Abstract
The bacterium Ralstonia eutropha forms cytoplasmic granules of polyhydroxybutyrate that are a source of biodegradable thermoplastic. While much is known about the biochemistry of polyhydroxybutyrate production, the cell biology of granule formation and growth remains unclear. Previous studies have suggested that granules form either in the inner membrane, on a central scaffold, or in the cytoplasm. Here we used electron cryotomography to monitor granule genesis and development in 3 dimensions (3-D) in a near-native, “frozen-hydrated” state in intact Ralstonia eutropha cells. Neither nascent granules within the cell membrane nor scaffolds were seen. Instead, granules of all sizes resided toward the center of the cytoplasm along the length of the cell and exhibited a discontinuous surface layer more consistent with a partial protein coating than either a lipid mono- or bilayer. Putatively fusing granules were also seen, suggesting that small granules are continually generated and then grow and merge. Together, these observations support a model of biogenesis wherein granules form in the cytoplasm coated not by phospholipid but by protein. Previous thin-section electron microscopy (EM), fluorescence microscopy, and atomic force microscopy (AFM) results to the contrary may reflect both differences in nucleoid condensation and specimen preparation-induced artifacts.
INTRODUCTION
Polyhydroxyalkanoates (PHAs) are linear polyoxoesters synthesized by many bacteria from water-soluble 3-hydroxyalkanoyl coenzyme A esters [R-CHOHCH2CO-S-CoA, where R = (CH2)nCH3 and n = 0 to 14] when in carbon-rich but otherwise nutrient-limited environments (44–46). As they grow, PHAs aggregate in the cytoplasm in the form of globular, insoluble granules and under optimized growth conditions can account for as much as 85% of the cell dry weight. Different PHAs have properties that vary from thermoplastics (R = CH3, CH2CH3) similar to oil-based polyethylenes and polypropylenes to elastomers [R = (CH2)nCH3, where n = 6 to 14]. In contrast to nonbiodegradable oil-based polymers, PHAs are “environmentally friendly” (and thus of high interest), since they can be hydrolyzed by extracellular depolymerases (24, 43).
The genetically manipulable bacterium Ralstonia eutropha (also Cupriavidus necator, formerly Wautersia eutropha) has served as a paradigm in studies of PHA biochemistry. R. eutropha uses a synthase, PhaC, to generate a specific PHA, polyhydroxybutyrate (PHB), from 3-hydroxybutyryl-CoA (HBCoA) via a covalent intermediate (54). HBCoA is in turn synthesized from acetyl-CoA using a thiolase (PhaB) and a reductase (PhaA) (32). The three biosynthetic genes phaABC are together on an operon whose expression is relatively constant during PHB production (27, 32). Chain elongation by PhaC is more rapid than chain initiation (16, 49), and chain termination occurs via a poorly understood mechanism once a remarkably uniform molecular mass (polydispersity of 1.2) of approximately 106 Da is reached (48, 49). PHB sequesters the repressing transcription factor PhaR to derepress production of PhaP1, a protein from the enigmatic phasin family (38, 53, 57), of which there are multiple paralogs within the R. eutropha genome (4, 22, 35, 36).
Compositional analysis of mature granules suggests that there are 1 or 2 PhaP1s for every PHB molecule but many fewer PhaCs (1 per 100 PHB molecules) (48). One possible role for PhaP1 and the other phasins may be to serve as a chaperone, preventing PHB from crystallizing (as it does in isolation), which makes it inaccessible to intracellular PHB depolymerases (20). PhaP1 has been shown to be present on the granule surface, and previous studies have shown that PhaP1 concentrations increase 10-fold during PHB production (48, 55, 56), sufficient to cover between approximately one-quarter to one-half of the total granule surface (31, 48). Deletion of the phaP1 gene results in reduction of the number of granules from 6 to 15 to a single giant granule per cell, with a concomitant 40% reduction in the amount of PHB (53, 56).
Early studies of PHB granules isolated by glycerol ultracentrifugation from Bacillus megatarium reported a composition of 98% PHB and 2% protein but also 0.5% lipid (17). Lipids were also found in PHB granules isolated from R. eutropha (20). A distinct “surface layer” was further noted in thin-section electron microscopy (EM) images of various stained cells whose thickness was consistent with a lipid monolayer (3 to 4 nm) (30). Western blots and immunogold labeling have shown that granule surfaces are proteinaceous, with PhaP1 (∼25%) predominating and smaller amounts of PhaC, PhaR, and depolymerases (PhaZ) (15, 38, 40, 53). Most recently, the possibility that granules are covered by both a smooth, outer “envelope” and an inner, proteinaceous “network” has been proposed based on atomic force microscopy (AFM) images of lysed cells (12).
The composition of granules has led to three models for granule genesis. In analogy to eukaryotic lipid body biogenesis (10), a “membrane-budding” model of granule formation in which hydrophobic PHB chains are initially synthesized within the cytoplasmic membrane, and granules subsequently bud from the membrane through an as-yet-unknown process, has been proposed (46). In support of this model, fluorescently tagged PhaC and PhaP1 and Nile Red-stained PHB have been reported to localize near the membrane and sometimes at the cell poles early in granule genesis (18, 21–23, 33, 34). In Escherichia coli, heterologously expressed R. eutropha PhaC localized at the pole with or without covalently bound PHB oligomers, but PhaP1 became pole localized only when PHB was produced (33, 34).
