −1.8 | 8.71E−3 | Respiratory nitrate reductase beta chain |
| azu | PA14_65000 | PA4922 | −1.9 | 2.39E−3 | Azurin precursor |
| moeA1 | PA14_13280 | PA3914 | −2.1 | 1.86E−3 | Molybdenum cofactor biosynthetic protein A1 |
| PA14_21630 | PA3278 | −2.1 | 4.00E−3 | Hypothetical protein | |
| rpmF | PA14_25630 | PA2970 | −2.6 | 1.90E−3 | 50S ribosomal protein L32 |
ID, identification number.
, expression of this gene is controlled by the PA2657 (bqsR) response regulator, as shown in Fig. 3.
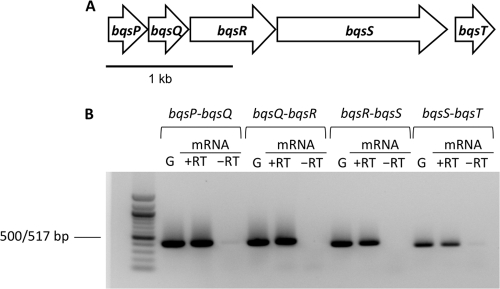
To determine whether these genes constitute an operon, we performed RT-PCR. All of the genes from PA14_29710 to PA14_ 29750 appear to be transcriptionally connected (Fig. 1), and thus we refer to this operon as the bqsPQRST operon, because homologs of BqsS and BqsR (exhibiting 100% amino acid identity) were previously identified in P. aeruginosa strain PAO1 for their role in biofilm development and quorum sensing (13). Whether bqsT is a bona fide member of this operon is less clear, as qRT-PCR results (see Fig. 3) show lower fold change in this gene than in other members of the operon, possibly reflecting transcriptional bleed-through. Moreover, whereas all of the other members of the operon possess overlapping reading frames, there are 90 bp between bqsS and bqsT. The five-gene cluster is conserved in its entirety only among P. aeruginosa strains. Orthologs of the sensor BqsS are encoded in similar predicted operons with a response regulator and one or two PepSY domain-containing proteins across other Pseudomonas and Azotobacter species, but the fifth hypothetical gene is excluded.
Fig 1.
The bqs operon. (A) Genomic organization of the bqs operon, which contains genes encoding two putative PepSY domain-containing membrane proteins (bqsP [PA14_29710] and bqsQ [PA14_29720]), a response regulator (bqsR [PA14_29730]), a histidine kinase (bqsS [PA14_29740]), and a hypothetical protein (bqsT [PA14_29750]). The length of each arrow represents the size of the gene relative to those of the other genes in the operon. (B) A 1% agarose gel confirming that bqs is an operon. The notation bqsP-bqsQ indicates, for example, that in lanes 2 to 4, PCR products were obtained using a forward primer within bqsP and a reverse primer within bqsQ. G, genomic DNA run as a positive control. mRNA −RT (reverse transcriptase) is a negative control to ensure there is no genomic DNA contamination in the mRNA. The presence of products in all of the cDNA lanes (mRNA +RT) reveals this is polycistronic mRNA that contains bqsP, bqsQ, bqsR, bqsS, and bqsT.
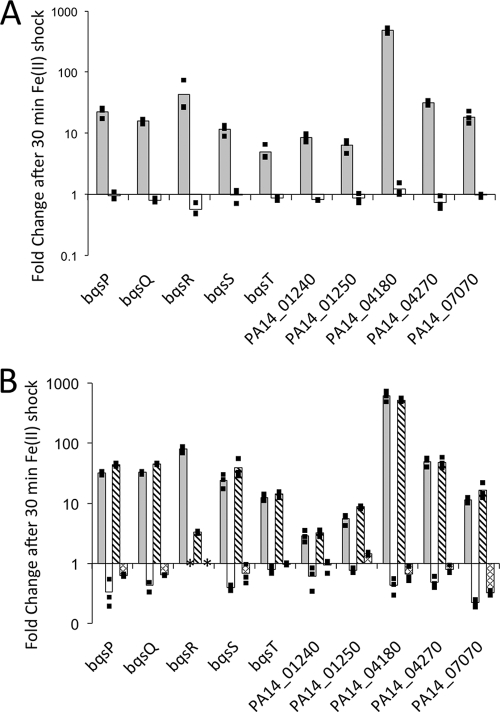
Fig 3.
