Abstract
Iron is an essential nutrient that is implicated in most cellular oxidation reactions. However, iron is a highly reactive element that, if not appropriately chaperoned, can react with endogenously and exogenously generated oxidants such as hydrogen peroxide to generate highly toxic hydroxyl radicals. Dps proteins (DNA-binding proteins from starved cells) form a distinct class (the miniferritins) of iron-binding proteins within the ferritin superfamily. Bacillus anthracis encodes two Dps-like proteins, Dps1 and Dps2, the latter being one of the main iron-containing proteins in the cytoplasm. In this study, the function of Dps2 was characterized in vivo. A B. anthracis Δdps2 mutant was constructed by double-crossover mutagenesis. The growth of the Δdps2 mutant was unaffected by excess iron or iron-limiting conditions, indicating that the primary role of Dps2 is not that of iron sequestration and storage. However, the Δdps2 mutant was highly sensitive to H2O2, and pretreatment of the cells with the iron chelator deferoxamine mesylate (DFM) significantly reduced its sensitivity to H2O2 stress. In addition, the transcription of dps2 was upregulated by H2O2 treatment and derepressed in a perR mutant, indicating that dps2 is a member of the regulon controlled by the PerR regulator. This indicates that the main role of Dps2 is to protect cells from peroxide stress by inhibiting the iron-catalyzed production of OH.
INTRODUCTION
Bacillus anthracis is a Gram-positive spore-forming bacterium that causes anthrax in humans and animals. Anthrax infects its host by the entry of spores through skin abrasions, by the ingestion of contaminated food, or via the respiratory system (9). Iron is an important nutrient for bacterial growth and for virulence, and consequently mammalian hosts have developed iron-withholding mechanisms as a line of defense against invading pathogens. The availability of free iron in the bloodstream is restricted by binding proteins such as transferrin and lactoferrin, which have binding constants of ∼10−20 M (13, 32). While transferrin severely limits the growth of B. anthracis in human serum, it does not inhibit other pathogens such as Staphylococcus aureus and Streptococcus pneumoniae (34). This indicates that B. anthracis has not evolved effective mechanisms to evade serum iron deprivation. Instead, the phagocytosis of B. anthracis spores and their subsequent germination and growth in macrophages serve both to protect this bacterium from the inhibitory effects of serum and to facilitate the accumulation of iron for the later systemic stage of infection (31, 34).
In addition to iron-withholding systems, host macrophages generate antimicrobial reactive oxygen species (ROS), such as superoxide radicals (O2·−) and hydrogen peroxide (H2O2), within macrophages to kill invading pathogens. The damaging effects of oxidative stress can be intensified by the reaction of these molecular species with intracellular iron (25). In particular, the combination of Fe(II) and H2O2 leads, via Fenton chemistry [H2O2 + Fe(II) → OH· + OH− + Fe(III)], to the production of highly toxic hydroxyl radicals (OH·) that damage most types of biological macromolecules (18). This means that pathogens must acquire adequate sources of iron while avoiding or reducing its availability for participation in OH· production. To this end, microorganisms have developed mechanisms that detoxify ROS and chaperone intracellular iron (41).
The connection between iron metabolism and oxidative stress in B. anthracis is emphasized by our recent observation that this organism takes up iron rather than manganese (2) in response to paraquat-induced superoxide stress (31, 39a). This seemingly counterintuitive response is puzzling since an increase in cellular iron would be expected to exacerbate the oxidative stress through iron-catalyzed formation of OH·. The compartmentalization of free iron into specialized iron storage proteins can neutralize iron toxicity, and consequently, bacteria encode iron-binding proteins such as ferritins, bacterioferritins, and Dps. Dps proteins belong to the ferritin superfamily but represent a distinct group within this family. Like ferritins, Dps proteins form a spherical protein complex (nanocage) that surrounds a central cavity that holds mineralized iron (8, 26). However, the ferroxidase center of Dps proteins, which is responsible for the oxidation of ferrous iron, generally consumes H2O2, whereas ferritins use dioxygen (O2) to generate H2O2. The resulting nonreactive ferric iron (hydrated ferric oxide) is stored in the central cavity of the proteins and can serve as a reserve of this element (8, 22, 23). Therefore, in addition to their role in iron storage, Dps proteins exert a protective effect by concurrently reducing the toxicity of ferrous iron and H2O2 (5, 19, 39, 40, 41).
B. anthracis encodes a predicted maxiferritin (BA5296) and two miniferritin Dps-like proteins, namely, Dps1 (BA2013) and Dps2 (BA5290). However, it should be noted that the nomenclature of the Dps proteins is somewhat confused. The original annotations for BA2013 (Dps1) and BA5290 (Dps2) (33) are used in this paper. However, Papinutto et al. (29) named BA2013 Dlp-2 and BA5290 Dlp-1, while Liu et al. (26) named BA2013 Dps2 and BA5290 Dps1. While the function of the B. anthracis ferritin has not been studied, both Dps1 (i.e., BA2013) and Dps2 (i.e., BA5290) have been shown to have ferritin-like activity when overexpressed in Escherichia coli (29). Dps1 and Dps2 have been extensively characterized in vitro (26, 29, 37). These studies have shown that their reactivity with H2O2 and O2 is in striking contrast to that of the Dps proteins characterized in other organisms (26), which generally use H2O2 as the preferred iron oxidant with an ∼100-to 1,000-fold preference over O2 (11, 38, 41). In vitro, Dps2 has no detectable ferroxidase activity with H2O2 and instead uses O2 as the iron oxidant. The absence of reactivity with H2O2 is unique among known Dps proteins and potentially points to a role in iron storage rather than detoxification. In contrast, Dps1 uses both H2O2 and O2 as iron oxidants, with just an ∼3-fold preference for H2O2 (26), and it may function as both a detoxifying enzyme and an iron storage protein.