These results conflict with fluorescence (22) and immuno-EM (15) studies in R. eutropha, however, indicating essentially random cytoplasmic localization for free PhaC in the absence of a carbon source. It is also unclear whether the lipids found in granule preparations may have simply been a contaminant acquired during cell lysis, given the hydrophobic nature of PHB. Granule “stickiness” is illustrated by nonspecific binding of many proteins to granules during isolation (22). If so, the surface layer may be proteinaceous instead of lipidic.
A time course study of granule formation using thin-sectioned, fixed, and stained cells gave rise to a different, “scaffold” model for PHB production because granules at all stages were observed in and around a dark-staining structure in the center of the cell designated a “mediation element” (50). In this model, synthases and phasins are localized by a central scaffold rather than a membrane as they produce PHB and coat the surface of growing granules, respectively. PHB crystallization would be prevented by either PhaP1 or nonspecific protein factors.
A third, “micelle” model predicts that PHB chains aggregate within the cytoplasm with covalently attached synthases on the surface (15, 17). This model is consistent with the random localization of synthase seen prior to granule initiation (15, 22) and does not require the existence of any as-yet-unidentified proteins for lipid manipulations or scaffolding. Water-soluble precursors are added to the growing hydrophobic PHB chain by surface-exposed synthases, and PHB is sequestered from the aqueous cytoplasm by both these synthases and, predominantly, PhaP1. Studies by Gerngross and Martin (14) and AFM studies by Hiraishi et al. (19) monitoring in vitro polymerization of PHB by PhaC support the formation of micelle-like structures. The latter study showed that during initial stages of PHB production, long fibrillar chains of PHB are found associated with synthases that later aggregate to form large granule-like structures (100 to 250 nm) with PhaC on the surface (19).
Some of the discrepancies in the literature may be due to cell perturbations such as lysis (during granule isolation), drying (before AFM), fixation (before traditional EM and sometimes fluorescent light microscopy), staining (before traditional EM), and overexpression of exogenous proteins and/or fusion with fluorescent proteins (for fluorescent light microscopy). In order to minimize cell perturbations, here we used whole-cell electron cryotomography (ECT), a technique that provides “macromolecular” resolution, three-dimensional (3-D) images of intact cells in a near-native, “frozen-hydrated” state. In ECT, living samples are suspended in a thin film of growth medium and vitrified by fast freezing, thus avoiding the need for fixatives (28). A series of images are then acquired as the sample is incrementally tilted in a cryogenic transmission electron microscope (TEM). Phase contrast arises from the scattering properties of the sample and chosen microscope settings, obviating the need for stains. The cryotomograms show that in R. eutropha, nascent granules are not seen within the membrane, granule localization is biased toward the central region of the cell, there is no evidence for a scaffold, and granule surfaces are only partially covered by a likely proteinaceous layer. Our results therefore strongly support a cytoplasmic micelle model of granule genesis.
MATERIALS AND METHODS
Bacterial growth and imaging.
A single colony of wild-type R. eutropha H16 was cultivated in 5 ml of tryptic soy broth (TSB) for 24 h to saturation. Culture media were supplemented with 10 μg/ml gentamicin; all growth was at 30°C and shaking at 200 rpm. Four milliliters of saturated culture was then transferred into 200 ml of TSB in a 1-liter baffled flask and grown for another 24 h. Cells harvested by centrifugation were washed and transferred into 500 ml of PHBHigh medium (minimal medium supplemented with 2% fructose and 0.01% ammonium chloride) (50) in a 2.8-liter baffled flask to obtain a culture with an initial optical density at 600 nm (OD600) of 0.5. Ten milliliters of cell suspension was removed after 2.5 h, 5 h, 9 h, and 24 h of cultivation in PHBHigh, harvested by centrifugation, and then resuspended to a final 20× concentration, 20 μl of which was mixed with 5× bovine serum albumin (BSA)-colloidal gold solution (Ted Pella, Redlands, CA). Four microliters of cell suspension was deposited on C-flat 2/2 grids that had been glow discharged for 4 min, blotted, and plunge frozen into a liquid ethane-propane mixture using a Vitrobot and stored in liquid nitrogen until imaging. Tilt series of whole cells were collected using Leginon (47) on a 300-kV FEI Polara G2 electron cryo-TEM using a total dose of approximately 100 e−/Å2, −12 μm defocus, and a tilt range of −61° to + 61° with 1° tilt increments at ×22,500 magnification and binned 4× using a post-GIF 4K × 4K Gatan UltraCam to generate final 1K × 1K images with 1.9-nm × 1.9-nm pixels; tilt series of mildly sonicated lysed cells were collected with an electron dosage of approximately 40e−/Å2, −8 μm defocus, 1.5° tilt increment, and ×34,000 magnification to generate images with a pixel size of 1.26 nm × 1.26 nm, with all other settings as for whole cells. Defocus values have a margin of error of approximately 1 μm. Tomograms were reconstructed with a combination of RAPTOR (2) and IMOD (26). Whole-cell tomograms were constructed using low-pass-filtered tilt series to an ∼13-nm resolution. Tomograms of isolated granules were generated using the SIRT algorithm implemented in tomo3d (1).
Data analysis.
3dmod (26) was used to visualize tomographic data and generate models. Granules were modeled as spheres and cells as cylinders capped with hemispheres, flattened as appropriate to correspond with cells. Models were analyzed and visualized using custom-written programs and POV-Ray. “Centrality” was defined as the fraction of the distance for the granule between the closest position the granule could take to the inner membrane and the long axis of the cell; the long axis of the cell passed along the length of the cell cylinder but did not continue into the hemispherical poles. “Polarity” was defined as the fraction of the distance from the center of the cell and the tip of the pole. To acquire rank-normalized values of each metric, 500 randomized data sets were generated. For each cell, all granules were removed and then replaced at random, disallowing overlap with another granule or the cell periphery. This resulted in a distribution of randomized centrality and polarity. Rank-normalized scores were subsequently calculated by ranking each metric's value within the random distribution. These models were subsequently used to project spherical granule surfaces onto a plane using the Sanson-Flamsteed method.