BqsS and BqsR are required for extracellular Fe(II) regulation. (A) WT (gray bars) and ΔbqsS mutant (white bars) transcriptional fold change after Fe(II) shock, as measured by qRT-PCR. (B) WT (gray bars), ΔbqsR mutant (white bars), ΔbqsR mutant with pMQ64-bqsR (bqsR complement) (diagonal bars), and ΔbqsR mutant with pMQ64 (vector-only control) (hashed bars) transcriptional fold change after Fe(II) shock, as measured by qRT-PCR. Both bqsR and bqsS are required for the Fe(II)-induced upregulation of the bqs operon and upregulation of the genes in the bqs regulon. ΔbqsR::pMQ64-bqsR shows WT levels of upregulation. The bars are the averages of biological triplicates, and the individual points indicate the fold changes of each replicate. *, values obtained within background error.
Extracellular Fe(II) upregulates the bqs operon.
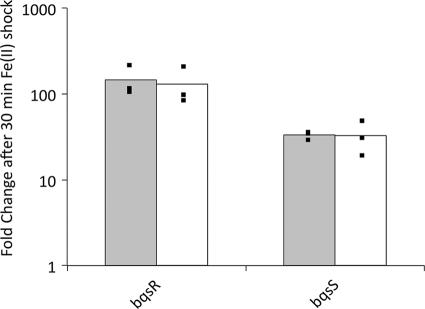
It was striking that exogenous Fe(II) induced a two-component system, because such systems are responsible for transmitting extracellular signals (e.g., the presence of ferrous iron) to the intracellular environment (55). This suggested that our two-component system, comprising a cytoplasmic response regulator (bqsR) and a sensor histidine kinase (bqsS) and identified by its conserved kinase domain and its predicted localization to the inner membrane, was sensing extracellular Fe(II). To test this, we used qRT-PCR to measure the expression of bqsR and bqsS in response to Fe(II) shock in the wild-type (WT) strain and in strain feoB::MAR2xT7 [which has a severe defect in Fe(II) uptake across the cytoplasmic membrane (31, 32, 61)]. If the cells were sensing extracellular Fe(II), we anticipated that bqsR and bqsS expression would be the same in both the wild type and the feoB mutant. Alternatively, if the cells were sensing cytoplasmic Fe(II), we expected bqsR and bqsS to be upregulated in the wild type compared to their expression in the feoB mutant. P. aeruginosa strains PA14 and PA14 feoB::MAR2xT7 were cultured under anaerobic conditions until early exponential phase, at which time 63 μM Fe(II) was added to the medium. After 30 min, the RNA was stabilized and extracted for qRT-PCR. As seen in Fig. 2, bqsR and bqsS were upregulated in both the wild type and the feoB mutant upon Fe(II) shock, which indicates their expression is activated in response to extracellular Fe(II).
Fig 2.
Extracellular Fe(II) upregulates bqsR and bqsS. Wild-type strain PA14 (gray bars) and mutant strain feoB::MAR2xT7 (white bars) were cultured anaerobically in MMM. The feoB mutant strain has a severe defect in Fe(II) uptake. A 30-min Fe(II) shock was performed, and qRT-PCR was performed to analyze expression of bqsR and bqsS. Since the wild type and the mutant show the same levels of transcriptional response, Fe(II) must be extracellular. The bars represent the averages of biological triplicates, and the individual points indicate the fold changes of each replicate.
Fe(II) response is due to the bqs operon.