The current studies are aimed at elucidating the role of Dps2 in iron homeostasis and oxidative stress resistance in B. anthracis and the regulation of its gene expression. In order to determine the role of this protein in defense against oxidative stress, we have created an isogenic mutant of B. anthracis in which the dps2 gene is deleted. The results of these in vivo studies complement previous in vitro analyses of the purified protein (26, 29) but point to a role for Dps2 in oxidative stress resistance.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study, together with their relevant characteristics, are described in Table 1. The avirulent Bacillus anthracis strain UM23C1-2 (pX01− pX02−) (17) was used to generate the Δdps2 and ΔperR mutants by replacement of their entire coding sequences with an omega element conferring kanamycin resistance (Ω-km DNA cassette [35]). The primers used to generate the upstream and downstream fragments are shown in Table 2. Escherichia coli strain TOP10 (Invitrogen, United Kingdom) was used as the host for cloning, and GM48 (dam dcm) (Stratagene, Netherlands) was used as an intermediate host to obtain nonmethylated plasmid DNA for electroporation into B. anthracis. Complementation studies were carried out using a pUTE583-based expression plasmid, pKG400. A SacI and SmaI fragment from pHT01 (MoBiTec, Göttingen, Germany), encoding the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pgrac promoter, ribosome-binding site (RBS), and lacI repressor gene, was cloned into the multiple-cloning site of pUTE583 to obtain pKG400. After amplification with primers Dps2-fwd and Dps2-rev (Table 2) and digestion with BamHI and EcoRI, the dps2 gene was cloned downstream of the Pgrac promoter and RBS in pKG400 to generate pKG400-dps2.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Trait or relevant genotypea | Source or reference |
|---|---|---|
| Strains | ||
| Bacillus anthracis | ||
| UM23C1-2 | pXO1− pXO2− Ura− Rifr | 17 |
| Δdps2 mutant | UM23C1-2 Δdps2::Kmr | This work |
| ΔperR mutant | UM23C1-2 ΔperR::Kmr | This work |
| Escherichia coli | ||
| TOP10 | F−mcrA (mrr-hsdRMS-mcrBC) ϕ80 lacZΔM15 lacX74 deoR recA1 araD139 (ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen Ltd., Paisley, United Kingdom |
| GM48 | JM110 dam dcm | Stratagene, Amsterdam, Netherlands |
| Plasmids | ||
| pUTE583 | Cmr in E. coli; Ermr in B. anthracis | 7 |
| pUTE618 | Cmr Kmr in E. coli | 7 |
| pGEM-T | Ampr in E. coli | Promega Corp. |
| pHT01 | Ampr Cmr, Pgrac promoter, lacI, E. coli/B. subtilis shuttle vector | MoBiTec, Germany |
| pKG400 | Expression vector for B. anthracis; derivative of pUTE583 containing Pgrac, lacI; Emr Cmr | This work |
| pKG400-dps2 | pKG400 with dps2 cloned downstream of the Pgrac promoter | This work |
Abbreviations: Kmr, kanamycin resistance; Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Ermr, erythromycin resistance; Spr, spectinomycin resistance.
Table 2.
Oligonucleotide primers used in this study
| Function and primer name | Sequencea (5′ → 3′) | PCR amplification product (bp) |
|---|---|---|
| Mutant construction | ||
| Dps2 up-F | GCCCCGCGGGGCTTGTTCAAGCATTAGGT | dps2 upstream region (760) |
| Dps2 up-R | CCGGAATTCTATGATCAATCTCCTTTTGAG | |
| Dps2 down-F | CCGGAATTCGGAAGCGGCTCGTTTAGAGTC | dps2 downstream region (582) |
| Dps2 down-R | GCCTCTAGAGGTTGTATTAGCGGTTTGAA | |
| PerR up-F | GCCTGCTCTAGAAATGGTTACAGTAGGAGAAATGGC | perR upstream region (428) |
| PerR up-R | GTACCCAAGCTTGGTGAGAATGAGCATTAAGTATTC | |
| PerR down-F | GTACCGGAATTCGCGTTTGTCCAGAGTGTCATAAGG | perR downstream region (315) |
| PerR down-R | GTACCGGAATTCTTTAATTCTTCTTTGACCACCGTC | |
| Northern blot probes | ||
| Dps2-For | ATGAACAAACAAGTAATCGAAG | dps2 probe (465) |
| Dps2-Rev-T7 | ctaatacgactcactatagggagaTAGCATCCAAGCGTGTTTTTC | |
| Complementation | ||
| Dps2-fwd | CAAGGATCCCATAATGAACAAACAAGTAATCGAAG | Complete dps gene (498) |
| Dps2-rev | CAAGAATTCAAGCGACTCTAAACGAGCC | |
| Primer extension | ||
| Dps2 | GAATAAAACGCTCCAGTCTG |
Primer sequences were derived from the B. anthracis genome sequence (33). Restriction sites (underlined) and additional bases to improve restriction enzyme binding (bold) are indicated. Lowercase letters indicate a 5′ extension with the T7 promoter sequence for the creation of an antisense RNA probe. Primers used for primer extension analysis were 5′-FAM labeled.