RESULTS
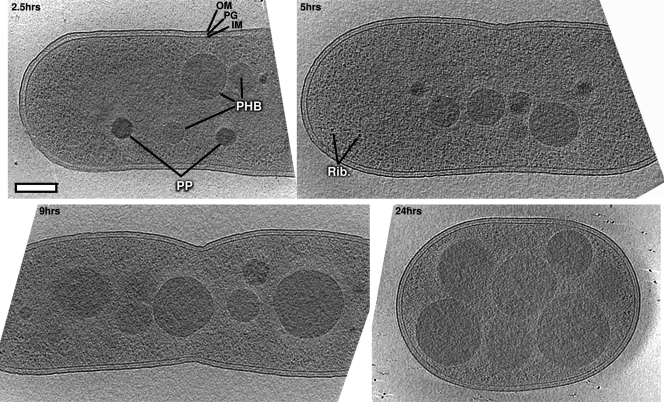
PHB production was induced by nitrogen limitation with fructose as a carbon source (50). Cells were plunge frozen 2.5, 5, 9, and 24 h after induction, and 14, 10, 9, and 12 tomograms were collected at each time point, respectively. Representative cells are shown in Fig. 1 (see also Fig. S1 in the supplemental material). Cells were rod shaped but became wider and shorter as granule formation progressed.
Fig 1.
Slices through representative tomograms of PHB granule formation in Ralstonia eutropha H16. Computational slices through the centers of representative cells from each of the four time points. Granules are clearly visible by their higher density, uniform texture, and circular cross sections. Note that not all granules from a cell are visible in these slices, as some may be above or below the cross section shown. Rib, ribosome; OM, outer membrane; IM, inner membrane; PHB, polyhydroxybutyrate granule (lighter in texture with defined surface); PP, polyphosphate granule (darker, with no surface layer). Centers of cells are lower contrast due to greater electron scattering there during the imaging process. Bar, 200 nm.
Granule identity.
A total of 463 granules were identified within the cytoplasms of 45 cells. Two varieties of granules were distinguishable by their surface, size, texture, and density (density being directly proportional to atomic mass density). The majority (325) had densities close to that of the cytoplasm, increased in size over the four time points, and possessed well-defined boundaries. The remainder (138) were darker, exhibited ∼100-nm diameters throughout the time course, and had less-pronounced boundaries. The lighter granules were clearly PHB, since their sizes increased dramatically under these conditions as established previously (50). Multiple lines of evidence indicate that the darker granules are polyphosphate: (i) as similarly light and dark granules were identified spectroscopically by Comolli et al. (9) in Caulobacter crescentus as carbon and phosphorous rich, respectively; (ii) as also found by Comolli et al., the darker granules were more resistant to electron beam damage than the PHB granules; (iii) the genes needed to produce and utilize polyphosphate granules are present in R. eutropha; and (iv) the darker granule appearance indicates higher atomic mass density, consistent with an abundance of phosphorus.
Quantification of cells and granules.
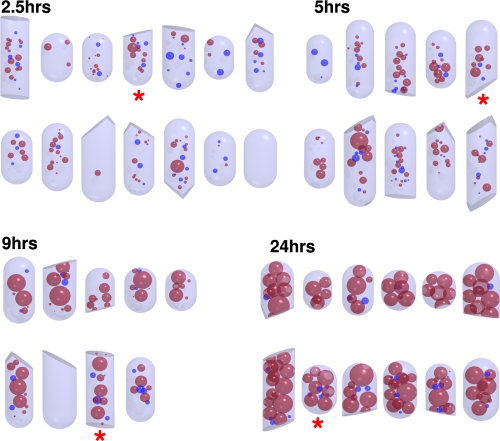
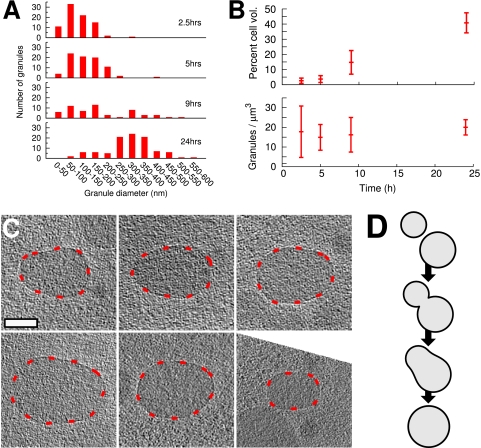
In order to quantify our findings, we developed simplified 3-D models of cells and granules (Fig. 2). While cells at time points at 2.5, 5, and 9 h were just over 0.7 μm in diameter (0.74 ± 0.04, 0.72 ± 0.04, and 0.74 ± 0.05 μm, respectively), cells from the 24-h time point were slightly wider at approximately 0.8 μm (0.80 ± 0.05 μm). The number of granules per cell varied, but the average per cell at each time point remained roughly constant at 10 ± 5 (corresponding to ∼20 ± 10 granules/μm3) for all four time points (additionally illustrated below [see Fig. 5B, bottom]). Small PHB granules (<100 nm) were observed at all time points, and the smallest we detected were ∼25 nm in diameter. While the cells appeared slightly flattened in the plane of the vitreous ice, nearly all the internal granules remained spherical. PHB granules observed in the debris of sonicated cells, however, were often distorted.