To gain insight into the mechanism of the extracellular Fe(II) shock transcriptional response, we examined expression of the nine genes most upregulated in the microarray (each having a fold change of ≥4) in strains lacking either the regulator (ΔbqsR mutants) or the sensor (ΔbqsS mutants) (Fig. 3). Four of these genes are components of the bqs operon (bqsPQRST). Under anaerobic conditions, the ΔbqsR mutant, ΔbqsS mutant, and wild type were grown to early exponential phase and exposed to Fe(II). RNA was extracted for qRT-PCR analysis before and after 30 min of exposure. Our results reveal that BqsR/BqsS is autoregulated. When both BqsR and BqsS are present, extracellular Fe(II) induces the upregulation of the entire bqs operon (Fig. 3). However, if either bqsR or bqsS is deleted, the bqs operon no longer responds to extracellular Fe(II). Complementation with bqsR expressed on pMQ64 resulted in an Fe(II) response similar to that of the wild type (Fig. 3). The exception to this is expression of bqsR itself, which does not exhibit the same dramatic increase in expression with Fe(II) shock; this can be explained by the fact that before Fe(II) shock, bqsR is upregulated 75-fold compared to wild-type levels because its expression is being driven off a multicopy plasmid. No data are presented for bqsR in both the ΔbqsR mutant and the vector-only control because no copies of bqsR are present and any values detected are within background error. The transcriptional responses of the five most highly upregulated genes outside this operon—PA14_04180, PA14_07070, PA14_01240, PA14_01250, and PA14_04270—also depended on bqsR and bqsS. Of these genes, PA14_04180 shows the highest upregulation by qRT-PCR (Fig. 3) and was the fourth most upregulated in the microarray. In general, the strength of the transcriptional response in our qRT-PCR experiments was significantly higher than what we observed in the array study (Table 2), which is not unexpected because qRT-PCR is a more sensitive technique than microarrays.
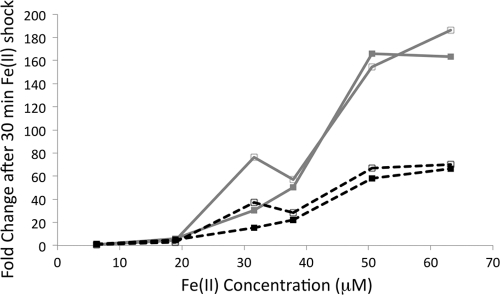
The bqs operon responds to low concentrations of extracellular Fe(II).
Intrigued by the differential fold changes we observed in our qRT-PCR experiments, we hypothesized that the BqsS/BqsR-mediated transcriptional response was tunable by the Fe(II) concentration. To test this, we performed an Fe(II) titration experiment, varying Fe(II) over an order of magnitude from ∼6 μM to ∼60 μM. A concentration of ∼20 μM was the lowest concentration of Fe(II) to elicit a response for bqsR or bqsS; the maximum response was seen for >60 μM Fe(II) (Fig. 4).
Fig 4.
Fe(II) titration curve. Duplicate trials showing induction of a transcriptional response for bqsR (gray lines) and bqsS (dashed black lines) in response to increasing Fe(II) concentrations. The minimum Fe(II) concentration to elicit a response was ∼20 μM, and the maximum fold change was seen with a concentration of ∼60 μM. Additional replicates show similar trends.
The bqs operon is specific for Fe(II).
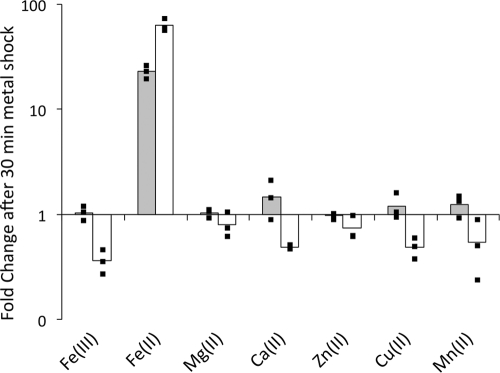
Because some two-component systems are induced by multiple external signals, we questioned whether BqsR/BqsS was specific for Fe(II). To determine this, we performed transcriptional shock experiments with five other dipositive cations that are commonly found in CF sputum (16). We also measured the transcriptional response to Fe(III) as a control. The wild type was cultured under either anaerobic or aerobic conditions and exposed to metal shock in early exponential phase. Fe(III), Ca(II), Cu(II), Mg(II), Mn(II), and Zn(II) do not significantly upregulate bqsS under either aerobic or anaerobic conditions (Fig. 5). From this, it appears that BqsR/BqsS is selective for ferrous iron. Notably, the extent of bqsS gene expression was attenuated in the aerobic Fe(II) shock experiment (Fig. 5). The lower degree of upregulation can be explained by rapid oxidation of Fe(II) to Fe(III): the initial concentration of Fe(II) was 100 μM, but after completion of the 30-min shock, the concentration was 4.9 μM.
