Abstract
Although successful iron acquisition by pathogens within a host is a prerequisite for the establishment of infection, surprisingly little is known about the intracellular distribution of iron within bacterial pathogens. We have used a combination of anaerobic native liquid chromatography, inductively coupled plasma mass spectrometry, principal-component analysis, and peptide mass fingerprinting to investigate the cytosolic iron distribution in the pathogen Bacillus anthracis. Our studies identified three of the major iron pools as being associated with the electron transfer protein ferredoxin, the miniferritin Dps2, and the superoxide dismutase (SOD) enzymes SodA1 and SodA2. Although both SOD isozymes were predicted to utilize manganese cofactors, quantification of the metal ions associated with SodA1 and SodA2 in cell extracts established that SodA1 is associated with both manganese and iron, whereas SodA2 is bound exclusively to iron in vivo. These data were confirmed by in vitro assays using recombinant protein preparations, showing that SodA2 is active with an iron cofactor, while SodA1 is cambialistic, i.e., active with manganese or iron. Furthermore, we observe that B. anthracis cells exposed to superoxide stress increase their total iron content more than 2-fold over 60 min, while the manganese and zinc contents are unaffected. Notably, the acquired iron is not localized to the three identified cytosolic iron pools.
INTRODUCTION
Iron is an essential micronutrient for almost all living organisms but can be toxic in excess due to its redox properties, making the control of its import, export, storage, and intracellular trafficking extremely important for the avoidance of cellular damage and the maintenance of viability. However, investigations of the subcellular distribution of iron and iron-proteins in the literature are rare (11, 12, 23), in part due to the technical challenges involved. Knowledge of the identities and quantities of iron-proteins and of the amount of iron with which they are associated under different growth conditions is a key factor in systems analyses of iron homeostasis (20).
Access to iron within the host is a prerequisite for the successful establishment of infection, and iron limitation is a well-characterized element of innate immunity (24, 34). Host systemic iron availability is restricted by the iron chelator transferrin, whose tight affinity for ferric iron is sufficient to inhibit the growth of the pathogen Bacillus anthracis, the causative agent of the mammalian disease anthrax, in human serum (32). Pathogens have evolved a variety of mechanisms to acquire and store sufficient quantities of this essential element to meet their needs, either through improving their ability to compete with the host for available iron or by reducing the host's ability to create iron-limiting conditions within their niche (13, 34).
Infection of a host by B. anthracis is initiated by the engulfment of bacterial spores by macrophages. Spore germination occurs within the maturing phagolysosome (15), an oxidative bactericidal compartment containing abundant superoxide anions (O2·−) produced by the host NOX2 NADPH oxidase, which are implicated in the mechanism of pathogen killing (4). Significantly, germination of B. anthracis spores is stimulated in vitro by O2·− (5). The cytotoxicity of phagosomal reactive oxygen species is augmented by the availability of redox-active metals such as iron and copper (26, 39, 46, 49). Despite the cytotoxicity of the phagolysosome, pathogens such as B. anthracis can persist and even thrive within this compartment, germinating and replicating before ultimately lysing the host cell and releasing vegetative bacteria to initiate the systemic stage of infection (1, 13). This suggests that bacterial defense against oxidative stress plays an important role in the survival of B. anthracis within the host niche.
We have initiated investigations of the intracellular distribution of iron in B. anthracis using established metalloproteomic methods with the aim of identifying the major cytosolic iron pools. Here we show that three of the detected iron pools are associated with ferredoxin, the miniferritin Dps2, and, surprisingly, the two predicted Mn-dependent superoxide dismutase (SOD) isozymes. This observation led us to investigate the functional metal specificity of the B. anthracis SODs. We find that SodA1 is cofactored primarily by manganese but contains a small amount of iron, whereas SodA2 is associated exclusively with iron in vivo, and we further demonstrate that all three of these metalated species are catalytically active in vitro.
A long-term goal in the field is to perform such metalloproteomic investigations using bacterial cells isolated from macrophages or from host organisms, in a manner akin to recent developments in transcriptomic studies (7, 35, 38). However, such studies will require significant technological advancements to achieve the required sensitivity. A first step in this direction involves the characterization of the intracellular iron distribution in cells cultured under standard laboratory conditions and, further, under culture conditions designed to mimic those predicted to be experienced by bacteria within the host environment. To this end, we have investigated the response of B. anthracis to superoxide stress and observed that B. anthracis cells doubled their cellular iron quota within 60 min of exposure. Importantly, this newly acquired iron is not associated with the major identified soluble pools.
MATERIALS AND METHODS
Chemicals, bacterial culture, and cell viability.
Reagents were from Sigma-Aldrich unless otherwise stated. Solutions were prepared in double-deionized water to minimize metal contamination. Bacillus anthracis strain UM23C1-2 was the parental strain (16), here designated wild-type B. anthracis. Escherichia coli strain TOP10 (Invitrogen) was used for cloning. Plasmids pUTE618 and pUTE583 were gifts from T. M. Koehler (University of Texas). Bacteria were cultured in Luria Bertani (LB) broth or brain heart infusion (BHI) (with 5% glycerol) broth (BD, France) at 37°C with vigorous agitation (220 rpm), supplemented with the antibiotic ampicillin (50 μg/ml), erythromycin (Em) (400 μg/ml for E. coli and 5 μg/ml for B. anthracis), kanamycin (Km) (20 μg/ml), or chloramphenicol (6 μg/ml) where necessary.
To monitor growth, overnight cultures were diluted into fresh LB broth without antibiotic to an optical density at 540 nm (OD540) of 0.05. Where required, paraquat (PQ) or 15 mM filter-sterilized desferrioxamine mesylate (DFM) was added to the culture to the indicated final concentration at log phase (OD540 of ∼0.3). Each growth curve was reproduced in three separate experiments on separate days.
For viability assays, dead cells were distinguished from live cells using specific binding of Sytox green nucleic acid stain (Invitrogen), which penetrates cells with compromised cell membranes. Aliquots (100 μl) of culture were mixed with 1 μl of Sytox green solution (0.5 μM final concentration). After a brief incubation, 2 μl of this mixture was mounted onto a microscope slide coated with 1.2% (wt/vol) agarose and immobilized using a 0.13- to 0.17-mm glass coverslip (VWR). Sytox green was excited at 504 nm and measured at 523 nm. Digital images were acquired using a 0.2-s exposure time, and live and dead bacteria were counted and statistically analyzed.
Construction of the B. anthracis ΔsodA1 mutant strain.