Fig 2.
Quantitative models. 3-D models of all 45 cells. Asterisks mark models of the cells depicted in Fig. 1. Red granules represent PHB; blue indicates polyphosphate.
Fig 5.
Granules are constantly being initiated and progressively fuse. (A) Distribution of granule sizes in cells from the four time points showing constant presence of small (<100-nm) granules. Note that cells at later time points are thicker, meaning that we may not have been able to detect the smallest granules in these cells. (B) Overall granule volume increases with time (top), and the number of granules per unit volume remains roughly constant over all time points (bottom). (C) Examples of nonspherical granules as candidates for fusion processes. Granules have been rotated so that the long axis of the nonspherical granules is horizontal in the plane of the figure; areas of indistinct detail are thus a result of the “missing wedge” of data in reciprocal space inherent to ECT. (D) Model of the time course of granule fusion. Granules depicted in C are caught at the third stage of fusion.
Granule surfaces.
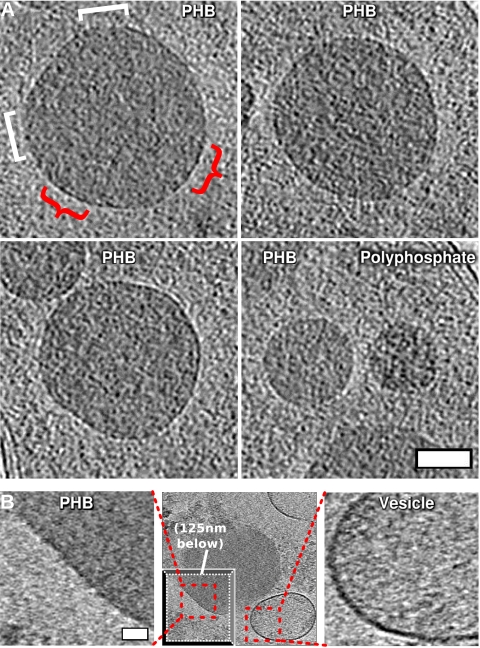
Although the PHB granules had a discernible surface layer at their interface with the cytoplasm (highlighted by red arcs for one granule in Fig. 3A), in contrast to the appearance of membranes, it never appeared continuous. Instead, segments of the granule appeared to be coated with a surface layer while other parts were exposed. Patches were observed randomly around the granule, ruling out an imaging artifact that might result in features always occurring at a specific orientation relative to the tomogram tilt axis. Where present, the surface layer appeared thinner and of lower contrast than either the inner or outer membrane of the cell (Fig. 3A). No periodicities or patterns (like those suggested by AFM [12]) were apparent. To further search for a lattice-like structure, we visualized granule surfaces by projecting the spherical surface onto a 2-D plane, but no pattern was apparent upon these projected surfaces (see Fig. S2 in the supplemental material). Additionally, although variable sample thickness made comparisons difficult, larger granules appeared to have more pronounced surface layers.
Fig 3.
Surface features of granules. (A) Computational slices through representative medium-sized PHB granules (top and bottom left) and, for comparison, PHB immediately to the left of a polyphosphate granule (bottom right) to illustrate difference in surface structure and density. Red curly brackets (top left) highlight the presence of a discernible, patchy surface layer; white square brackets the absence. Bar, 100 nm. (B). Slices (12.6 nm thick) through a granule isolated from a lysed cell (middle; “drop-away” inset at bottom left depicts another Z-slice through the tomogram so as to depict the optimal cross section through PHB granule, 125 nm beneath the main slice with an optimal cross section through vesicle). Close-ups of PHB surface (left) and vesicle membrane (right) demonstrate by direct comparison the lack of membrane on the granule surface. Note that the close-up of the granule is of the area with most pronounced surface. Bar, 50 nm. Scales of PHB and vesicle close-up panels are the same.
To obtain higher contrast, we also imaged granules outside of mildly sonicated R. eutropha cells. Lysate was plunge frozen and imaged immediately, without subsequent purification steps as used in previous studies of isolated granules. These granules had increased sensitivity to electron beam damage, necessitating even lower-dose reconstructions. Granules in the lysate nevertheless appeared essentially identical to granules in intact cells except that they were less spherical. The surface layers of granules next to vesicles were considerably less dense than immediately adjacent lipid bilayer membranes (Fig. 3B).
Granule localization.
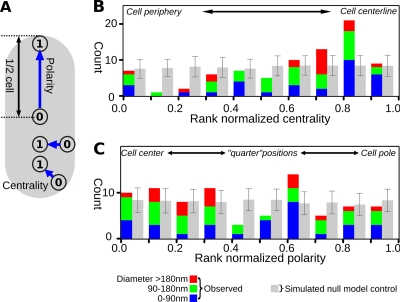
In order to analyze the locations of the granules more quantitatively, centrality and polarity metrics varying from 0 to 1 representing positions between the membrane and the central axis, or the pole and the cell center, respectively, were developed and applied to all the granules in cells where both poles were visible (128 granules in 22 cells) (Fig. 4A). To compensate for the contributions of granule size and other granules in the cell, centrality and polarity scores were normalized by ranking them against 500 randomized placements. For each randomization of each cell, all the granules were removed and sequentially reinserted into the empty cell body at random positions, but without overlapping either the cell boundary or any other previously (re)inserted granule. The ranked position of the observed granule relative to this randomized distribution was then plotted in Fig. 4. For example, if the raw centrality measurement of a given granule was 0.63, but this value was higher than 450 out of 500 randomizations of that granule's position, its rank-normalized centrality would be designated as 0.9 (i.e., 450/500). High rank-normalized ratings thus indicate that the granule is closer to the centerline or pole of the cell than the expectation from a randomized distribution. Data from the 24-h time point were omitted, since these cells were nearly half full of granules and granule location was most likely dominated by packing forces.