Fig 5.
bqsS responds specifically to Fe(II). Expression of bqsS under both anaerobic (gray bars) and aerobic (white bars) conditions after shock with Fe(II) and other cations. Metal concentrations of 100 μM FeCl3, 100 μM Fe(NH4)2(SO4)2, 10 mM MgCl2, 5 mM CaCl2, 100 μM ZnCl2, 100 μM CuCl2, and 100 μM MnCl2 were used in the 30-min metal shocks for Fe(III), Fe(II), Mg(II), Ca(II), Zn(II), Cu(II), and Mn(II), respectively. The bars are the averages of biological triplicates, and the individual points indicate the fold changes of each replicate.
The bqs operon is transcribed as Fe(II) is produced during growth.
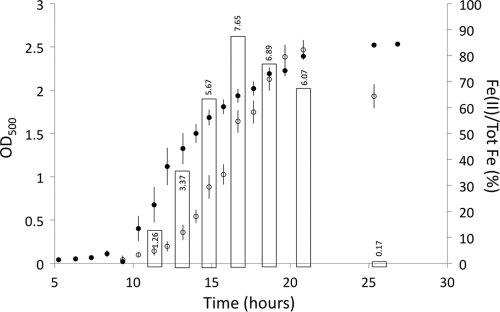
To explore the connection between bqs operon transcriptional induction and biological iron reduction in a wild-type culture, a 27-h growth experiment was conducted in duplicate using PA14 grown in MOPS medium with an initial iron allocation of almost exclusively Fe(III) (Fig. 6). OD500 measurements show a classical growth curve exhibiting exponential growth between approximately 10 and 18 h after inoculation. Pyocyanin, a type of phenazine, was detected in the culture starting in mid-exponential phase and continuing through the end of the experiment. The culture's iron oxidation state shifted from entirely Fe(III) at the beginning of the experiment to >85% Fe(II) by the end; this shift coincided with the exponential phase of growth and the presence of phenazines. qRT-PCR showed upregulation of bqsS during exponential phase, when extracellular Fe(II) was at a concentration of ∼125 μM (well above the concentration needed to induce the bqs operon), and a repression of transcription in stationary phase.
Fig 6.
bqsR is transcribed over the course of growth as Fe(II) accumulates. Correlation between growth, phenazine production, iron reduction, and bqsR transcription in PA14. Growth curves (OD500) averaged for triplicate cultures of PA14 are plotted alongside the percentages of total Fe that were Fe(II). Bars indicate the average upregulation of bqsS (relative to recA) compared to that in cells grown without Fe and normalized to OD500. For the complete qPCR data set, see Table S2 in the supplemental material. Filled circles, PA14 OD500; open circles, percentage of total Fe that was Fe(II).
DISCUSSION
In this study, we identified a two-component system in P. aeruginosa strain PA14, BqsR/BqsS, that controls a specific transcriptional response to extracellular Fe(II). To our knowledge, this is the first extracellular Fe(II) sensor described in bacteria. BqsR/BqsS autoregulates its own expression, is turned on by Fe(II) concentrations of >10 μM, and does not respond to other physiologically relevant dipositive ions. Previously, BqsR/BqsS was shown to play a role in the dispersal of P. aeruginosa strain PAO1 biofilms (13). This appears to be due to the fact that biofilms lacking BqsR or BqsS produce lower concentrations of rhamnolipids, which are known to promote biofilm dispersal (7).
Interestingly, bqsR and bqsS expression peaks during late exponential growth, in which cells are thought to experience conditions similar to those in biofilms (52, 58). The physiological context of bqsR and bqsS expression thus provides a satisfying link between our results and those of Dong et al.: as cell densities and phenazine concentrations rise, Fe(II) is generated, inducing bqsR and bqsS expression, which promotes biofilm dispersal. While several previous studies have indicated that moderate concentrations of iron are necessary to stimulate biofilm formation regardless of whether iron is in the ferric or ferrous form (5, 48), it has also been established that this phenomenon is concentration dependent: at sufficiently high concentrations, iron can promote biofilm dispersal (37). It thus seems likely that BqsR/BqsS acts as a “gatekeeper” to sense when Fe(II) concentrations have crossed the threshold where conditions are no longer favorable for biofilm maintenance.
