Abstract
Spx is a global regulator that is widespread among the low-G+C-content Gram-positive bacteria. Spx has been extensively studied in Bacillus subtilis, where it acts as an activator and a repressor of transcription in response to disulfide stress. Under nonstress conditions, Spx is rapidly degraded by the ClpXP protease. This degradation is enhanced by the YjbH adaptor protein. Upon disulfide stress, the amount of Spx rapidly increases due to a decrease in degradation. In the opportunistic pathogen Staphylococcus aureus, Spx is a global regulator influencing growth, biofilm formation, and general stress protection, and cells lacking the spx gene exhibit poor growth also under nonstress conditions. To investigate the mechanism by which the activity of Spx is regulated, we identified a homolog in S. aureus of the B. subtilis yjbH gene. The gene encodes a protein that shows approximately 30% sequence identity to YjbH of B. subtilis. Heterologous expression of S. aureus yjbH in a B. subtilis yjbH mutant restored Spx to wild-type levels both under nonstress conditions and under conditions of disulfide stress. From these studies, we conclude that the two YjbH homologues have a conserved physiological function. Accordingly, inactivation of yjbH in S. aureus increased the level of Spx protein and transcription of the Spx-regulated gene trxB. Notably, the yjbH mutant exhibited reduced growth and increased pigmentation, and both phenotypes were reversed by complementation of the yjbH gene.
INTRODUCTION
All aerobic organisms encounter oxidative stress and have evolved different ways to reduce stress-induced damage. Oxidative stress can lead to the formation of unwanted disulfide bonds, a phenomenon known as disulfide stress (2). Spx is a transcriptional regulator that has been thoroughly studied in Bacillus subtilis, where it acts both as an activator and a repressor of transcription in response to disulfide stress by interacting with the C-terminal domain of the RNA polymerase α subunit (31, 42). Among the genes that are repressed by Spx are many that are involved in cellular metabolism during normal growth, such as biosynthesis of amino acids, vitamins, purines, and pyrimidines, while some of the induced genes are involved in maintenance of the cellular thiol-redox homeostasis (29). By inducing the Spx regulon, the cell can repair damage caused by disulfide stress and return the cytoplasm to its normal reducing state, while not spending energy on the biosynthesis of cellular components.
The nature of the Spx regulon, where several genes needed for vegetative growth are repressed, calls for a tight regulation of Spx. Under nonstress conditions, Spx is kept at a very low concentration by the ClpXP protease, and upon disulfide stress, there is a decrease in degradation, thereby increasing the amount of Spx in the cell (29, 42). ClpXP is a multisubunit protease complex, where ClpP acts as a protease that works together with the AAA+ ClpX unfoldase to degrade misfolded and truncated proteins (8, 13). Substrate specificity is provided by ClpX (3, 6, 13), which utilizes ATP for unfolding and translocation of the substrate into the ClpP proteolytic chamber (13). Mutations in B. subtilis clpP and clpX give pleiotropic phenotypes with respect to stress tolerance, competence for DNA uptake, high-temperature tolerance, sporulation, morphology, and motility, and the clpP and clpX mutants display an extended lag phase (10, 19). Originally, the spx gene was discovered as the site for mutations that could suppress the pleiotropic phenotype of a clpXP mutation in B. subtilis (suppressor of clpP and clpX) (27). It was demonstrated that the accumulation of Spx in the clpXP mutants is what causes the observed pleiotropic phenotype (30). A similar phenotype is also seen in a strain lacking the yjbH gene, which encodes a 34-kDa cytosolic protein that acts as an adaptor protein to enhance Spx degradation by binding to Spx, thereby making it more available for ClpX recognition (9, 21). YjbH has not been structurally characterized; however, bioinformatics analysis predicts that it is a member of the thioredoxin-like superfamily. It is not fully understood how the adaptor activity of YjbH is regulated. However, recently is has been reported that in B. subtilis the 54-amino-acid protein YirB may function as an antiadaptor protein (18). It is suggested that YirB functions by interacting with YjbH, causing a subsequent release of Spx from YjbH (18). However, it is not known under which conditions YirB might be functional. Moreover, YirB is not conserved in Firmicutes and it is, for example, not present in Staphylococcus, suggesting that alternative mechanisms are used to control the activity of YjbH. In addition to the proteolytic control, the activity of Spx is regulated by a disulfide redox switch involving a CXXC present in Spx that affects the interaction between Spx and the RNA polymerase (28).