Fig 4.
Granule localization is biased toward the central region of the cell. (A) Cartoon to illustrate the centrality and polarity metrics. (B) Distribution of rank-normalized centrality of observed granules (colored bars) and a data set randomized 100 times (gray bars; error bars are standard deviation across all 100 randomizations to illustrate the significance of the deviation of observed granules from the null model). Note that granules occur around the cell periphery less than would be expected at random and more toward the center than would be expected. (C) Polarity of granules as a function of size across the first three time points, alongside mean and standard deviations of 100 further randomizations of the observed data set (gray).
The results indicated that granule positioning was biased toward the central region of the cell (Fig. 4B). Although there is a small peak toward the cell edge (rank-normalized centrality between 0 and 0.1), there are still fewer granules in this position than expected at random. Those granules were proximal to the membrane but were of various sizes (not just small ones), showed no evidence of special association with the membrane, and were not in the process of budding. At no time point was there any indication that smaller granules tended to be less central than larger granules, as would be expected if genesis were at the cell periphery. Biases in granule distribution along the length of the cell were less clear (Fig. 4C): the “one-quarter” and “three-quarters” length positions may have been depleted, but the data were too noisy for confident conclusions. It was nevertheless clear that granules were not polar localized. To ensure that this analysis was robust to the possibility that we misidentified some of the granules, we repeated it including all previously rejected polyphosphate granules and also by discarding granules at random from the combined population of PHB and polyphosphate. The result that granule location is biased toward the center of the cell was unaffected in both cases (results not shown).
Cytoplasmic structures.
The membrane-budding model predicts a budding process, while the scaffold model predicts a cytoplasmic scaffold structure (50). In no case did we observe a scaffold, a granule within the membrane, or a granule in the process of budding from a membrane. Although four cytoplasmic filament bundles were observed (three at the 2.5-h time point and one at the 5-h time point), in no case were they associated with a granule. In addition, 10 cytoplasmic vesicles were observed (of which eight were double-bilayered vesicles), all in cells from the 2.5-h time point, but in none of these cases was a granule observed associated with, or within, a vesicle.
Granule growth over time.
PHB granules occupied increasingly greater proportions of the cytoplasm over time (Fig. 5B). Despite this increase in granule size with time (Fig. 3 and 5A), the number of granules per unit volume remained roughly constant (Fig. 5B). While the size distribution shifted toward larger granules over time (Fig. 5A), small granules (<100 nm) were nevertheless visible at all time points (Fig. 5A). Additionally, several granules were nonspherical (Fig. 5C), as if two spherical granules of different radii had just fused.
DISCUSSION
We used ECT to image PHB granules in a near-native state to “macromolecular” resolution in the model system R. eutropha. While previous studies have suggested that PHB granules are generated either in the membrane (21, 22), upon a scaffold (50), or as micelles in the cytoplasm (15), our results support the last suggestion.
Concerning the membrane-budding model, we did not observe a nascent granule within the membrane, a granule-budding event, or any special bias of small granules to localize close to a membrane or a cell pole in this study. Furthermore, we have never seen a storage granule embedded in a membrane in any of the now thousands of PHB-producing bacterial cells imaged in the Jensen lab in recent years (e.g., references 5, 8, and 25). We are confident that had these structures been present, we would have observed them, since membranes are very clear in cryotomograms and buds would have been as clear as, for instance, magnetosomes (∼50-nm-diameter membrane invaginations) were in previous cryotomograms (25). Additionally, the membrane-budding model predicts that granules should be at least partially covered in lipids. Looking closely at the surface of granules both within intact cells and outside of gently lysed cells, we observed a thin (∼2-nm), discontinuous (patchy) surface layer. This is inconsistent with either a continuous mono- or bilayer, and it is doubtful that even the patches are ordered lipidic monolayers due to resulting instability. Moreover, if the granules were coated with lipids as previous studies have suggested (12, 30), extensive as-yet-undiscovered protein machinery would be required to first bud granules from the membrane, later add additional lipid to the surface of growing granules after they left the membrane, and finally fuse granules in the cytoplasm. If the granules were coated with lipids, one might also expect that lipid biosynthetic genes would be upregulated during PHB production, but recent microarray data indicate the contrary: phosphatidylethanolamine and phosphatidylglycerol/cardiolipin (the major lipids identified in R. eutropha membranes [20]) biosynthesis is downregulated 4- and 2-fold, respectively, during PHB production [6]; GEO series accession number GPL10276). Fixation and/or staining artifacts may have caused the surface layer to appear more continuous in previous thin-section EM studies (37). The smooth outer “envelope” seen by AFM (12) may have been a fragment of cell membrane deposited on the granules during suctioning. Indeed, the layers observed were reported to depend on sonication power. Additionally, the resolution of the AFM tip, the possible interaction of the tip with the sample, and the softness of the sample could have made it difficult to discern the patchy nature of the surface layer. Finally, the above presupposes that the lipid layer is maintained after budding. The alternative, an unmaintained lipid layer that becomes incrementally diluted, is also at odds with our observations, as granule surfaces appear to become more distinct as granules enlarge.