That the BqsR/BqsS system responds specifically to extracellular Fe(II) but not Fe(III) was initially surprising, given that the PmrA/PmrB two-component system is specific for extracellular Fe(III) but not Fe(II) (67). In a detailed study by Wösten et al. (67), it was shown that PmrB directly interacts with Fe(III) through glutamate residues in two HExxE motifs and a serine outside these motifs in the periplasmic region. BqsS contains a similar motif that could be responsible for directly sensing Fe(II), a theory which we plan to test biochemically. Of the 23 BqsS orthologs we identified across currently sequenced pseudomonads, only 14 are predicted to contain the putative Fe(II) binding motif; 7 of these are strains of P. aeruginosa. Once the BqsR Fe(II) binding motif is biochemically confirmed, it will be interesting to determine how the BqsR/BqsS regulon differs among these species.
Not only does BqsS differentiate Fe(II) from Fe(III), it also distinguishes Fe(II) from other dipositive ions that Pseudomonas might encounter: when challenged with Ca(II), Cu(II), Mg(II), Mn(II), and Zn(II), the wild type did not upregulate bqsS. This differs from the Fe(III)-sensing PmrA/PmrB system, which can also be activated by low extracellular Mg(II) (51) and mild acid (41). In addition, BqsR/BqsS are highly sensitive to Fe(II). The minimum Fe(II) level found to elicit a transcriptional response was ∼20 μM; the response rapidly increased over a small concentration range, with the maximum response achieved by a concentration of ∼60 μM. These levels of Fe(II) are well within physiologically relevant Fe(II) concentrations, as the Fe(II) concentration has been measured up to 110 μM in CF sputum samples (R.C. Hunter and D. K. Newman, unpublished data). Other studies have obtained sputum samples and measured total Fe concentrations which range from approximately 20 μM to 140 μM (45).
Both BqsP/BqsQ contain PepSY domains (Pfam accession number PF03413) (68) and are predicted to be single-pass transmembrane proteins by TMpred (18) and DAS (11). BqsP is predicted to share structural homology with the protein YycI from Bacillus subtilis (75% estimated precision using the Phyre homology modeling program) (23). YycI, along with another auxiliary protein, is known to regulate the kinase activity of a two-component system, YycFG, through its transmembrane domain (56a). Deleting YycI caused a 10-fold increase in expression of a lacZ kinase promoter fusion (54). Given the similarity between BqsP and YycI, it seems likely that BqsP/BqsQ play a similar role in modulating the kinase activity of BqsS. bqsT encodes a hypothetical protein that lacks conserved domains and has only very weak similarity to any known proteins. Orthologs to bqsT are only present in other P. aeruginosa strains, implying that, if it is a true member of the bqs operon, it has been integrated into the operon recently and may not serve a critical function.
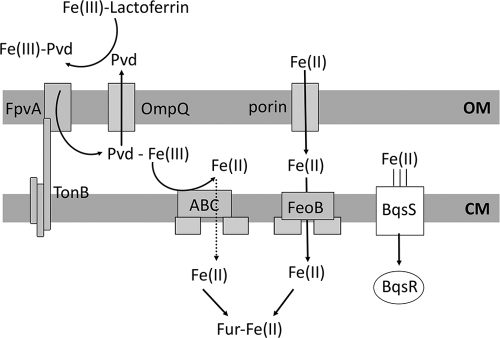
Strikingly, only a small number of genes in our microarray were upregulated in the Fe(II) shock condition compared to their levels in the Fe(III) shock condition, and we did not observe upregulation of pqsA, phnA (PQS biosynthesis), and rhlA (rhamnolipid production), as reported by Dong et al. (13). This is likely because we performed our shock experiment on cells harvested from early exponential phase. If BqsR/BqsS are involved in the regulation of genes that are also quorum regulated (i.e., pqsA, phnA, and rhlA), we would not have detected them. Another possibility is that the BqsR/BqsS regulon is different in PA14 than in PAO1. Regardless, the “core” Fe(II)-sensitive BqsR/BqsS regulon is quite limited. This may be due to the fact that only Fe(II) enters and is sensed in the cytoplasm, no matter whether Fe(III) or Fe(II) is extracellular (Fig. 7). Thus, the primary network of iron-responsive genes would not need to distinguish between the two forms. Perhaps instead, extracellular Fe(II) is sensed to assess environmental conditions that foster Fe(II) formation. Fe(II) exists in reducing and acidic environments. Consistent with this, genes that respond to Fe(II) shock include PA14_01240, encoding carbonic anhydrase, and iscA, an acid response gene involved in iron-sulfur cluster biogenesis (62). Their protein products can help the cell adjust to an external decrease in pH. If the extracellular environment becomes more reducing, the disulfide bonds of periplasmic proteins are reduced to free sulfhydryl groups, greatly decreasing their stability. The upregulated dsbB encodes a disulfide bond-forming protein with predicted periplasmic localization which could ameliorate these effects.