The B. anthracis ΔsodA1 mutant strain was created by replacement of the sodA1 coding sequence, open reading frame BA4499, with an omega element conferring Km resistance (Ω-km DNA cassette). B. anthracis chromosomal DNA was extracted with the DNeasy blood and tissue kit (Qiagen). Plasmid DNA was isolated from E. coli using a miniprep kit (Promega). Recombinant DNA techniques were performed using standard procedures (33). DNA sequences (600 to 800 bp) flanking the sodA1 gene were PCR amplified (Platinum Pfx DNA polymerase; Invitrogen) using primer pairs 5′-GCCCCGCGGTCTTCTTTTTGAATAGGTTC-3′ and 5′-CCGCCCGGGTGTTATATTCCTCCCAGTTT-3′ (upstream flank) and 5′-CCGCCCGGGAAAAAGCTAGGAGAATGCT-3′ and 5′-GCCAAGCTTTACGCGAAATTGTGTATCCA-3′ (downstream flank), and the products were digested with SmaI, ligated together with T4 DNA ligase, and then cloned into the pGEM-T vector (Promega). The Ω-km cassette from plasmid pUTE618 was inserted between the flanking fragments using the SmaI site. This construct was cloned into the shuttle vector pUTE583 to create pUTE583::ΔsodA1, which was passaged through E. coli GM48 (dam) to obtain nonmethylated plasmid DNA for electroporation into B. anthracis (18). To isolate a double-crossover recombinant in which sodA1 was replaced with the Ω-km cassette, Em-resistant transformants were grown in BHI medium with Em for 2 days, diluted 1:1,000 every 12 h, and then shifted to BHI medium without antibiotic to facilitate clearance of autonomous plasmids. The culture was diluted 1:1,000 in fresh medium every 12 h for several days and then plated onto BHI agar with Km. Colonies were patch plated to score clones for Km resistance and Em resistance. Chromosomal DNA from the Kmr Ems isolates was purified and verified by PCR using primers 5′-GTTGTATTTGGGTCAAATACG-3′ and 5′-TAGCGAGGGCTTTACTAAGC-3′, which are complementary to sequences upstream of the sodA1 flanking region and within the kanamycin gene, respectively.
RNA extraction and Northern blotting.
Aliquots of cultures (10 ml, representing ∼108 cells) were mixed with an equal volume of RNA stabilization solution (RNAlater; Ambion) and were harvested by centrifugation at 4°C (5,000 × g, 10 min). Total RNA was isolated using a RiboPure bacterial kit (Ambion). RNA purity and integrity were determined by ensuring an A260/A280 ratio of 1.8 to 2.0 and by comparing the 16S and 23S rRNA bands following gel electrophoresis on a formaldehyde denaturing gel (data not shown).
For Northern blotting, samples containing 3 μg of RNA were resolved on a 1.2% (wt/vol) agarose gel containing 6% (wt/vol) formaldehyde and transferred onto a nylon membrane (Hybond-N; Amersham Biosciences). The membranes were fixed by UV treatment and hybridized with digoxigenin (DIG)-labeled RNA probes specific for fer and fdx. Probes were synthesized by in vitro transcription with T7 RNA polymerase and PCR-generated DNA fragments as templates using primer pairs 5′-ATGGCAAAATATACAATCGT-3′ and 5′-ctaatacgactcactatagggagaCTATTCGAATTTTAAAGCGT-3′ for fer and 5′-GCCAAAATTAACAATTGAAG-3′ and 5′-ctaatacgactcactatagggagaAGCATCAAGACCTGAACTTT-3′ for fdx, where lowercase letters indicate a 5′ extension with the T7 promoter sequence for the creation of an antisense RNA probe. Hyperfilm-ECL autoradiography film (Kodak) was used for chemiluminescent detection of the hybridized probe. Equal amounts of RNA prepared from cells exposed to 1 mM H2O2 and 0.8 mM PQ were analyzed in parallel with cultures grown under control conditions.
A spot assay was used to determine the efficiency of the DIG-labeling reaction for the synthesis of DIG-labeled RNA probes specific for fer and fdx. One microliter of each serial dilution of the DIG-labeled transcripts was spotted onto a nylon membrane and fixed to the membrane by UV treatment, followed by chemiluminescent detection. The efficiency of the DIG labeling was determined semiquantitatively by comparison with dilution of a standard DIG-labeled control RNA of known concentration.
Measurement of total cellular metal content.
Aliquots (25 ml) of parallel cultures (control and PQ treated), prepared as described above, were removed immediately before addition of PQ (at an OD540 of ∼0.3) and after 60 min exposure. Cell densities were recorded, and then equal culture volumes were harvested by centrifugation (6,000 × g, 10 min, 4°C) and cells were washed twice with 25 ml of chilled 10 mM HEPES (pH 8.8) containing 1 mM EDTA and then twice with 25 ml of the same buffer without EDTA. Washed pellets were digested in 1 ml of 65% (wt/vol) HNO3 (Merck) and incubated for >48 h at room temperature. The triplicate digested samples were centrifuged (13,000 × g, 20 min), and the supernatants were diluted 1:10 with analytical grade water and analyzed for iron and manganese by inductively coupled plasma mass spectrometry (ICP-MS). Metal concentrations were normalized according to the OD540 recorded for each culture. It should be noted that a small decrease in growth was observed in PQ-treated cultures (see Fig. 5A), reaching an optical density ∼10% lower than that of untreated cultures after 60 min.
Fig 5.
Time-dependent growth and metal accumulation during exposure of B. anthracis to superoxide stress. (A) Growth analysis of B. anthracis cultured in LB medium with (open symbols) or without (closed symbols) addition of 800 μM PQ at an OD540 of ∼0.3 (arrow). (B) Paired cultures of B. anthracis were grown to mid-log phase and aliquots normalized by optical density removed. PQ (800 μM) was added to culture B, with an equivalent volume of water added to culture A as a control, and further incubated for 60 min. After this period, aliquots of each culture were removed. All four samples were digested in 65% HNO3 and analyzed by ICP-MS for manganese and iron.
Anaerobic 2D-LC metal profiling.
Whole-cell protein extracts were prepared from 400-ml cultures, grown in 2-liter flasks with or without 60 min of PQ treatment, harvested, and washed as described above. The cells, suspended in 10 ml of 10 mM HEPES (pH 8.8), were transferred to a 20-ml Teflon disruption vessel (Braun, Germany) and filled with liquid N2. Cells were pulverized with a chromium steel ball (10-mm diameter, 7.85 g ml−1; Braun) using a Mikro-Dismembrator (Braun) (2,600 rpm 3 times for 5 min, with 5-min cooling intervals in liquid N2). The resulting frozen powder was transferred to an N2 chamber (Belle Technologies), and all subsequent steps were conducted under a stringent anaerobic atmosphere (<1 ppm O2). Thawed lysates were clarified by centrifugation (8,000 × g, 20 min, 4°C) followed by ultracentrifugation (100,000 × g, 30 min, 4°C). Soluble protein content was determined using Coomassie Plus reagent (Thermo-Fisher) calibrated with bovine serum albumin (BSA). Cell densities differed slightly between control and PQ-treated cells (see Fig. 5A), and protein yields varied to a similar extent. For each 2D-LC metal profile, a total of 50 mg protein was used. Profiles were repeated in triplicate to ensure reproducibility. Aliquots of the centrifugation pellets were dissolved in 1 ml 65% HNO3 and subjected to ICP-MS analysis. 2D-LC/ICP-MS metal profiling and identification of metalloproteins were conducted as previously described (26, 40).