Due to the unlikelihood of a lipid coat, the patchy surface layer is most likely proteinaceous, and based on biochemical studies, these proteins are at least in part composed of the phasin PhaP1 and the PHB-metabolic enzymes. Immuno-EM has shown, for instance, that PhaP1 localizes to granule surfaces (53), though abundance studies predict that PhaP1 could cover only a part of the granule surface (31, 48). We did not observe, however, the semiordered, 2- to 4-nm-thick, ∼20-nm-wide network reported by Dennis et al. (12). Based on the clarity of the patchy surface in the images, as well as previous ECT studies that have resolved proteinaceous networks within other bacteria (5, 25, 29), we believe such a network would have been resolved here, if present. Thus, while Dennis et al. convincingly showed that the network was dependent upon PhaP1, it may have been a denatured form, particularly given the unusual physiochemical characteristics of PhaP1 (31) and the nonnative (isolated and air-dried) conditions under which the granules were imaged, which also may have caused the PHB to crystallize (12).
Our results are also at odds with certain fluorescence microscopy studies which showed that granule-associated proteins and granules themselves localize to cell poles and membranes (21, 22, 33, 34). While these results may be explained at least in part by the limited resolution of conventional light microscopy, overexpression and/or fluorescent tagging are also known to perturb localization (52). Misfolded proteins in E. coli sometimes localize to the cell poles (39), as do physiologically active fluorescently tagged proteins (3). Given the unusual biophysical properties of phasins (31) and PhaC (which requires a small amount of nonionic detergent to allow chromatography in vitro [16, 54]), fluorescently tagged fusion proteins may have misfolded and aggregated at the cell pole. Nile Red stain was also seen at the poles, presumably locating granules (18, 21), but Nile Red is also known to bind protein aggregates (11). The strongest contribution to explaining polar localization, however, comes from Peters and Rehm (34), who demonstrated that the polar localization of granules in E. coli was due to nucleoid exclusion: while GFP-PhaC localized to the poles, anucleate mutants with the same GFP-PhaC displayed a random distribution of foci along the cell length. ECT studies of different bacteria have revealed that nucleoid compaction can vary dramatically and can vary between species and growth conditions (51). It is also known that fixing and staining procedures, as well as the expression of fluorescently tagged proteins, can all affect nucleoid compaction (7, 37). It is therefore possible that the polar localization patterns that have been seen in other studies were driven by changes in nucleoid compaction.
Our results also fail to support the scaffold model. This model was originally proposed based on earlier thin-section EM images showing granules clustered around a dark-stained “mediation element” (50). We observed, however, nothing like the mediation element (and have not in other PHB-producing species imaged by ECT in the Jensen lab). Furthermore, while ECT has revealed many cytoskeletal elements in many different bacteria (e.g., references 5 and 8), and some filaments were seen here in R. eutropha, they were not granule associated. Our results therefore also do not support a proteinaceous scaffold model, where the scaffold would be filaments or repeated protein units. Instead the “mediation element” was most likely the fixed, dehydrated, and stained nucleoid. Interestingly, recent studies of granule formation in both R. eutropha (36) and Pseudomonas putida (13) have shown that a member of the phasin family in each organism (PhaM and PhaF, respectively) binds nonspecifically to DNA. Thus, in analogy to carboxysome segregation in Synechococcus elongatus (41), granules are equally segregated between daughter cells upon cell division (13), in this case utilizing nonspecific nucleoid binding by granules. Thus, nonspecific interaction of the phasin PhaM with the nucleoid in R. eutropha may explain why granules localized toward the center of the cell.
We conclude that granules are initiated in the cytoplasm and remain cytoplasmic throughout growth. Supporting this, we have recently shown that soluble PHB can be copurified from cell lysate with PhaC and PhaP1, indicating a soluble precursor to insoluble granules (M. Cho and J. Stubbe, submitted for publication). As discussed above, previous data showing granules at the membrane are consistent with this model if the nucleoid pushed granules to the cell periphery. Further support for cytoplasmic granules comes from the evidence of granule fusion. Small granules were seen at all time points. While these may have been granules that simply failed to grow, the observation of (i) apparently fusing, pear-shaped granules, (ii) the progressive increase in net granule volume per cell, and (iii) the roughly constant number of granules per cell across all time points together suggest that granules are constantly being formed but fuse as they grow larger. Spherical postfusion granules require that PHB remain fluid, as supported by their nonstructured texture in the tomograms and solid-state nuclear magnetic resonance (NMR) studies (42).
A patchy-surface cytoplasmic genesis model with granule fusion suggests mechanisms for how phasin proteins could maintain multiple granules and maximize the amount of PHB produced (53, 56). Phasin coating, even though sparse, may discourage granule fusion yet allow it if granule growth rate exceeds the rate of phasin synthesis, which would expose PHB. A steady state of approximately 10 granules per cell would provide an approximately 2-fold increase in surface area relative to a single, giant granule of the same volume (as seen in the absence of PhaP1). This suggests that PhaP1 concentrations may therefore tune the total surface area-to-volume ratio of PHB granules in the cell and thereby help regulate both the growth and use of granular PHB (56). Phasins may also act as “emulsifiers” that moderate granule surface hydrophobicity, preventing them from clustering in the cytoplasm. Fluorescent tags may reduce this effect, contributing to the clustering of granules at the poles through hydrophobic interactions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH GM49171 to J. Stubbe and Howard Hughes Medical Institute funding to G. J. Jensen.
We thank Wesley Chen for assistance in manual reconstruction of tomograms.