Homologs of Spx are widespread among the low-G+C-content Gram-positive bacteria (17, 39, 42). In the opportunistic pathogen Staphylococcus aureus, Spx has been identified as a global regulator influencing growth, biofilm formation, and general stress protection (34). A proteomic analysis comparing S. aureus wild-type and spx mutant cells indicated that Spx acts both as a negative and a positive regulator of genes encoding proteins involved in DNA metabolism, protein synthesis, cell division, and thiol homeostasis. An spx mutation causes severely impaired growth in S. aureus, and it has been shown that the transcription of the essential trxB gene, encoding thioredoxin reductase, is virtually undetectable in an spx mutant, possibly causing the growth defect of the mutant. The spx mutant is hypersensitive to a variety of stresses, including high and low temperatures as well as oxidative and disulfide stress (34). The B. subtilis and S. aureus Spx proteins share 79% sequence identity (see Fig. S1 in the supplemental material), and Spx was suggested to be a substrate of ClpXP in S. aureus, because Spx accumulates strongly in clpP and clpX mutants despite that transcription of the gene is reduced (34).
In B. subtilis, the efficient degradation of Spx by ClpXP under nonstress conditions requires the Spx adaptor protein YjbH (21). In S. aureus, a gene encoding a protein that exhibits some similarity (∼30% identity) to B. subtilis YjbH (hypothetical protein SAOUHSC_00938) is present, here referred to as S. aureus YjbH (Fig. 1). In B. subtilis, yjbH is located in an operon together with the yjbI gene, encoding a truncated hemoglobin (35). The S. aureus yjbH gene is located downstream of a gene encoding a protein with a high sequence similarity (∼47% identity) to B. subtilis YjbI, which has been experimentally shown to encode a truncated hemoglobin (P. Kjelgaard and C. von Wachenfeldt, unpublished data). This chromosomal arrangement suggests that S. aureus YjbH may have a role similar to YjbH in B. subtilis. Interestingly, global mutagenesis screens designed to identify virulence genes in S. aureus and Listeria monocytogenes have identified transposon insertions in the yjbH gene (14, 37, 41). However, the connection to virulence remains to be clarified.
Fig 1.
Alignment of YjbH from B. subtilis and S. aureus. White letters on black background represent identical residues. The asterisks indicate the cysteine residues of the CXXC motif in B. subtilis YjbH. Cysteine residues are encircled.