Determination of metal concentrations by ICP-MS.
Metal concentrations were determined by ICP-MS (Thermo X-series) essentially as previously described (40), monitoring 55Mn, 56Fe (with H2/He collision gas, 3.5 ml min−1), 57Fe (without collision gas), and 66Zn (3 × 100 reads, 20-ms integration time, 5 channels, 0.02-amu separation).
Mass spectrometric identification of proteins.
Proteins were excised from sodium dodecyl sulfate (SDS)-polyacrylamide gels after staining with Sypro Ruby (Molecular Probes) and trypsin digested, and peptides were analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF-MS) using a Voyager DE-STR instrument (Applied Biosystems) (40) or by liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) using an LTQ-FT instrument (Thermo) with NanoACQUITY ultraperformance liquid chromatography (UPLC) (Waters) and a custom column (21). Proteins were identified using peptide mass fingerprint data and the Mascot search engine program (Matrix Science Ltd.), with searching against the latest NCBI nonredundant protein sequence database.
Preparation of apo-SodA1 and apo-SodA2 and in vitro metal reconstitution.
Recombinant proteins SodA1 and SodA2 were gifts provided by M. J. Fogg and A. J. Wilkinson, University of York, United Kingdom (9). On receipt, aliquots were diluted to a 1 μM final concentration in 2% HNO3 and analyzed by ICP-MS. Apo, Mn2+, and Fe2+ forms were prepared using published protocols (37, 47). To obtain apoproteins, native enzymes (1.5 ml at 25 μM) were denatured in 50 mM Tris (pH 7.8)–8 M urea–10 mM EDTA and then renatured by dialysis twice against 1 liter of 50 mM Tris buffer (pH 7.8)–10 mM EDTA. EDTA was removed by dialysis against 1 liter of 50 mM Tris (pH 7.8) overnight at 4°C. The resulting apo-SodA1 and apo-SodA2 were stored at 4°C.
For in vitro iron reconstitution, apo-SodA1 or apo-SodA2 (25 μM in 20 mM Tris, pH 7.8) was incubated overnight anaerobically with 200 μM Fe(II)SO4 · 7H2O (BDH) at room temperature in the presence of 2 mM ascorbate to maintain the reduced iron. For manganese reconstitution of SodA1, the unfolded protein was renatured by dialysis in buffer containing 1 mM MnCl2 and lacking EDTA. Any precipitate formed was removed by centrifugation. Excess metal was removed from metal-reconstituted SODs by gel filtration on Sephadex G-25 (GE Healthcare). Proteins were concentrated with Centricon YM-10 centrifugal filter units (Millipore). The protein concentration was determined using the Coomassie Plus reagent (Thermo-Fisher) with BSA as standard, and bound metals were quantified by ICP-MS. Several unsuccessful attempts were made to generate Mn2+-SodA2, including incubation of apo-SodA2 (25 μM in 20 mM Tris, pH 7.8) with 0.5 mM MnCl2 at room temperature, 37°C, and 50°C or with 0.5 mM MnCl2 and 2 mM ascorbate at room temperature and renaturation of SodA2 in the presence of 10 mM MnCl2 or 0.5 mM MnCl2 and 1 mM dithiothreitol (37). No significant Mn content was detected under any conditions tested after size exclusion chromatography.
SOD activity assays.
SOD activity was determined in gel by staining native polyacrylamide gels (6) or spectrophotometrically with a SOD assay kit according to the manufacturer's instructions (Fluka Analytical, Sigma).
RESULTS
Ferredoxin, Dps2, and two superoxide dismutases represent the most abundant detected cytosolic iron pools.
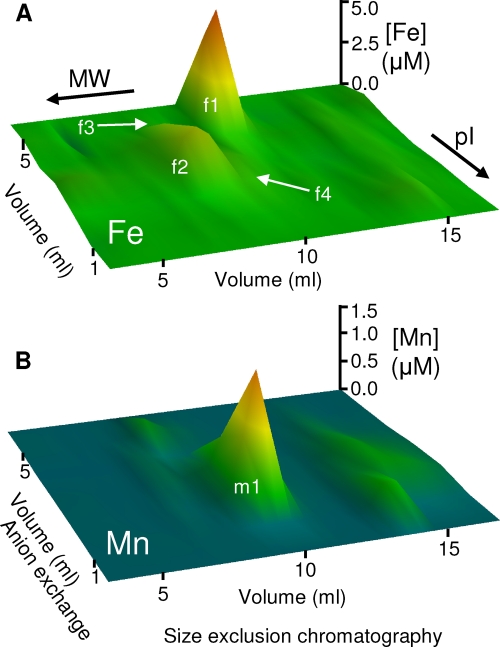
To investigate the cytosolic distribution of iron, we prepared anaerobic cytosolic extracts of B. anthracis by freeze-grinding cells in liquid nitrogen and resolved the resulting soluble extracts by native two-dimensional liquid chromatography (anion-exchange chromatography and size exclusion chromatography) under anaerobic conditions. The resulting fractions were analyzed for metal content by ICP-MS. Several major iron pools were observed in the resulting metal profiles, as well as one major manganese pool (Fig. 1). All of these metal pools eluted from size exclusion chromatography consistent with their being high-molecular-weight (high-MW) complexes. Importantly, no low-MW iron pools were detected, suggesting that putative intracellular low-MW iron chelators, such as the siderophores bacillibactin and petrobactin, are not present in this fraction. Fractions of eluant across each iron peak were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with a fluorescent dye. All protein species observed in the gel were quantified by densitometry and their distribution across the fractions compared with that of the metal by principal-component analysis (PCA) (26, 40).
Fig 1.
Cytosolic metal profiles of wild-type Bacillus anthracis showing the distribution of the major detectable metal pools. Soluble extracts (50 mg total protein) prepared from wild-type B. anthracis cells were resolved by native 2D-LC (anion exchange followed by size exclusion chromatography) under anaerobic conditions. The resulting fractions were analyzed by ICP-MS for iron (A) and manganese (B) abundances. The z axis represents metal concentration, with color scales set at arbitrary levels. Data represent mean values for three independent biological replicates. The four major iron pools (f1 to f4) and one major manganese pool (m1) are labeled, though note that pools f3 and f4 are partially obscured by the more abundant pool f2 at this resolution (see Fig. 2 and 3).
The most abundant iron pool (f1) was eluted from the anion-exchange column by 500 mM NaCl. After resolution of this fraction on a size exclusion column by high-performance liquid chromatography (HPLC), SDS-PAGE analysis (Fig. 2A, left) showed that few proteins were eluted in the fractions that contained iron (Fig. 2B, left). Application of PCA identified the distribution of a single protein species as best matching that of iron (Fig. 2C, left). This protein was excised from the gel and subjected to peptide mass fingerprinting (PMF) by LC-ESI-MS, identifying it as the product of the fer gene (open reading frame [ORF] BA1503; see Tables S1 and S2 in the supplemental material), one of two putative [2Fe-2S] ferredoxin isozymes encoded by the B. anthracis genome (31). It is worth noting that ferredoxin was previously identified as the most abundant detected iron-protein in soluble cytosolic extracts from the cyanobacterium Synechocystis strain PCC 6803 using the same approach (45). No iron pool could be identified as being bound to the alternative ferredoxin, Fdx (ORF BA2790), and Northern blotting failed to detect any significant expression of the fdx gene under these conditions (see Fig. S1 in the supplemental material).