Bacterial cultures were routinely grown at 37°C in Luria-Bertani (LB) broth with vigorous agitation (220 rpm) or on LB agar. Bacterial growth was monitored spectrophotometrically at 540 nm. The following antibiotics (Sigma-Aldrich, United Kingdom) were used to supplement media as required: ampicillin (50 μg/ml), erythromycin (400 μg/ml for E. coli and 5 μg/ml for B. anthracis), kanamycin (20 μg/ml), and chloramphenicol (6 μg/ml).
DNA manipulations and general techniques.
Restriction enzymes and T4 DNA ligase were used according to the manufacturer's instruction (New England BioLabs). A miniprep system (Promega, United Kingdom) was used to isolate plasmid DNA from E. coli. PCR was carried out with Platinum Pfx DNA polymerase (Invitrogen, United Kingdom) using primers (Table 2) purchased from Invitrogen (United Kingdom) or Eurogentec (United Kingdom). PCR products were purified with the QIAquick PCR purification kit (Qiagen, CA). Extraction of chromosomal DNA from bacterial strains was carried out with the DNeasy blood and tissue kit (Qiagen, CA).
Liquid culture growth assays.
Overnight precultures of B. anthracis strains were diluted into fresh LB medium without a selective antibiotic to an optical density at 540 nm (OD540) of 0.05 and then grown at 37°C with agitation (220 rpm). To determine the effect of iron limitation on the growth of B. anthracis strains, deferoxamine mesylate (DFM) (Sigma-Aldrich, United Kingdom) was added to the exponentially growing cultures (OD540 of 0.3) at a final concentration of 15.0 mM and growth monitored with respect to an untreated control. Similarly, to achieve the conditions of iron overload, Fe(II)SO4 · 7H2O was added to the cultures at 1.0 and 2.0 mM. Sensitivities to oxidative stress compounds were determined by the addition of either H2O2 (1.0 mM) or paraquat (0.8 mM), and when necessary, cultures were pretreated for 10 min with Fe(II)SO4 · 7H2O or DFM, respectively, to overload cells with or restrict them of iron.
RNA extraction and Northern blot analysis.
Total RNA was isolated using a RiboPure bacterial kit (Ambion) according to the manufacturer's protocol with modifications to optimize RNA yield or using the hot-phenol method as described previously (10). A digoxigenin (DIG)-labeled RNA probe specific for dps2 was synthesized in vitro with T7 RNA polymerase, using a PCR template synthesized using the oligonucleotide primers in Table 2 and the DIG RNA labeling kit (Roche, United Kingdom). Northern blot analysis was conducted as described by Eymann et al. (10).
Primer extension analysis.
Primer extension analysis, used to determine the transcriptional start sites, was performed using a 5′-6-carboxyfluorescein (FAM)-labeled oligonucleotide primer (Eurogentec, United Kingdom) (Table 2) for the reverse transcriptase reaction. The primer extension reaction was completed in two stages. In the initial reverse transcription step, total RNA (40 to 50 μg) was heated at 70°C for 5 min in the presence of 100 pmol primer, followed by the addition of 100 U Moloney murine leukemia virus (MMLV) reverse transcriptase (Ambion, TX), 1 mM deoxynucleoside triphosphates (dNTPs) (Fermentas, United Kingdom), and 40 U of recombinant RNasin RNase inhibitor (Promega, United Kingdom) in a total volume of 30 μl. The reaction mixtures were incubated at 42°C for 60 min. The initial reverse transcription reaction mixture was replenished with the above reagents and then subject to a second extension reaction by incubation at 42°C for further 60 min. The FAM-labeled cDNA products were treated with 10 ng RNase A (Ambion, TX) and purified with a MinElute reaction cleanup kit (Qiagen, CA). In addition, a target DNA fragment was sequenced with dideoxynucleotide-based reactions utilizing the Thermo Sequenase primer cycle sequencing kit (GE Healthcare, United Kingdom) and the same FAM-labeled primer. Both sequencing and extension products were loaded in a capillary-based electrophoresis DNA sequencer (Applied Biosystems 3730, CA) and the resulting electropherograms aligned using Peak Scanner software version 1.0 (Applied Biosystems, CA) to determine the sizes of the primer extension products.
RESULTS
The major role of Dps2 is not that of iron storage.
We recently identified Dps2 as one of four major iron-binding proteins present in the cytoplasm of B. anthracis (39a). In order to investigate the role of this protein, we determined the growth characteristics of a Δdps2 mutant in LB broth in the presence of the ferrous iron chelator deferoxamine mesylate (DFM), which reduces the availability of iron, and excess iron (Fig. 1). The concentrations of DFM (15 mM) and iron (1 and 2 mM) that were used visibly limited the growth of wild-type B. anthracis, mildly in the case of DFM, transiently in the case of 1 mM iron, and severely in the case of 2 mM iron (Fig. 1). Irrespective of the conditions tested, the growth of the Δdps2 mutant was similar to that of the wild-type strain (Fig. 1A and B). In addition, a plate diffusion assay was performed to test the sensitivity of the Δdps2 mutant to Fe(II)SO4, but again no significant changes in iron sensitivity, as determined by the size of the zone of inhibition, were observed in comparison to the wild type (data not shown). If Dps2 were important for iron storage, the growth of the Δdps2 mutant would be expected to be differentially affected by iron excess or limitation in comparison to the wild type.