Footnotes
Published ahead of print 16 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Agulleiro JI, Fernandez JJ. 2011. Fast tomographic reconstruction on multicore computers. Bioinformatics 27:582–583 [DOI] [PubMed] [Google Scholar]
- 2. Amat F, et al. 2008. Markov random field based automatic image alignment for electron tomography. J. Struct. Biol. 161:260–275 [DOI] [PubMed] [Google Scholar]
- 3. Bardy SL, Maddock JR. 2007. Polar explorations: recent insights into the polarity of bacterial proteins. Curr. Opin. Microbiol. 10:617–623 [DOI] [PubMed] [Google Scholar]
- 4. Beeby M, Bobik TA, Yeates TO. 2009. Exploiting genomic patterns to discover new supramolecular protein assemblies. Protein Sci. 18:69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Briegel A, et al. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. U. S. A. 106:17181–17186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brigham CJ, et al. 2010. Elucidation of beta-oxidation pathways in Ralstonia eutropha H16 by examination of global gene expression. J. Bacteriol. 192:5454–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Butan C, et al. 2011. Spiral architecture of the nucleoid in Bdellovibrio bacteriovorus. J. Bacteriol. 193:1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen S, et al. 2011. Structural diversity of bacterial flagellar motors. EMBO J. 30:2972–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Comolli L, Kundmann M, Downing K. 2006. Characterization of intact subcellular bodies in whole bacteria by cryo-electron tomography and spectroscopic imaging. J. Microsc. 223:40–52 [DOI] [PubMed] [Google Scholar]
- 10. Czabany T, Athenstaedt K, Daum G. 2007. Synthesis, storage and degradation of neutral lipids in yeast. Biochim. Biophys. Acta 1771:299–309 [DOI] [PubMed] [Google Scholar]
- 11. Demeule B, Gurny R, Arvinte T. 2007. Detection and characterization of protein aggregates by fluorescence microscopy. Int. J. Pharm. 329:37–45 [DOI] [PubMed] [Google Scholar]
- 12. Dennis D, Sein V, Martinez E, Augustine B. 2008. PhaP is involved in the formation of a network on the surface of polyhydroxyalkanoate inclusions in Cupriavidus necator H16. J. Bacteriol. 190:555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galán B, et al. 2011. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol. Microbiol. 79:402–418 [DOI] [PubMed] [Google Scholar]
- 14. Gerngross TU, Martin DP. 1995. Enzyme-catalyzed synthesis of poly[(R)-(-)-3-hydroxybutyrate]: formation of macroscopic granules in vitro. Proc. Natl. Acad. Sci. U. S. A. 92:6279–6283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gerngross TU, Reilly P, Stubbe J, Sinskey AJ, Peoples OP. 1993. Immunocytochemical analysis of poly-beta-hydroxybutyrate (PHB) synthase in Alcaligenes eutrophus H16: localization of the synthase enzyme at the surface of PHB granules. J. Bacteriol. 175:5289–5293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gerngross TU, et al. 1994. Overexpression and purification of the soluble polyhydroxyalkanoate synthase from Alcaligenes eutrophus: evidence for a required posttranslational modification for catalytic activity. Biochemistry 33:9311–9320 [DOI] [PubMed] [Google Scholar]
- 17. Griebel R, Smith Z, Merrick JM. 1968. Metabolism of poly-beta-hydroxybutyrate. I. Purification, composition, and properties of native poly-beta-hydroxybutyrate granules from Bacillus megaterium. Biochemistry 7:3676–3681 [DOI] [PubMed] [Google Scholar]
- 18. Hermawan S, Jendrossek D. 2007. Microscopical investigation of poly(3-hydroxybutyrate) granule formation in Azotobacter vinelandii. FEMS Microbiol. Lett. 266:60–64 [DOI] [PubMed] [Google Scholar]
- 19. Hiraishi T, et al. 2005. Atomic force microscopic observation of in vitro polymerized poly[(R)-3-hydroxybutyrate]: insight into possible mechanism of granule formation. Biomacromolecules 6:2671–2677 [DOI] [PubMed] [Google Scholar]
- 20. Horowitz DM, Sanders JKM. 1994. Amorphous, biomimetic granules of polyhydroxybutyrate: preparation, characterization, and biological implications. J. Am. Chem. Soc. 116:2695–2702 [Google Scholar]
- 21. Jendrossek D. 2005. Fluorescence microscopical investigation of poly(3-hydroxybutyrate) granule formation in bacteria. Biomacromolecules 6:598–603 [DOI] [PubMed] [Google Scholar]
- 22. Jendrossek D. 2009. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J. Bacteriol. 191:3195–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jendrossek D, Selchow O, Hoppert M. 2007. Poly(3-hydroxybutyrate) granules at the early stages of formation are localized close to the cytoplasmic membrane in Caryophanon latum. Appl. Environ. Microbiol. 73:586–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keshavarz T, Roy I. 2010. Polyhydroxyalkanoates: bioplastics with a green agenda. Curr. Opin. Microbiol. 13:321–326 [DOI] [PubMed] [Google Scholar]
- 25. Komeili A, Li Z, Newman DK, Jensen GJ. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242–245 [DOI] [PubMed] [Google Scholar]
- 26. Kremer J, Mastronarde D, McIntosh J. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 27. Lawrence AG, et al. 2005. Transcriptional analysis of Ralstonia eutropha genes related to poly-(R)-3-hydroxybutyrate homeostasis during batch fermentation. Appl. Microbiol. Biotechnol. 68:663–672 [DOI] [PubMed] [Google Scholar]