Fig 2.
Identification of metalloproteins that constitute the cytosolic iron pools f1 and f3. (A) To identify the proteins binding the iron in the major detectable cytosolic pools (Fig. 1), aliquots of fractions across the metal peaks f1 (left) and f3 (right) were resolved by SDS-PAGE, visualized with Sypro Ruby, and scanned for fluorescence intensity. Abundances of all proteins in all fractions were estimated by densitometry. Proteins excised for PMF are indicated (arrows). The migration pattern of protein standards is shown on the left. (B) Protein abundance profile (closed symbols), with arbitrary units, of a single protein species (Fer [left] or Dps2 [right]) that most closely correlated with the corresponding iron distribution (open symbols). (C) PCA comparing variations in iron distribution (triangles) of the f1 pool (left) and the f3 pool (right) compared with each protein observed in the fractions (circles). The protein whose distribution most closely matched that of the iron is shown by open symbols.
An abundant iron pool (f2) was eluted from the anion-exchange column in both the 300 and 400 mM NaCl fractions. SDS-PAGE analysis of the distribution of proteins in the iron-containing fractions after resolution by size exclusion chromatography revealed a large number of protein species (data not shown). PCA was unable to identify a single protein species that best matched the distribution of iron across these fractions, and thus the metalloprotein(s) associated with the f2 iron pool is as yet unknown.
A third, low-abundance iron pool (f3) was eluted from the anion-exchange column by 500 mM NaCl. SDS-PAGE analysis of the resulting fractions (Fig. 2A, right) and subsequent PCA (Fig. 2C, right) identified a single prominent protein as the best match (Fig. 2B, right). This band was excised and subjected to PMF by MALDI-TOF-MS, identifying it as the product of the dps2 gene (BA5290; see Table S1 in the supplemental material). Dps2 is a miniferritin that has been shown to bind up to 500 atoms of iron per protein dodecamer in vitro (19). It should be noted that the nomenclature used by Liu and colleagues (19) is the opposite to that used for the genome annotation for Dps proteins in this species (31). This represents the first direct observation of iron binding to B. anthracis Dps2 in cell extracts.
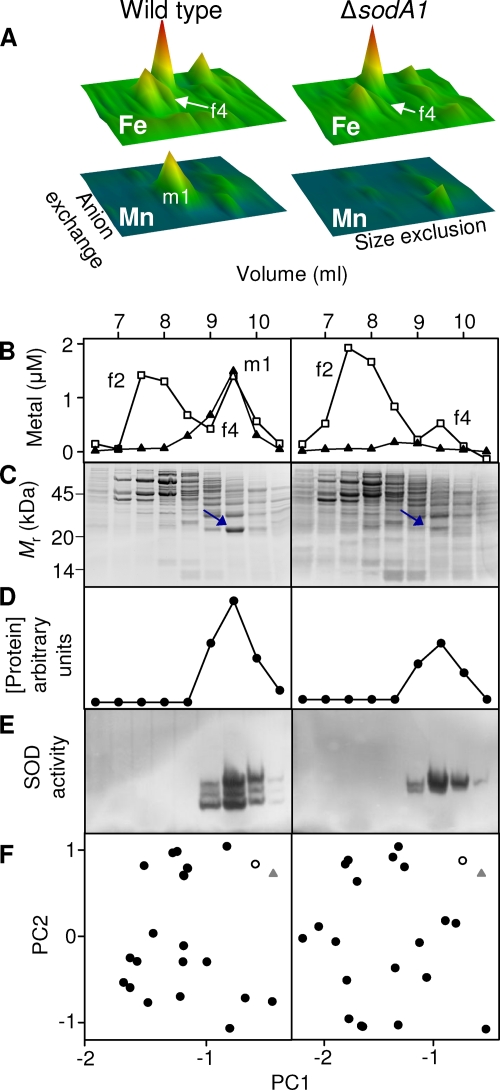
A single major manganese pool (m1) (Fig. 1) was eluted in the 200 mM and 300 mM NaCl fractions from the anion-exchange column (Fig. 3A, left panel). Resolution of the 300 mM NaCl fraction by HPLC size exclusion chromatography demonstrated that this manganese pool (m1) (Fig. 3B, left) comigrated with an equivalent pool of iron (f4) (Fig. 3B, left). Aliquots of these fractions analyzed by SDS-PAGE (Fig. 3C, left) and PCA (Fig. 3F, left) identified a single protein as the best match to both the manganese and iron distributions (Fig. 3D, left). PMF using MALDI-TOF-MS identified this protein as SodA1 (ORF BA4499; see Table S1 in the supplemental material) (predicted MW = 22,664; pI = 5.31), one of four SOD enzymes encoded by the B. anthracis genome (31), which is predicted to utilize Mn2+ as a cofactor (9, 27, 28). A number of unmatched peptides in this sample suggested that a mixture of proteins was present. The mass spectrum was therefore reanalyzed after removal of the peaks that were matched to SodA1, and this revealed the presence of SodA2 (ORF BA5696) (predicted MW = 24,023; pI = 5.40), which is also predicted to utilize Mn2+ as a cofactor (9, 27, 28). The presence of a mixture of SodA1 and SodA2 was confirmed by LC-ESI-MS (see Table S1 in the supplemental material). Aliquots of the same fractions were resolved by native PAGE and stained in gel for the qualitative detection of SOD activity (6). Enzyme activity comigrated with the manganese (m1) and iron (f4) pools (Fig. 3E, left). The triplet pattern of enzyme activity has been observed previously in B. anthracis and was proposed to represent a heterodimeric enzyme formed between SodA1 and SodA2, in addition to the two homodimers (27).
Fig 3.