Fig 7.
Iron acquisition model. Fe(III). Outside of the cell, Fe(III) binds to the siderophore pyoverdine, and this complex enters the periplasm via the TonB-dependent outer membrane transporter FpvA (59). The reduction of Fe(III) to Fe(II) frees the iron from pyoverdine. A pyoverdine efflux pump, OmpQ, recycles free pyoverdine by transporting it out of the cell (69). The Fe(II) then enters the cytoplasm through an unidentified ABC transporter. Upon entering the cytoplasm, Fe(II) binds the ferric uptake regulator (Fur), which subsequently represses multiple iron acquisition and iron storage genes (17). Fe(II). Fe(II) enters the periplasm through a nonspecific porin in the outer membrane. While in the periplasm, Fe(II) activates the histidine kinase BqsS, which subsequently activates the response regulator BqsR. BqsR then goes on to induce its regulon. Fe(II) enters the cytoplasm through the ferrous iron transporter FeoB (31). Once in the cytoplasm, Fe(II) binds Fur, which results in the repression of iron acquisition genes. Solid arrows indicate known pathways, dashed arrows indicate putative pathways, and three parallel lines indicates binding. Pvd, pyoverdine; OM, outer membrane; CM, cytoplasmic membrane.
One clear result is that the PA14_04180 gene is highly upregulated by BqsR/BqsS. Bioinformatic predictions suggest it is periplasmically localized and has a bacterial oligonucleotide/oligosaccharide-binding domain (OB-fold). The distinguishing feature of bacterial OB-fold proteins compared to other OB-fold proteins is that the bacterial ligand-binding domain binds cationic ligands rather than nucleotides (15). Proteins with such domains can serve a variety of functions and have been implicated in bacterial pathogenesis (36). In fact, the Fe(III)-binding PmrA/PmrB regulates a similar periplasmic OB-fold protein (OmdA) that confers resistance to polymyxin B, a cationic antimicrobial peptide (42). Whether PA14_04180 confers a similar resistance in Pseudomonas species is currently unknown.
The identification of an extracellular Fe(II)-sensing system that is specific for Fe(II), can distinguish between Fe(II) and Fe(III), and is insensitive to other dipositive ions opens up several avenues for future research. It reminds us that the concentration of Fe(II) can be sufficiently high in some habitats for bacteria to have evolved specific mechanisms to sense it. What their transcriptional response is and how it helps them adapt to these conditions remain to be determined. Once the molecular basis of Fe(II) specificity is understood, it may be possible to identify other proteins that have a similar specificity for Fe(II). Finally, monitoring the transcription of genes regulated by BqsR/BqsS may provide an opportunity to indirectly gauge whether P. aeruginosa is experiencing Fe(III) or Fe(II) in its environment. For example, knowing the iron oxidation state in CF sputum or biofilm microenvironments would inform the proper administration of antimicrobial therapies targeting iron uptake.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Howard Hughes Medical Institute (HHMI) for supporting this work. D.K.N. is an HHMI investigator. N.K. was supported by an NIH training grant (GM07616). J.C.W. and J.J.M. were supported by NSF graduate fellowships.