Fig 1.
Response of B. anthracis to excess- or low-iron conditions. The B. anthracis UM23C1-2 wild-type strain (■ and □) and its Δdps2 mutant (▲ and △) were grown to exponential phase (OD540 of 0.3) in LB medium at 37°C and treated with Fe(II)SO4 · 7H2O (1 mM and 2 mM) (A) and the iron chelator DFM (15 mM) (B). The arrows indicate the time points at which the Fe(II)SO4 · 7H2O and DFM were added to the culture. Similar growth kinetics were obtained from a minimum of two independent experiments.
Dps2 promotes tolerance to peroxide but not superoxide stress.
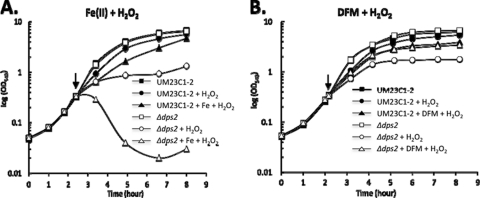
To establish whether Dps2 has a role in protecting B. anthracis against oxidative stress, we determined the sensitivity of the Δdps2 mutant to exogenous H2O2 and endogenous superoxide stress, the latter generated by treatment with paraquat. Cells were grown to exponential phase (OD540 0.3) in LB medium and either H2O2 (1 mM) or paraquat (0.8 mM) added. The Δdps2 mutant was significantly more sensitive to peroxide stress than the wild type (Fig. 2), while superoxide stress had no observable influence on its growth (Fig. 3). These data indicate that Dps2 is a component of the oxidative stress defense system of B. anthracis that is specific to H2O2.
Fig 2.
Response of B. anthracis to oxidative stress. The B. anthracis UM23C1-2 wild-type strain (■, ●, and ▲) and its Δdps2 mutant (□, ○, and △) were grown to exponential phase (OD540 of 0.3) in LB medium at 37°C with or without H2O2 (1 mM), and with or without pretreatment with Fe(II)SO4 · 7H2O (1 mM) (A) or DFM (15 mM) (B). The arrows indicate the time points at which the cultures were pretreated with DFM or Fe(II)SO4 · 7H2O, which was followed, after 10 min, by the addition of H2O2. Similar growth kinetics were obtained from a minimum of two independent experiments.
Fig 3.
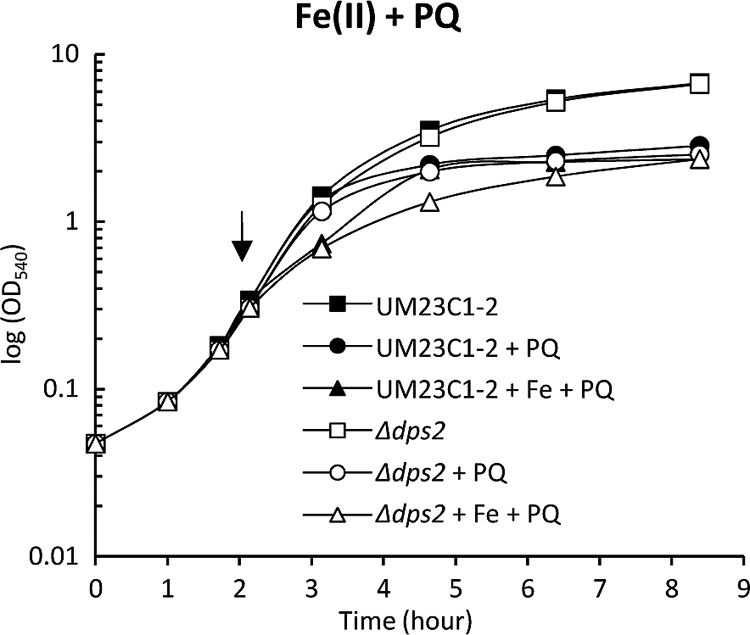
Effect of the addition of iron [1 mM Fe(II)SO4 · 7H2O)] on the response of wild-type B. anthracis UM23C1-2 and the Δdps2 mutant to paraquat (PQ). Exponentially growing (OD540 of 0.3) cultures were pretreated with Fe(II)SO4 · 7H2O for 10 min before being challenged with 0.8 mM paraquat. The arrow indicates the time point at which the cultures were pretreated with Fe(II)SO4 · 7H2O, followed by the addition of paraquat. Similar growth kinetics were obtained from a minimum of two independent experiments.
To confirm the role of Dps2 in peroxide stress resistance, we constructed a strain in which the dps2 mutation was complemented with an IPTG-inducible copy of dps2 on plasmid pKG400-dps2 (Fig. 4). Cells grown to exponential phase (OD540 of 0.3) in the presence or absence of IPTG (10 mM) were treated with H2O2 (1 mM). Neither the presence of pKG400-dps2 nor the addition of IPTG alone affected growth. However, when the complementation strain grown in the absence of IPTG was challenged with H2O2, growth was severely inhibited. The presence of IPTG in the medium to induce the expression of dps2 from pKG400-dps2 significantly limited the inhibitory affect of H2O2 on growth, confirming that the inhibition of the Δdps2 mutant by peroxide was due to the absence of Dps2.