SodA1 is associated with manganese and iron in vivo, whereas SodA2 is associated only with iron. (A) Cytosolic iron (upper panels) and manganese (lower panels) profiles, analogous to those shown in Fig. 1, from soluble extracts (50 mg total protein) prepared from wild-type (left panels) and ΔsodA1 mutant (right panels) B. anthracis. The data shown here are from single biological replicates, and all axes are identical to those illustrated in Fig. 1. The major manganese pool (m1) is absent in the ΔsodA1 profile. (B) Chromatographs showing comigration of the manganese pool (m1) with an iron pool (f4) on size exclusion chromatography of the 300 mM NaCl anion-exchange fraction in wild-type extract (left) and the absence of the manganese pool (m1) and a decrease in the associated iron pool (f4) in ΔsodA1 extract (right). (C) SDS-PAGE analyses of size exclusion fractions from panel B, stained with Sypro Ruby, as in Fig. 2. The migration pattern of protein standards is shown on the left. (D) Abundance of the single protein species (circles) that most closely correlated with the corresponding manganese distribution (m1) and iron distribution (f4) in fractions from the wild type (left) and with the iron distribution (f4) in fractions from the ΔsodA1 mutant (right). (E) In-gel SOD activity assays using the same fractions indicate the presence of SodA1 and SodA2 in wild-type extracts (left) and the presence of SodA2 in extracts from the ΔsodA1 strain (right). (F) PCA comparing variation in the distribution of iron in f3 (triangles) of wild-type extract (left) or the ΔsodA1 mutant (right) with each protein observed in the gel (circles). The protein whose distribution best matched that of the corresponding metal is shown by open symbols. Although the wild-type data set shown here utilized the iron distribution for PCA, the same protein species was identified as the best match when the manganese distribution was used (data not shown).
Metal occupancy of SOD isozymes in vivo.
The presence of iron (f4) in addition to manganese (m1) in the 2D-LC fractions that exhibit superoxide dismutase activity (Fig. 3) raised the possibility that one or both of these SOD isozymes is at least partly cofactored with iron in vivo, whether functionally active or not. For example, a subpopulation of Mn-SOD has been suggested to be associated with an inactive iron cofactor in E. coli in the absence of oxidative stress (3, 8).
To investigate the metal occupancy of SodA1 and SodA2, we created an isogenic mutant strain of B. anthracis in which the sodA1 gene was replaced with a kanamycin resistance cassette (see Fig. S2A in the supplemental material). Anaerobic soluble extracts prepared from the ΔsodA1 strain were resolved by native 2D-LC and fractions analyzed for iron and manganese by ICP-MS (Fig. 3A, right). Comparison of the results for ΔsodA1 (Fig. 3A, right) and wild-type (Fig. 3A, left) extracts standardized by the amount of total protein loaded (50 mg), showed the complete loss of the manganese pool (m1) in the mutant but only a reduction in the iron pool (f4) (Fig. 3B). Approximately 53% ± 9% of the f4 iron pool was retained in the ΔsodA1 mutant, as calculated using the contributions after size exclusion chromatography from both the 300 mM NaCl (Fig. 3) and 200 mM NaCl anion-exchange fractions, both of which exhibit superoxide dismutase activity (data not shown). Aliquots of fractions obtained from the ΔsodA1 strain after 2D-LC, analyzed by SDS-PAGE (Fig. 3C, right) and PCA (Fig. 3F, right), identified a single band as the best match to the iron distribution. The molecular weight of the protein was consistent with that of SodA2, but the protein could not be identified by PMF, presumably due to its low abundance. However, in-gel activity staining confirmed that the residual SOD activity in the ΔsodA1 strain comigrated with the f3 iron pool (Fig. 3E, right). Taken together, these data demonstrate that the cofactor associated with SodA1 in vivo is predominantly manganese, with only a minor fraction associated with iron, whereas SodA2 is associated exclusively with iron. Furthermore, the SOD activity observed using extracts from the ΔsodA1 mutant (Fig. 3E, right) suggests that SodA2, predicted to be manganese dependent, is catalytically active when cofactored with iron. These results conflict with previous publications predicting that SodA1 and SodA2 were manganese-dependent enzymes (28, 31), and the metal ion bound at the active site in published crystal structures of both enzymes was previously modeled as Mn2+ (9).
The overall abundance of each metal in the m1 and f4 pools in each strain was calculated by integrating the metal abundances under each peak. Assuming that all of the f4 iron pool in the ΔsodA1 mutant represents SodA2-associated iron and that the expression/metalation of SodA2 is unaffected by the sodA1 mutation (27), we can estimate the proportion of the f4 iron pool that is associated with SodA1 in wild-type cells. Subtraction of the ΔsodA1 f4 pool from that in the wild-type strain, across three independent biological replicates, showed that the SodA1 enzyme is cofactored predominantly by manganese, with ∼13% ± 1% iron occupancy.
SodA1 is a cambialistic SOD, whereas SodA2 uses exclusively iron as a cofactor.
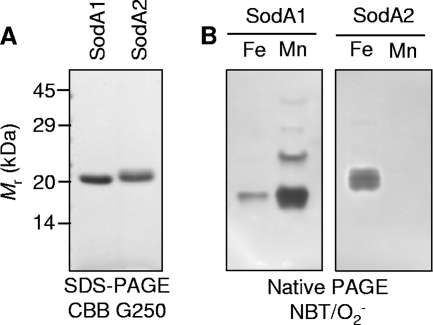
To investigate metal specificity and enzyme activity, samples of purified, recombinant SodA1 and SodA2 (Fig. 4A) previously used for structural determination (9) were analyzed for metal content by ICP-MS (Table 1). Both proteins contained significant amounts of iron, with negligible manganese content, demonstrating that the published crystal structures of both SodA1 and SodA2 (9) likely contain an iron cofactor. In order to determine the metal specificities of the enzymatically active proteins, Mn2+- and Fe2+-loaded forms of each enzyme were prepared by denaturation/renaturation using established protocols (37, 47). In-gel activity analyses (Fig. 4B) and in vitro SOD activity assays (Table 2) demonstrated that both the Mn2+-SodA1 and Fe2+-SodA1 isoforms are enzymatically active, though activity is ∼30-fold greater with Mn2+ than with Fe2+ as a cofactor. The activity of Fe2+-SodA2 is intermediate between those of Fe2+-SodA1 and Mn2+-SodA1. Unexpectedly, despite rigorous attempts to reconstitute SodA2 with Mn2+ using various protocols, we were unable to obtain detectable yields of Mn2+-SodA2, precluding determination of whether SodA2 is enzymatically active with Mn2+. Low-affinity binding of Mn2+ would seem unlikely given that members of this enzyme family typically exhibit kinetically irreversible metal binding (37, 47). Therefore, our data suggest that SodA2 is in fact an iron-dependent SOD, whereas SodA1 is a cambialistic SOD with a physiological and catalytic preference for Mn2+.
Fig 4.
SodA1 is a cambialistic iron- or manganese-dependent enzyme, and SodA2 is an iron-dependent enzyme. (A) SDS-PAGE of purified recombinant SodA1 and SodA2 preparations (2 μg) previously used for crystal structure determinations (9), visualized with Coomassie brilliant blue G-250. The migration pattern of protein standards is shown. (B) In-gel SOD activity assays of recombinant SodA1 (left) and SodA2 (right) proteins (2 μg) after metal reconstitution demonstrated that both Mn2+-SodA1 and Fe2+-SodA1 are enzymatically active. Despite multiple attempts to reconstitute SodA2 with Mn2+, we were unable to obtain any yield of Mn2+-SodA2, with this lane representing resolution of a sample of apo-SodA2 in excess Mn2+.
Table 1.