Footnotes
Published ahead of print 22 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Aisen P, Leibman A, Zweier J. 1978. Stoichiometric and site characteristics of the binding of iron to human transferrin. J. Biol. Chem. 253:1930–1937 [PubMed] [Google Scholar]
- 2. Baker BJ, Banfield JF. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139–152 [DOI] [PubMed] [Google Scholar]
- 3. Banin E, et al. 2008. The potential of desferrioxamine-gallium as an anti-Pseudomonas therapeutic agent. Proc. Natl. Acad. Sci. U. S. A. 105:16761–16766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Banin E, Vasil ML, Greenberg EP. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 102:11076–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beinert H, Holm RH, Munck E. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653–659 [DOI] [PubMed] [Google Scholar]
- 6. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300 [Google Scholar]
- 7. Boles BR, Thoendel M, Singh PK. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 57:1210–1223 [DOI] [PubMed] [Google Scholar]
- 8. Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 9. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 10. Cox CD. 1986. Role of pyocyanin in the acquisition of iron from transferrin. Infect. Immun. 52:263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673–676 [DOI] [PubMed] [Google Scholar]
- 12. Dietrich LEP, Price-Whelan A, Petersen A, Whiteley M, Newman DK. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61:1308–1321 [DOI] [PubMed] [Google Scholar]
- 13. Dong Y-H, Zhang X-F, An S-W, Xu J-L, Zhang L-H. 2008. A novel two-component system BqsS-BqsR modulates quorum sensing-dependent biofilm decay in Pseudomonas aeruginosa. Commun. Integr. Biol. 1:88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gentleman R, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ginalski K. 2004. BOF: a novel family of bacterial OB-fold proteins. FEBS Lett. 567:297–301 [DOI] [PubMed] [Google Scholar]
- 16. Gray RD, et al. 2010. Sputum trace metals are biomarkers of inflammatory and suppurative lung disease. Chest 137:635–641 [DOI] [PubMed] [Google Scholar]
- 17. Hassett DJ, et al. 1996. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J. Bacteriol. 178:3996–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hofmann K, Stoffel W. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe Seyler. 374:166 [Google Scholar]
- 19. Huebers HA, Josephson B, Huebers E, Csiba E, Finch CA. 1984. Occupancy of the iron binding sites of human transferrin. Proc. Natl. Acad. Sci. U. S. A. 81:4326–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hugo WB, Turner M. 1957. A soil bacterium producing an unusual blue pigment. J. Bacteriol. 73:154–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hunter RC, Beveridge TJ. 2005. Application of a pH-sensitive fluoroprobe (C-SNARF-4) for pH microenvironment analysis in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:2501–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Irizarry RA, et al. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264 [DOI] [PubMed] [Google Scholar]
- 23. Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 24. Kimata N, Nishino T, Suzuki S, Kogure K. 2004. Pseudomonas aeruginosa isolated from marine environments in Tokyo Bay. Microb. Ecol. 47:41–47 [DOI] [PubMed] [Google Scholar]
- 25. Koley D, Ramsey MM, Bard AJ, Whiteley M. 2011. Discovery of a biofilm electrocline using real-time 3D metabolite analysis. Proc. Natl. Acad. Sci. U. S. A. 108:19996–20001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lam J, Chan R, Lam K, Costerton JW. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28:546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levy J, Smith AL, Kenny MA, Ramsey B, Schoenknecht FD. 1983. Bioactivity of gentamicin in purulent sputum from patients with cystic fibrosis or bronchiectasis: comparison with activity in serum. J. Infect. Dis. 148:1069–1076 [DOI] [PubMed] [Google Scholar]
- 28. Li J, Bishop PL. 2003. Monitoring the influence of toxic compounds on microbial denitrifying biofilm processes. Water Sci. Technol. 47(5):211–216 [PubMed] [Google Scholar]
- 29. Liang L, Korte N, Gu B, Puls R, Reeter C. 2000. Geochemical and microbial reactions affecting the long-term performance of in situ ‘iron barriers’. Adv. Environ. Res. 4:273–286 [Google Scholar]
- 30. Litwin CM, Calderwood SB. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. U. S. A. 99:16243–16248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marshall B, Stintzi A, Gilmour C, Meyer J-M, Poole K. 2009. Citrate-mediated iron uptake in Pseudomonas aeruginosa: involvement of the citrate-inducible FecA receptor and the FeoB ferrous iron transporter. Microbiology 155:305–315 [DOI] [PubMed] [Google Scholar]
- 33. Mashburn LM, Jett AM, Akins DR, Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187:554–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moreau-Marquis S, O'Toole GA, Stanton BA. 2009. Tobramycin and FDA-approved iron chelators eliminate P. aeruginosa biofilms on cystic fibrosis cells. Am. J. Respir. Cell Mol. Biol. 41:305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morel F, Hering JG. 1993. Principles and applications of aquatic chemistry. Wiley, New York, NY [Google Scholar]