Fig 4.

Complementation of the Δdps2 mutant with a plasmid-encoded copy of Dps2. The Δdps2 mutant was transformed with pKG400-dps2, encoding an IPTG-inducible dps2 gene. Cultures were grown with (open symbols) or without (closed symbols) IPTG (10 mM) to exponential phase (OD540 of 0.3 [arrow]) in LB medium at 37°C (squares) and treated as required with H2O2 (1 mM) (circles). Similar growth kinetics were obtained from a minimum of two independent experiments.
To some extent our in vivo data are in apparent conflict with the in vitro data of Liu et al. (26) that indicate that Dps2 (Dps-1 according to their terminology) uses O2 rather than H2O2 as an iron oxidant; the use of the latter would have provided an explanation for the role of Dps2 in the detoxification of to H2O2.
Iron incorporation in vivo exerts tolerance to peroxide stress.
Since iron homeostasis is especially important under conditions of oxidative stress, we determined the impact of peroxide and superoxide stresses on the Δdps2 mutant in the presence of excess iron. The B. anthracis wild-type strain and Δdps2 mutant were grown in LB with and without excess iron to exponential phase (OD540 of 0.3) and then treated with H2O2 to induce peroxide stress. The combination of H2O2 and excess iron resulted in a severe growth inhibition of the Δdps2 mutant strain followed by cell lysis, while this combination of treatments only moderately affected the wild type (Fig. 2A). In contrast to the dramatic effects of H2O2, iron excess had little or no effect on the growth of the wild type or the Δdps2 mutant under paraquat stress (Fig. 3). This suggests that Fenton chemistry does not play a major role in the paraquat-induced killing of B. anthracis, and this view is reinforced by array date that shows that the LexA regulon is induced in response to peroxide but not superoxide stress (31).
Next we tested the growth of the wild type and the Δdps2 mutant under H2O2 stress under iron-depleted conditions (Fig. 2B). Pretreatment of wild-type B. anthracis with DFM alone has a relatively small influence on cell growth under H2O2 stress (Fig. 2B), presumably due to iron limitation and a requirement of iron-utilizing enzymes involved in respiration and resistance to endogenous oxidative stress. However, pretreatment of the culture medium with DFM reduces the sensitivity of the Δdps2 mutant to H2O2, and consequently its response to peroxide stress is indistinguishable from that in the wild type (Fig. 2B). Taken together, these results clearly implicate ferrous iron in the sensitivity of the Δdps2 mutant to H2O2. They indicate that Dps2 has a role in protecting cells from superoxide stress, presumably by limiting the production of OH· through its ability to mitigate the effects of the toxic combination of ferrous iron and H2O2.
Dps2 is a member of the PerR peroxide stress regulon.
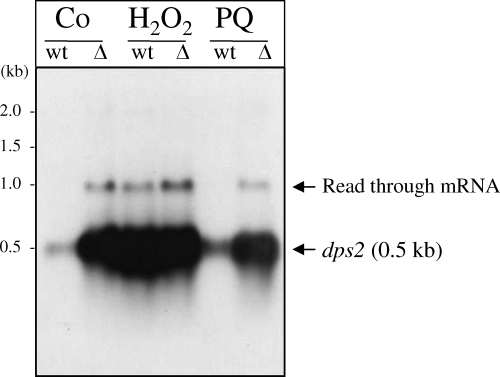
We analyzed the expression of the dps2 gene in response to H2O2 and paraquat by measuring the abundance of the dps2 transcript by Northern blotting. The dps2 gene was transcribed as a monocistronic transcript of ∼0.5 kb. While the expression of dps2 is not changed in response to superoxide stress, the abundance of dps2 mRNAs was significantly increased in response to H2O2 (Fig. 5). The second, very-low-abundance transcript of approximately 1.1 kb is likely to be the result of read-through into the downstream gene, BA5291. PerR is an H2O2- and metal-responsive regulatory protein that regulates the peroxide stress response in Gram-positive bacteria. As dps2 is induced in response to H2O2, we investigated whether its expression was affected in a ΔperR mutant. Northern analysis of the total RNA from the ΔperR mutant showed a high level of dps2 transcription in both the presence and absence of oxidative stress (Fig. 5), which suggests that dps2 expression is negatively regulated by PerR.
Fig 5.
Regulation of dps2 in response to oxidative stress and the role of PerR. Northern blot analysis was performed using 3 μg total RNA of B. anthracis UM23C1-2 (wild type [wt]) and the ΔperR mutant (Δ) using a DIG-labeled probe internal to the dps2 gene fragment. Cells were grown aerobically in Luria-Bertani (LB) broth to an OD540 of 0.3, treated with hydrogen peroxide (H) or paraquat (PQ) to final concentrations of 1 mM and 0.8 mM, respectively, and compared to the untreated control (C). The dps2 mRNA was expressed as a 0.5-kb monocistronic transcript. Expression of dps2 is induced by H2O2 but not by paraquat, while deletion of perR did not significantly affect the expression of dps2 following oxidative stress. Basal levels of the dps2 transcript were significantly increased in the ΔperR mutant, indicating the repression of dps2 expression by PerR. The second, very weak transcript at 1.1 kb is likely to be a read-through mRNA extending into the downstream gene, BA5291.