Metal contents of recombinant SodA1 and SodA2 enzyme preparations used previously (9) for X-ray crystallography
| Sample | mol equivalents |
||
|---|---|---|---|
| Mn | Fe | Zn | |
| SodA1 | 0.02 | 0.34 | 0.15 |
| SodA2 | <LODa | 0.52 | <LOD |
Limit of detection (LOD) (3σ) = 0.003 mol equivalents Mn and 0.002 mol equivalents Zn.
Table 2.
Specific activities and metal contents of recombinant SodA1 and SodA2 enzymes reconstituted with Fe or Mn
| Sample | Sp act (U · mol protein−1) | Metal content (mol equivalents) |
|
|---|---|---|---|
| Fe | Mn | ||
| SodA1-Fe | 4.8 × 104 | 0.65 | <LODa |
| SodA1-Mn | 176 × 104 | 0.17 | 0.75 |
| SodA2-Fe | 59 × 104 | 0.83 | <LOD |
Limit of detection (LOD) (3σ) = 0.002 mol equivalents.
Superoxide stress induces functional iron import systems in B. anthracis.
B. anthracis is predicted to experience significant oxidative stress within the phagolysosome of the host macrophage. Thus, we have been investigating the responses of B. anthracis to such stresses (30, 41). Transcriptomic studies have shown the induction of a number of genes encoding putative iron import and siderophore biosynthetic systems during PQ treatment (28, 30), which contrasts with the response of E. coli, which represses iron transport in favor of manganese import (3). In light of our identification of the major cytosolic iron pools, we investigated whether this induced gene expression gave rise to increased iron import and whether iron occupancy of these known iron pools was altered under superoxide stress.
B. anthracis was cultured to log phase (OD540 of ∼0.3) prior to the addition of 800 μM PQ, and time-dependent growth was monitored by measuring changes in optical density relative to that of untreated control cultures. This concentration was selected because our previous data showed that it perturbs but does not prevent growth of B. anthracis and because it was the condition that we previously used for a combined proteomic and transcriptomic study (30). The cultures treated with PQ showed a mild impairment of growth, consistent with previous data (30), and were unable to achieve the same stationary-phase density as that of untreated cultures (Fig. 5A). Cell viability, as determined using a viability stain, was not affected by PQ treatment (data not shown). Aliquots of parallel cultures, normalized by optical density to achieve approximately equal cell numbers, were removed prior to PQ addition and at intervals after PQ addition, washed, digested in 65% nitric acid, and subjected to elemental analysis by ICP-MS. The iron content of PQ-treated cells steadily increased in a time-dependent manner (data not shown). After 60 min of exposure to PQ, the iron content had increased more than 2-fold, whereas the manganese content was unaffected (Fig. 5B). Untreated cultures showed no increase in cellular metal content (Fig. 5B).
Accumulated iron is localized to insoluble, not cytosolic pools.
Given the observed increase in cellular iron content under superoxide stress, we next sought to quantify cytosolic iron pools in extracts prepared from B. anthracis cells after exposure to PQ to determine if the magnitude of these pools was increased. Anaerobic cytosolic extracts of B. anthracis cells cultured in the presence of 800 μM PQ for 1 h were resolved by native 2D-LC under anaerobic conditions and the resulting fractions analyzed for iron and manganese contents by ICP-MS (Fig. 6). Unexpectedly, none of the soluble iron pools (f1 to f4) were significantly increased in extracts prepared from cultures treated with PQ (Fig. 6), despite the increased cellular iron content under these conditions (Fig. 5B). This suggests that the additional iron accumulated by these cells is in a nonanionic form and/or is associated with the cell membrane/wall. This result was confirmed by elemental analysis of whole cells (Fig. 7A) and subcellular fractions (Fig. 7B) from the cultures used for metal profiling, in which increased iron content was detected predominantly in the insoluble fraction. No significant increase was observed in manganese or zinc content of whole cells or subcellular fractions used as controls (Fig. 7).
Fig 6.
Cytosolic metal profiles of wild-type Bacillus anthracis exposed to superoxide stress. Soluble extracts prepared from wild-type B. anthracis cells after 60 min of exposure to PQ were resolved by native 2D-LC (anion-exchange chromatography followed by size exclusion chromatography) under anaerobic conditions. The resulting fractions were analyzed by ICP-MS for iron (A) and manganese (B) abundances. The z axis represents metal concentration, with color scales set at arbitrary levels. Data represent mean values for three independent biological replicates. All scales and labels are as in Fig. 1.
Fig 7.
Iron accumulated by B. anthracis is localized to insoluble, not cytosolic pools. Whole cells (A) and the insoluble (B) and soluble (C) subcellular fractions of cultures used for cytosolic metal profiles (Fig. 1 and 6) were acid digested and analyzed for metal by ICP-MS. Each data set represents the mean of triplicate samples, with error bars representing standard deviations (SD).
DISCUSSION
Despite an increasing appreciation of the abundance, diversity, and importance of metalloproteins (2, 44, 50), the intracellular distribution of essential metal ions in vivo remains largely unknown, due at least in part to the technical challenges involved in identifying metalloproteins and quantifying their associated metal from complex crude cell extracts (10, 12, 36, 40). We have previously used liquid chromatographic separations of proteins under native conditions followed by PCA and PMF to identify metalloproteins in periplasmic extracts of Synechocystis PCC 6803 (40) and Salmonella enterica serovar Typhimurium (26). Here we have used the same approach to investigate the distribution of metals in cytosolic extracts of the pathogen B. anthracis and to identify the detected cellular iron pools.
We were able to identify three of the major iron pools observed in extracts prepared from wild-type cells, which represent metal associated with Fer, Dps2, and SodA1/SodA2. Only the last of these was surprising; whereas all three belong to known iron-binding protein families, both SodA1 and SodA2 were predicted to be manganese dependent (27, 28), and the metal ions bound in the active site of each protein in published crystal structures were modeled as Mn2+ (9). The SOD enzymes comigrate with an equivalent pool of manganese in extracts from wild-type cells (Fig. 3B, left) but not in extracts from a ΔsodA1 mutant strain (Fig. 3B, right), showing that SodA1 is cofactored predominantly with manganese in vivo, with only minor iron occupancy. In vitro analysis of recombinant protein demonstrates that both of these forms of SodA1 catalyze superoxide dismutation, albeit with ∼30-fold-higher activity using manganese rather than iron (Fig. 4B and Table 2). Conversely, SodA2 is associated exclusively with iron in vivo (Fig. 3), and this form is active in vitro (Fig. 4B and Table 2). Thus, we suggest redefining these enzymes to reflect their respective metal specificities, with SodA1 renamed SodA and SodA2 renamed SodB in accordance with the E. coli nomenclature. Our results highlight the challenge that members of this ubiquitous protein family present to bioinformatics (17, 48). The manganese- and iron-dependent SODs share sequence and structural homology and bind their cofactor using identical ligands within a similar active site (17, 48). Yet, while each enzyme is generally competent to bind the “wrong” metal in vitro (25), enzyme activity is usually specific to the “correct” metal. In E. coli, the manganese-dependent SodA is inactive when associated with Fe2+ (25), whereas the iron-dependent SodB is likewise devoid of activity when associated with Mn2+ (42). Genome annotation is further complicated by the discovery of members of this family that are cambialistic, i.e., display activity with either Mn2+ or Fe2+ cofactor (22, 29). These difficulties emphasize the importance of empirical data to definitively assign metalloprotein cofactors in vivo, particularly but not exclusively those of SOD enzymes.