- 28. Li Z, Jensen GJ. 2009. Electron cryotomography: a new view into microbial ultrastructure. Curr. Opin. Microbiol. 12:333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z, Trimble M, Brun Y, Jensen G. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26:4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mayer F, Hoppert M. 1997. Determination of the thickness of the boundary layer surrounding bacterial PHA inclusion bodies, and implications for models describing the molecular architecture of this layer. J. Basic Microbiol. 37:45–52 [Google Scholar]
- 31. Neumann L, et al. 2008. Binding of the major phasin, PhaP1, from Ralstonia eutropha H16 to poly(3-hydroxybutyrate) granules. J. Bacteriol. 190:2911–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peoples OP, Sinskey AJ. 1989. Poly-beta-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J. Biol. Chem. 264:15298–15303 [PubMed] [Google Scholar]
- 33. Peters V, Becher D, Rehm B. 2007. The inherent property of polyhydroxyalkanoate synthase to form spherical PHA granules at the cell poles: the core region is required for polar localization. J. Biotechnol. 132:238–245 [DOI] [PubMed] [Google Scholar]
- 34. Peters V, Rehm B. 2005. In vivo monitoring of PHA granule formation using GFP-labeled PHA synthases. FEMS Microbiol. Lett. 248:93–100 [DOI] [PubMed] [Google Scholar]
- 35. Pfeiffer D, Jendrossek D. 2011. Interaction between poly(3-hydroxybutyrate)granule-associated proteins as revealed by two-hybrid analysis and identification of a new phasin in Ralstonia eutropha H16. Microbiology (Reading, Engl.) 157:2795–2807 [DOI] [PubMed] [Google Scholar]
- 36. Pfeiffer D, Wahl A, Jendrossek D. 2011. Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol. Microbiol. 82:936–951 [DOI] [PubMed] [Google Scholar]
- 37. Pilhofer M, Ladinsky MS, McDowall AW, Jensen GJ. 2010. Bacterial TEM: new insights from cryo-microscopy. Methods Cell Biol. 96:21–45 [DOI] [PubMed] [Google Scholar]
- 38. Pötter M, Madkour M, Mayer F, Steinbüchel A. 2002. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology (Reading, Engl.) 148:2413–2426 [DOI] [PubMed] [Google Scholar]
- 39. Rokney A, et al. 2009. E. coli transports aggregated proteins to the poles by a specific and energy-dependent process. J. Mol. Biol. 392:589–601 [DOI] [PubMed] [Google Scholar]
- 40. Saegusa H, Shiraki M, Kanai C, Saito T. 2001. Cloning of an intracellular poly[D(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183:94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Savage D, Afonso B, Chen A, Silver P. 2010. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327:1258–1261 [DOI] [PubMed] [Google Scholar]
- 42. Shaw GL, Melby MK, Horowitz DM, Keeler J, Sanders JK. 1994. Nuclear magnetic resonance relaxation studies of poly(hydroxybutyrate) in whole cells and in artificial granules. Int. J. Biol. Macromol. 16:59–63 [DOI] [PubMed] [Google Scholar]
- 43. Snell KD, Peoples OP. 2009. PHA bioplastic: a value added coproduct for biomass biorefineries. Biofuels Bioproducts Biorefining 3:456–467 [Google Scholar]
- 44. Steinbüchel A, Valentin HE. 1995. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol. Lett. 128:219–228 [Google Scholar]
- 45. Stubbe J, Tian J. 2003. Polyhydroxyalkanoate (PHA) homeostasis: the role of PHA synthase. Nat. Prod. Rep. 20:445–457 [DOI] [PubMed] [Google Scholar]
- 46. Stubbe J, et al. 2005. Nontemplate-dependent polymerization processes: polyhydroxyalkanoate synthases as a paradigm. Annu. Rev. Biochem. 74:433–480 [DOI] [PubMed] [Google Scholar]
- 47. Suloway C, et al. 2009. Fully automated, sequential tilt-series acquisition with Leginon. J. Struct. Biol. 167:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tian J, et al. 2005. Analysis of transient polyhydroxybutyrate production in Wautersia eutropha H16 by quantitative Western analysis and transmission electron microscopy. J. Bacteriol. 187:3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tian J, Sinskey AJ, Stubbe J. 2005. Class III polyhydroxybutyrate synthase: involvement in chain termination and reinitiation. Biochemistry 44:8369–8377 [DOI] [PubMed] [Google Scholar]
- 50. Tian J, Sinskey A, Stubbe J. 2005. Kinetic studies of polyhydroxybutyrate granule formation in Wautersia eutropha H16 by transmission electron microscopy. J. Bacteriol. 187:3814–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Travers A, Muskhelishvili G. 2005. Bacterial chromatin. Curr. Opin. Genet. Dev. 15:507–514 [DOI] [PubMed] [Google Scholar]
- 52. Werner J, et al. 2009. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc. Natl. Acad. Sci. U. S. A. 106:7858–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wieczorek R, Pries A, Steinbüchel A, Mayer F. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177:2425–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wodzinska J, et al. 1996. Polyhydroxybutyrate synthase: evidence for covalent catalysis. J. Am. Chem. Soc. 118:6319–6320 [Google Scholar]
- 55. York G, Junker B, Stubbe J, Sinskey A. 2001. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J. Bacteriol. 183:4217–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. York G, Stubbe J, Sinskey A. 2001. New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J. Bacteriol. 183:2394–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. York GM, Stubbe J, Sinskey AJ. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 184:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.