Primer extension analysis was carried out to map the transcriptional start site of dps2. The analysis showed that dps2 was expressed primarily from transcripts initiated at 49 and 50 nucleotides (AT) upstream of the ATG translation initiation codon (Fig. 6). Analysis of the region upstream of the transcriptional start site revealed the presence of putative −10 (TATAAT) and −35 (TTGACA) boxes, corresponding to Bacillus subtilis σA-dependent consensus promoter sequence (5′ TTGGCA-16/18 bp-TATAAT 3′ [16]). A consensus sequence identical to the well-characterized recognition sites of B. subtilis PerR-regulated genes (TTATAATnATTATAA [12]), with a nucleotide identity of 15/15, was found overlapping the mapped transcriptional start sites and the −10 promoter element (Fig. 6). Taken together, these data indicate that dps2 mRNA is initiated from a single promoter and is a member of the B. anthracis PerR regulon.
Fig 6.
(A) Primer extension analysis of the dps2 promoter. Two adjacent peaks were detected in the primer extension analysis using 50 μg total RNA from B. anthracis UM23C1-2 (wild-type) cells. (B) Nucleotide sequence of the dps2 promoter region. The mapped transcription start sites are in bold and marked by dots, the potential −10 and −35 regions and ribosome-binding site (RBS) of the dps2 promoter are indicated in bold lowercase, and the start (ATG) and stop (TAA) codons for the dps2 translation sequence are underlined. The putative PerR-binding site is boxed. The 23-bp palindromic sequence with a calculated ΔG value of −7.2 kcal mol−1 (IDT OligoAnalyzer 3.1) predicted as the transcriptional terminator for dps2 is as indicated by a dotted line. The FAM-labeled primer for primer extension analysis is shown by an arrow. The primer extension reaction mixtures were loaded in a capillary-based electrophoresis DNA sequencer (Applied Biosystems 3730) and compared with the corresponding sequencing reaction mixtures using the same labeled primer. Results were analyzed with the Peak Scanner (version 1) program (Applied Biosystems).
DISCUSSION
Iron acquisition is essential for bacterial pathogenesis (4). However, due to its propensity to participate in redox chemistry, iron is also potentially toxic to living cells (23, 24). Therefore, concomitant with iron uptake, cells need to avoid toxicity by storing intracellular iron in a less reactive form. It has been suggested that Dps1 and Dps2 of B. anthracis are important for iron binding and sequestration, and overexpression of either Dps1 or Dps2 in E. coli increased its resistance to the presence of excess iron in the growth medium (29). To determine the role of Dps2 in B. anthracis, we constructed a dps2 mutant and analyzed its response to iron excess and deficiency. Growth of the Δdps2 mutant was unaffected by either relative to that of the wild type, suggesting that the primary role of Dps2 is not that of iron storage.
In addition to the Dps-like miniferritins, B. anthracis also encodes the maxiferritin BA5296, and this might explain why Dps2 does not appear to be required for growth under iron-depleted conditions. In Listeria monocytogenes, which does not encode a maxiferritin homologue, the Dps-like protein Fri has been shown to be required for iron storage (28). Dps family proteins are important components of the prokaryotic defense against oxidative stress. In B. subtilis, dpsA, which is a homologue of both dps1 and dps2 of B. anthracis, is required for oxidative stress resistance during stationary growth phase (3). However, despite the presence of a weak putative B. subtilis-like Fur recognition sequence (TGATAATnATTATCA [12]) within the dps1 promoter region (with an identity 11 of 15 nucleotides), this gene is not induced along with other putatively Fur-regulated genes in response to superoxide stress (31).
Dps proteins generally utilize H2O2 as an iron oxidant for their ferroxidase activity, thereby removing H2O2 and reducing the potential for the formation of OH· radicals (8, 14). It has been proposed that Dps proteins also protect DNA against oxidative stress by binding nonspecifically to DNA. For example, the E. coli Dps protein is expressed under both oxidative and nutritional stress conditions, binding to DNA in a nonspecific manner to form stable Dps-DNA complexes that shield the DNA from the resulting oxidative environment (1, 27). Previous studies have shown that B. anthracis Dps1 and Dps2 do not bind to DNA (29). However, despite the lack of interaction, in vitro DNA damage assays clearly demonstrate that both Dps1 and Dps2 protect DNA from oxidative damage, although Dps1 provides complete DNA protection from degradation from a combination of Fe2+ and H2O2, while Dps2 provides only partial protection (26). This is consistent with in vitro data that shows that Dps1 uses both H2O2 and O2 as iron oxidants, with an ∼3-fold preference for the former, whereas Dps2 uses only O2 (26). It is worth emphasizing that the latter observation is exceptional, as other Dps family members generally use H2O2 as the physiological iron oxidant and with an ∼1,000-fold greater efficiency than O2 (8). The physiological relevance of this observation in B. anthracis is not yet clear, particularly since, in contrast to dps2, dps1 is not induced in response to either peroxide or superoxide stress (31).
The B. anthracis Δdps2 mutant has much lower tolerance to H2O2 (Fig. 2A), which suggests a significant role for Dps2 in peroxide stress resistance. The presence of DFM abolishes the sensitivity of the Δdps2 mutant to H2O2 (Fig. 2A and B). DFM is a membrane-permeative iron chelator that interferes with the intracellular free iron pool, but it does not chelate iron from proteins (20, 21). Thus, complementing the in vitro study by Liu et al. (26), in vivo Dps2 is likely to confer protection against H2O2 by inhibiting the iron-mediated production of OH· through sequestration of ferrous iron rather than its detoxification.