Our investigations of the response of B. anthracis to oxidative stress (30, 41), which relates to the environment predicted to be experienced within the macrophage phagolysosome, led us to investigate the effects of superoxide stress on iron accumulation and its intracellular distribution. Transcriptomic analyses have previously shown an upregulation of transcripts encoding the siderophore biosynthetic enzymes and putative iron transporters in B. anthracis cells in response to the superoxide-generating reagent paraquat (28, 30). Our data suggest that this transcriptional response gives rise to the synthesis of functional iron import systems, which leads to the hyperaccumulation of iron, with no effect on manganese or zinc content (Fig. 5 and 7). This contrasts with the situation in E. coli, in which exposure to oxidative stress induces uptake of manganese, which is able to substitute for iron in divalent metal-dependent enzymes (3). Importantly, however, the increased cellular iron content of B. anthracis does not significantly affect the soluble iron pools (Fig. 6). Instead, the accumulated iron is associated predominantly with the insoluble fraction after extraction (Fig. 7). The nature of the ligand(s) with which this iron is associated is currently unclear and is a focus of ongoing investigation. It is noteworthy that 4 NEAT domain-containing proteins, which are associated with the cell wall, as well as 25 putative iron-binding or iron-transporting membrane-associated proteins, are transcriptionally induced by PQ treatment (30). One or more of these proteins may represent a reservoir for iron in the insoluble fraction of the cell.
At first sight, the import of iron in response to oxidative stress appears to be counterintuitive, as increased cellular iron would be expected to exacerbate the potential damage through iron-mediated redox cycling. In E. coli, constitutive iron import caused by mutation of the ferric uptake regulator gene fur increases the toxicity of endogenous peroxide, increasing DNA damage (43). It is worth noting that the additional iron accumulated by B. anthracis under superoxide stress conditions leads to only slight growth inhibition after 60 min (Fig. 5A). We previously observed that the regulon controlled by LexA, a global regulator of the DNA damage response, was not induced in B. anthracis by superoxide stress (30). E. coli responds to oxidative stress by importing manganese through the induction of the manganese transporter encoded by mntH (3, 43). Mn2+ is used as a cofactor by the stress-inducible SodA as well as by other divalent metal-binding proteins, sparing bioavailable iron for its essential functions (3). No effect on the manganese content of B. anthracis cells exposed to superoxide stress was observed (Fig. 5B), consistent with the lack of induction of known or putative manganese transporters observed by microarray analysis (28, 30). Despite multiple attempts, we have been unable to generate a strain of B. anthracis in which the putative fur gene (BA4313) is inactivated (data not shown), consistent with a previous report (14), precluding investigation of the role of Fur in the observed iron accumulation.
We propose that the iron uptake observed here relates to the germination and outgrowth of B. anthracis in the phagolysosomal compartment (15). Superoxide stress might represent an environmental signal that triggers an iron accumulation response that facilitates the detoxification of the vacuolar redox insult by removing the iron catalyst of the Fenton reaction and/or aids acquisition and storage of iron for later stages of infection where iron acquisition is known to be limiting (32). The data presented are consistent with a model proposed by Baillie and coworkers (5), whereby superoxide is used by B. anthracis as a signal of phagosomal localization that triggers responses required by B. anthracis for a switch to a pathogenic lifestyle. The determined SOD metal specificities are consistent with an ability to maintain sufficient levels of SOD activity under oxidative stress conditions where iron content is dramatically increased. Though neither SodA1 nor SodA2 is induced in B. anthracis by oxidative stress or iron deficiency in culture (28, 30), it is significant that expression of the iron-dependent SodA2 is upregulated in macrophages (7).
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Fogg and A. Wilkinson, Structural Biology Lab, University of York, United Kingdom, for generously providing recombinant SodA1 and SodA2 proteins.
W.Y.T., S.P., and C.R.H. were supported by the BaSysBio project funded by the European Commission (LSHG-CT-2006-037469). In addition, W.Y.T. was supported by the Overseas Research Student Award Scheme (ORSAS) from United Kingdom higher education funding bodies and by the Sarawak Higher Education Scheme, Sarawak Foundation, Malaysia. K.J.W. and N.J.R. were supported by the BBSRC, United Kingdom (BB/E001688/1). Preliminary experiments were performed by Duncan Harvie, supported by the BBSRC (P18581).
Footnotes
Published ahead of print 16 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Amer AO, Swanson MS. 2002. A phagosome of one's own: a microbial guide to life in the macrophage. Curr. Opin. Microbiol. 5:56–61 [DOI] [PubMed] [Google Scholar]
- 2. Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. 2008. Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorg. Chem. 13:1205–1218 [DOI] [PubMed] [Google Scholar]
- 3. Anjem A, Varghese S, Imlay JA. 2009. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 72:844–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Babior B,. 2004. NADPH oxidase. Curr. Opin. Immunol. 16:42–47 [DOI] [PubMed] [Google Scholar]
- 5. Baillie L, Hibbs S, Tsai P, Cao G-L, Rosen GM. 2005. Role of superoxide in the germination of Bacillus anthracis endospores. FEMS Microbiol. Lett. 245:33–38 [DOI] [PubMed] [Google Scholar]
- 6. Beauchamp C, Fridovich I. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44:276–287 [DOI] [PubMed] [Google Scholar]
- 7. Bergman NH, et al. 2007. Transcriptional profiling of Bacillus anthracis during infection of host macrophages. Infect. Immun. 75:3434–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beyer WF, Fridovich I. 1991. In vivo competition between iron and manganese for occupancy of the active site region of manganese-superoxide dismutase of Escherichia coli. J. Biol. Chem. 266:303–308 [PubMed] [Google Scholar]
- 9. Boucher IW, et al. 2005. Structures of two superoxide dismutases from Bacillus anthracis reveal a novel active centre. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 61:621–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cvetkovic A, et al. 2010. Microbial metalloproteomes are largely uncharacterized. Nature 466:779–782 [DOI] [PubMed] [Google Scholar]
- 11. Expert D, Boughammoura A, Franza T. 2008. Siderophore-controlled iron assimilation in the enterobacterium Erwinia chrysanthemi: evidence for the involvement of bacterioferritin and the Suf iron-sulfur cluster assembly machinery. J. Biol. Chem. 283:36564–36572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferrer M, Golyshina OV, Beloqui A, Golyshin PN, Timmis KN. 2007. The cellular machinery of Ferroplasma acidiphilum is iron-protein-dominated. Nature 445:91–94 [DOI] [PubMed] [Google Scholar]