- 36. Murzin AG. 1993. OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for nonhomologous sequences. EMBO J. 12:861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Musk DJ, Banko DA, Hergenrother PJ. 2005. Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chem. Biol. 12:789–796 [DOI] [PubMed] [Google Scholar]
- 38. Naikare H, Palyada K, Panciera R, Marlow D, Stintzi A. 2006. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74:5433–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ochsner UA, Johnson Z, Vasil ML. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185–198 [DOI] [PubMed] [Google Scholar]
- 40. Patriquin GM, et al. 2008. Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 190:662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perez JC, Groisman EA. 2007. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol. Microbiol. 63:283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pilonieta MC, Erickson KD, Ernst RK, Detweiler CS. 2009. A protein important for antimicrobial peptide resistance, YdeI/OmdA, is in the periplasm and interacts with OmpD/NmpC. J. Bacteriol. 191:7243–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ratledge C, Dover LG. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881–941 [DOI] [PubMed] [Google Scholar]
- 44. Reid DW, Anderson GJ, Lamont IL. 2009. Role of lung iron in determining the bacterial and host struggle in cystic fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 297:L795–L802 [DOI] [PubMed] [Google Scholar]
- 45. Reid DW, Withers NJ, Francis L, Wilson JW, Kotsimbos TC. 2002. Iron deficiency in cystic fibrosis. Chest 121:48–54 [DOI] [PubMed] [Google Scholar]
- 46. Satoh H, et al. 2004. Macroscale and microscale analyses of nitrification and denitrification in biofilms attached on membrane aerated biofilm reactors. Water Res. 38:1633–1641 [DOI] [PubMed] [Google Scholar]
- 47. Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singh PK, Parsek MR, Greenberg EP, Welsh MJ. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552–555 [DOI] [PubMed] [Google Scholar]
- 49. Singh PK, et al. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764 [DOI] [PubMed] [Google Scholar]
- 50. Smyth G. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3 [DOI] [PubMed] [Google Scholar]
- 51. Soncini FC, Vescovi EG, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092–5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, Dancis A. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552–1557 [DOI] [PubMed] [Google Scholar]
- 54. Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6:199–210 [DOI] [PubMed] [Google Scholar]
- 55. Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 56. Stookey LL. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal. Chem. 42:779–781 [Google Scholar]
- 56a. Szurmant H, Mohan MA, Imus PM, Hoch JA. 2007. YycH and YycI interact to regulate the essential YcyFG two-component system in Bacillus subtilis. J. Bacteriol. 189:3280–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Velayudhan J, et al. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274–286 [DOI] [PubMed] [Google Scholar]
- 58. Waite RD, Papakonstantinopoulou A, Littler E, Curtis MA. 2005. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J. Bacteriol. 187:6571–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wandersman C, Delepelaire P. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611–647 [DOI] [PubMed] [Google Scholar]
- 60. Wang Y, Newman DK. 2008. Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ. Sci. Technol. 42:2380–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang Y, et al. 2011. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J. Bacteriol. 193:3606–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. White S, Tuttle FE, Blankenhorn D, Dosch DC, Slonczewski JL. 1992. pH dependence and gene structure of InaA in Escherichia coli. J. Bacteriol. 174:1537–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Whiteley M, et al. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864 [DOI] [PubMed] [Google Scholar]
- 64. Whiteley M, Lee KM, Greenberg EP. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:13904–13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wilson R, et al. 1988. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56:2515–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Worlitzsch D, et al. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wösten MMSM, Kox LFF, Chamnongpol S, Soncini FC, Groisman EA. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113–125 [DOI] [PubMed] [Google Scholar]
- 68. Yeats C, Rawlings ND, Bateman A. 2004. The PepSY domain: a regulator of peptidase activity in the microbial environment? Trends Biochem. Sci. 29:169–172 [DOI] [PubMed] [Google Scholar]
- 69. Yeterian E, Martin LW, Lamont IL, Schalk IJ. 2010. An efflux pump is required for siderophore recycling by Pseudomonas aeruginosa. Environ. Microbiol. Rep. 2:412–418 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.