The intracellular levels of ROS are detected by regulatory molecules in the cell that trigger a homeostatic response, leading to an increase in the activities of oxidative stress defense systems and a faster removal of the oxidant. B. subtilis also encodes two Dps-like proteins, known as DpsA and MrgA, but with a different patterns of regulation. Liu et al. (26) have argued that B. anthracis Dps1 shows more sequence similarity with B. subtilis MrgA, which is regulated in log phase by H2O2 and PerR (6), while Dps2 shows more sequence similarity with B. subtilis DpsA, which is regulated by the general stress sigma factor, σB, in the stationary phase (3). However, we find that Dps1 and Dps2 are both more closely related to DpsA (62% and 58% identity, respectively) than to MrgA (48% and 59% identity, respectively). Primer extension analysis revealed that dps2 is expressed from a single σA-dependent promoter and includes a consensus PerR-like binding site (Fig. 6). We observed an increase in dps2 expression under H2O2, and its expression is constitutive in the absence of PerR (Fig. 5). Therefore, like that of the B. subtilis homologue mrgA, the expression of B. anthracis dps2 is PerR regulated.
The role, if any, of Dps2 with respect to resistance to endogenous superoxide stress is not yet clear. The link between iron metabolism and superoxide stress is of significance in light of recent evidence that B. anthracis takes up iron following treatment with paraquat (30, 31, 39a). It is recognized that endogenous superoxide stress can lead to oxidative damage by increasing the relative amounts of intracellular free iron (20), and consequently, free cytoplasmic iron may need to be neutralized to avoid iron-mediated oxidative stress. However, the abundance of dps2 mRNA did not change in response to superoxide stress (Fig. 5), while the growth of the Δdps2 mutant was only marginally reduced compared with that of the wild type (Fig. 4). This suggests either that the cytoplasmic free iron levels are not a major factor in the toxicity of endogenous superoxide stress in this organism or that free iron is rapidly sequestrated. Taken together, our results show that Dps2 does not play a significant role in superoxide stress resistance, a view that is consistent with our observation that the abundance of iron in Dps2 is largely unaffected by exposure to paraquat (39a). In the same study, we did not detect significant iron pools associated with either Dps1 or ferritin, nor is there an increase in their gene expression in response to paraquat (30, 31). The importance of these proteins for iron storage and oxidative stress resistance has not been determined, and it remains to be seen of how B. anthracis safely accumulates iron in response to paraquat.
We conclude that B. anthracis Dps2 contributes to peroxide stress resistance by sequestering cellular free iron to prevent the production of OH·. This protective mechanism may be important for virulence, as has been shown for Dps homologues in other intracellular pathogens, including L. monocytogenes (28), Helicobacter pylori (NapA) (36), and Salmonella enterica serovar Typhimurium (15). Further characterization of B. anthracis dps2 to understand the importance of the Dps2 protein in the interaction between this pathogen and its host is necessary.
ACKNOWLEDGMENTS
This study was supported by the EU-funded BaSysBio project LSHG-CT-2006-037469. W.Y.T. was supported by the Overseas Research Student Award Scheme (ORSAS) from the United Kingdom Higher Education Funding Council and the Sarawak Higher Education Scheme, Sarawak Foundation, Malaysia, and K.G. was supported by a studentship from the United Kingdom Biotechnology and Biological Sciences Research Council.
Footnotes
Published ahead of print 9 December 2011
REFERENCES
- 1. Almiron M, Link AJ, Furlong D, Kolter R. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646–2654 [DOI] [PubMed] [Google Scholar]
- 2. Anjem A, Varghese S, Imlay JA. 2009. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 72:844–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antelmann H, et al. 1997. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor σB in Bacillus subtilis. J. Bacteriol. 179:7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown JS, Holden DW. 2002. Iron acquisition by Gram-positive bacterial pathogens. Microbes Infect. 11:1149–1156 [DOI] [PubMed] [Google Scholar]
- 5. Chasteen ND, Harrison PM. 1999. Mineralization in ferritin: an efficient means of iron storage. J. Struct. Biol. 126:182–194 [DOI] [PubMed] [Google Scholar]
- 6. Chen L, Helmann JD. 1995. Bacillus subtilis MrgA is a Dps (PexB) homologue: evidence for metalloregulation of an oxidative-stress gene. Mol. Microbiol. 18:295–300 [DOI] [PubMed] [Google Scholar]
- 7. Chen Y, Tenover FC, Koehler TM. 2004. β-Lactamase gene expression in a penicillin-resistant Bacillus anthracis strain Antimicrob. Agents Chemother. 48:4873–4877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chiancone E. 2008. Dps proteins, an efficient detoxification and DNA protection machinery in the bacterial response to oxidative stress. Rendiconti Lincei 19:261–270 [Google Scholar]
- 9. Dixon TC, Meselson M, Guillemin J, Hanna PC. 1999. Medical progress: anthrax. N. Engl. J. Med. 341:815–826 [DOI] [PubMed] [Google Scholar]
- 10. Eymann C, Homuth G, Scharf C, Hecker M. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Franceschini S, Ceci P, Alaleona F, Chiancone E, Ilari A. 2006. Antioxidant Dps protein from the thermophilic cyanobacterium Thermosynechococcus elongatus. FEBS J. 273:4913–4928 [DOI] [PubMed] [Google Scholar]