- 13. Flannagan RS, Cosío G, Grinstein S. 2009. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7:355–366 [DOI] [PubMed] [Google Scholar]
- 14. Gat O, et al. 2008. Characterization of Bacillus anthracis iron-regulated surface determinant (Isd) proteins containing NEAT domains. Mol. Microbiol. 70:983–999 [DOI] [PubMed] [Google Scholar]
- 15. Guidi-Rontani C, Weber-Levy M, Labruyère E, Mock M. 1999. Germination of Bacillus anthracis spores within alveolar macrophages. Mol. Microbiol. 31:9–17 [DOI] [PubMed] [Google Scholar]
- 16. Hoffmaster AR, Koehler TM. 1997. The anthrax toxin activator gene atxA is associated with CO2-enhanced non-toxin gene expression in Bacillus anthracis. Infect. Immun. 65:3091–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jackson SMJ, Cooper JB. 1998. An analysis of structural similarity in the iron and manganese superoxide dismutases based on known structures and sequences. BioMetals 11:159–173 [DOI] [PubMed] [Google Scholar]
- 18. Koehler TM, Dai Z, Kaufman-Yarbray M. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one or two promoters. J. Bacteriol. 176:586–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu X, Kim K, Leighton T, Theil EC. 2006. Paired Bacillus anthracis Dps (mini-ferritin) have different reactivities with peroxide. J. Biol. Chem. 281:27827–27835 [DOI] [PubMed] [Google Scholar]
- 20. Lopes TJS, et al. 2010. Systems analysis of iron metabolism: the networks of iron pools and fluxes. BMC Syst. Biol. 4:112–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malik R, et al. 2009. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J. Proteome Res. 9:4553–4563 [DOI] [PubMed] [Google Scholar]
- 22. Martin ME, et al. 1986. A Streptococcus mutans superoxide dismutase that is active with either manganese or iron as a cofactor. J. Biol. Chem. 261:9361–9367 [PubMed] [Google Scholar]
- 23. Miao R, Holmes-Hampton GP, Lindahl PA. 2011. Biophysical investigation of the iron in Aft1-1(up) and Gal-YAH1 Saccharomyces cerevisiae. Biochemistry 50:2660–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ong ST, Ho JZS, Ho B, Ding JL. 2006. Iron-withholding strategy in innate immunity. Immunobiology 211:295–314 [DOI] [PubMed] [Google Scholar]
- 25. Ose DE, Fridovich I. 1976. Superoxide dismutase. Reversible removal of manganese and its substitution by cobalt, nickel or zinc. J. Biol. Chem. 251:1217–1218 [PubMed] [Google Scholar]
- 26. Osman D, et al. 2010. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 285:25259–25268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Passalacqua KD, Bergman NH, Herring-Palmer A, Hanna P. 2006. The superoxide dismutases of Bacillus anthracis do not cooperatively protect against endogenous superoxide stress. J. Bacteriol. 188:3837–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Passalacqua KD, Bergman NH, Lee JY, Sherman DH, Hanna PC. 2007. The global transcriptional responses of Bacillus anthracis Sterne (34F2) and a delta sodA1 mutant to paraquat reveal metal ion homeostasis imbalances during endogenous superoxide stress. J. Bacteriol. 189:3996–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pennington CD, Gregory EM. 1986. Isolation and reconstitution of iron- and manganese-containing superoxide dismutases from Bacteroides thetaiotamicron. J. Bacteriol. 166:528–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pohl S, et al. 2011. Combined proteomic and transcriptomic analysis of the response of Bacillus anthracis to oxidative stress. Proteomics 11:3036–3055 [DOI] [PubMed] [Google Scholar]
- 31. Read TD, et al. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81–86 [DOI] [PubMed] [Google Scholar]
- 32. Rooijakkers SHM, et al. 2010. Human transferrin confers serum resistance against Bacillus anthracis. J. Biol. Chem. 285:27609–27613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sambrook J, Russell DW. 2000. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34. Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946–953 [DOI] [PubMed] [Google Scholar]
- 35. Schnappinger D, et al. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sevcenco AM, et al. 2009. Development of a generic approach to native metalloproteomics: application to the quantitative identification of soluble copper proteins in Escherichia coli. J. Biol. Inorg. Chem. 14:631–640 [DOI] [PubMed] [Google Scholar]
- 37. Tabares LC, Bittel C, Carrillo N, Bortolotti A, Cortez N. 2003. The single superoxide dismutase of Rhodobacter capsulatus is a cambialistic, manganese-containing enzyme. J. Bacteriol. 185:3223–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Talaat AM, Lyons R, Howard ST, Johnston SA. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. U. S. A. 101:4602–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taylor CM, Osman D, Cavet JS. 2009. Differential expression from two iron-responsive promoters in Salmonella enterica serovar Typhimurium reveals the presence of iron in macrophage-phagosomes. Microb. Pathog. 46:114–118 [DOI] [PubMed] [Google Scholar]
- 40. Tottey S, et al. 2008. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature 455:1138–1142 [DOI] [PubMed] [Google Scholar]
- 41. Tu WY, Pohl S, Gizynski K, Harwood CR. 2012. The iron-binding protein Dps2 confers peroxide stress resistance on Bacillus anthracis. J. Bacteriol. 194:925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vance CK, Miller AF. 2001. Novel insights into the basis for Escherichia coli superoxide dismutase's metal ion specificity from Mn-substituted FeSOD and its very high Em. Biochemistry 40:13079–13087 [DOI] [PubMed] [Google Scholar]
- 43. Varghese S, Wu A, Park S, Imlay KRC, Imlay JA. 2007. Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol. Microbiol. 64:822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Waldron KJ, Rutherford JC, Ford D, Robinson NJ. 2009. Metalloproteins and metal sensing. Nature 460:823–830 [DOI] [PubMed] [Google Scholar]
- 45. Waldron KJ, Tottey S, Yanagisawa S, Dennison C, Robinson NJ. 2007. A periplasmic iron-binding protein contributes to inward copper supply. J. Biol. Chem. 282:3837–3846 [DOI] [PubMed] [Google Scholar]
- 46. White C, Lee J, Kambe T, Fritsche K, Petris MJ. 2009. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 284:33949–33956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Whittaker MM, Whittaker JW. 1999. Thermally triggered metal binding by recombinant Thermus thermophilus manganese superoxide dismutase, expressed as the apo-enzyme. J. Biol. Chem. 274:34751–34757 [DOI] [PubMed] [Google Scholar]
- 48. Wintjens R, et al. 2004. Specificity and phonetic relationships of iron- and manganese-containing superoxide dismutases on the basis of structure and sequence comparisons. J. Biol. Chem. 279:9248–9254 [DOI] [PubMed] [Google Scholar]
- 49. Wolschendorf F, et al. 2011. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 108:1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Y, Gladyshev VN. 2011. Comparative genomics of trace element dependence in biology. J. Biol. Chem. 286:23623–23629 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.