- 12. Fuangthong M, Helmann JD. 2003. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J. Bacteriol. 185:6348–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goswami T, Rolfs A, Hediger MA. 2002. Iron transport: emerging roles in health and disease. Biochem. Cell Biol. 80:679–689 [DOI] [PubMed] [Google Scholar]
- 14. Haikarainen T, Papageorgiou AC. 2010. Dps-like proteins: structural and functional insights into a versatile protein family. Cell. Mol. Life Sci. 67:341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. 2004. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect. Immun. 72:1155–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Helmann JD, Moran CP., Jr 2002. RNA polymerase and sigma factors, p 289–312 In Sonenshein AL, Hoch JH, Losick R. (ed), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 17. Hoffmaster AR, Koehler TM. 1997. The anthrax toxin activator gene atxA is associated with CO2-enhanced non-toxin gene expression in Bacillus anthracis. Infect. Immun. 65:3091–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imlay JA. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395–418 [DOI] [PubMed] [Google Scholar]
- 19. Ishikawa T, et al. 2003. The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J. Bacteriol. 185:1010–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keyer K, Imlay JA. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. U. S. A. 93:13635–13640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kicic A, Chua AC, Baker E. 2001. Effect of iron chelators on proliferation and iron uptake in hepatoma cells. Cancer 92:3093–3110 [DOI] [PubMed] [Google Scholar]
- 22. Le Brun NE, et al. 1995. Identification of the ferroxidase centre of Escherichia coli bacterioferritin. Biochem. J. 312:385–392 [PMC free article] [PubMed] [Google Scholar]
- 23. Lewin A, Moore GR, Le Brun NE. 2005. Formation of protein-coated iron minerals. Dalton Trans. 22:3597–3610 [DOI] [PubMed] [Google Scholar]
- 24. Liochev SI, Fridovich I. 2007. The effects of superoxide dismutase on H2O2 formation. Free Radic. Biol. Med. 42:1465–1469 [DOI] [PubMed] [Google Scholar]
- 25. Liochev SI, Fridovich I. 1999. Superoxide and iron: partners in crime. IUBMB Life 48:157–161 [DOI] [PubMed] [Google Scholar]
- 26. Liu X, Kim K, Leighton T, Theil EC. 2006. Paired Bacillus anthracis Dps (mini-ferritin) have different reactivities with peroxide. J. Biol. Chem. 281:27827–27835 [DOI] [PubMed] [Google Scholar]
- 27. Martinez A, Kolter R. 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179:5188–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olsen KN, et al. 2005. The Dps-like protein of Listeria monocytogenes promotes stress tolerance and intracellular multiplication in macrophage-like cells. Microbiology 151:925–933 [DOI] [PubMed] [Google Scholar]
- 29. Papinutto E, et al. 2002. Structure of two iron-binding proteins from Bacillus anthracis. J. Biol. Chem. 277:15093–15098 [DOI] [PubMed] [Google Scholar]
- 30. Passalacqua KD, Bergman NH, Lee JY, Sherman DH, Hanna P. 2007. The global transcriptional responses of Bacillus anthracis Sterne (34F2) and a ΔsodA1 mutant to paraquat reveal metal iron homeostasis imbalances during endogenous superoxide stress. J. Bacteriol. 189:3996–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pohl S, et al. 2011. Combined proteomic and transcriptomic analysis of the response of Bacillus anthracis to oxidative stress. Proteomics 11:3036–3055 [DOI] [PubMed] [Google Scholar]
- 32. Raupach B, Kaufmann SH. 2001. Immune responses to intracellular bacteria. Curr. Opin. Immunol. 13:417–428 [DOI] [PubMed] [Google Scholar]
- 33. Read TD, et al. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81–86 [DOI] [PubMed] [Google Scholar]
- 34. Rooijakkers SH, et al. 2010. Human transferrin confers serum resistance against Bacillus anthracis. J. Biol. Chem. 285:27609–27613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saile E, Koehler TM. 2006. Bacillus anthracis multiplication, persistence, and genetic exchange in the rhizosphere of grass plants. Appl. Environ. Microbiol. 72:3168–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Satin B, et al. 2000. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J. Exp. Med. 191:1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwartz JK, et al. 2010. CD and MCD spectroscopic studies of the two Dps miniferritin proteins from Bacillus anthracis: role of O2 and H2O2 substrates in reactivity of the diiron catalytic centers. Biochemistry 49:10516–10525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Su M, Cavallo S, Stefanini S, Chiancone E, Chasteen ND. 2005. The so-called Listeria innocua ferritin is a Dps protein, iron incorporation, detoxification, and DNA protection properties. Biochemistry 44:5572–5578 [DOI] [PubMed] [Google Scholar]
- 39. Tsou CC, et al. 2008. An iron-binding protein, Dpr, decreases hydrogen peroxide stress and protects Streptococcus pyogenes against multiple stresses. Infect. Immun. 76:4038–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a. Tu WY, et al. 2012. Cellular iron distribution in Bacillus anthracis. J. Bacteriol. 194:932–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang G, Hong Y, Olczak A, Maier SE, Maier RJ. 2006. Dual roles of Helicobacter pylori NapA in inducing and combating oxidative stress. Infect. Immun. 74:6839–6846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao G, et al. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 277:27689–27696 [DOI] [PubMed] [Google Scholar]