The low overall conservation of YjbH, and the finding that the N-terminal redox-sensitive region of B. subtilis YjbH is absent altogether in S. aureus YjbH, prompted us to investigate if the function of YjbH is conserved between the two organisms. Heterologous expression of S. aureus yjbH in a B. subtilis yjbH mutant restored Spx levels to wild-type levels both under nonstress conditions and under conditions of disulfide stress. From these studies, we conclude that the two YjbH homologues have a conserved physiological function. Accordingly, inactivation of yjbH in S. aureus increased the level of Spx protein and transcription of the Spx-regulated gene trxB. Notably, the yjbH mutant exhibited reduced growth and increased pigmentation, and both phenotypes were reversed by complementation of the yjbH gene.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains used in this study are listed in Table 1. The plasmids used in this work are listed in Table 2. Escherichia coli strain TOP10 was used for construction and maintenance of plasmids and was grown in LB or on LB agar plates (36). S. aureus strains were grown in tryptic soy broth (TSB) or on tryptic soy agar (TSA). B. subtilis strains were grown on tryptose blood agar base (TBAB) or in nutrient sporulation medium with phosphate supplemented with 0.5% glucose (NSMPG) (40). All bacterial cultures were grown at 37°C with shaking at 200 rpm. The following antibiotics were added to the growth medium when required: kanamycin (50 μg ml−1), ampicillin (100 μg ml−1), spectinomycin (150 μg ml−1), and erythromycin (100 μg ml−1) for E. coli strains; spectinomycin (150 μg ml−1), erythromycin (5 μg ml−1), chloramphenicol (20 μg ml−1), and tetracycline (4 μg ml−1) for S. aureus strains; spectinomycin (150 μg ml−1), chloramphenicol (15 μg ml−1), tetracycline (20 μg ml−1), and a mixture of erythromycin (0.5 μg ml−1) and lincomycin (12.5 μg ml−1) for B. subtilis strains. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added to solid medium (50 μg ml−1) as an indicator of β-galactosidase activity. When appropriate, isopropyl-β-d-thiogalactoside (IPTG) was added to a final concentration of 100 μg ml−1.
Table 1.
Bacterial strains used in this study
| Strain | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| S. aureus | ||
| NCTC 8325-4 | Wild type, derivative of NCTC 8325 cured of all known prophages | 33 |
| RN4220 | Restriction-deficient derivative of NCTC 8325-4 | 33 |
| 8325-4 Δspx mutant | Δspx | 34 |
| 8325-4 ΔclpX mutant | ΔclpX | 7 |
| 8325-4 ΔclpP mutant | ΔclpP | 7 |
| Newman | Human clinical isolate | 5 |
| LUSA1 | NCTC 8325-4 ΔyjbH::spc, Spr | This study |
| LUSA2 | Newman ΔyjbH::spc, Spr | This study |
| LUSA3 | RN4220 pCL25, Tcr | This study |
| LUSA4 | RN4220 pCL25_yjbIH, Tcr | This study |
| LUSA7 | Newman pCL25, Tcr | This study |
| LUSA8 | Newman pCL25_yjbIH, Tcr | This study |
| LUSA9 | Newman pCL25 ΔyjbH::spc, Spr Tcr | This study |
| LUSA10 | Newman pCL25_yjbIH ΔyjbH::spc, Spr Tcr | This study |
| B. subtilis | ||
| 1A1 | trpC2 | BGSCb |
| LUW272 | ΔyjbH::spc, Spr | 35 |
| LUW297 | spxΩpMUTIN2, Emr | 21 |
| LUW362 | spxΩpMUTIN2 amyE::spxBs spc, Emr Spr | This study |
| LUW400 | ΔyjbH::spc amyE::cat, Cmr Spr | This study |
| LUW428 | spxΩpMUTIN2 amyE::spxSA cat, Emr Cmr | This study |
| LUW448 | ΔyjbH::spc amyE::yjbHSA cat, Spr Cmr | This study |
| LUW421 | ΔyjbH::spc amyE::yjbHBS cat, Spr Cmr | This study |
| LUW442 | ΔyjbH::spc spxΩpMUTIN2 amyE::spxSA tet/pCW7_ yjbHSA, Spr Cmr Tcr Emr | This study |
| LUW458 | ΔyjbH::spc amyE::yjbHBS-Cys cat, Spr Cmr | This study |
| LUW454 | ΔyjbH::spc amyE::pCW101_ yjbHSA, Cys-free, Spr Cmr | This study |
All B. subtilis LUW strains are derivatives of 1A1 and thus carry the trpC2 auxotrophic marker. Apr, ampicillin resistance; Emr, erythromycin resistance; Kmr, kanamycin resistance; Spr, spectinomycin resistance; Cmr, chloramphenicol resistance; Tcr, tetracycline resistance.
BGSC, Bacillus Genetic Stock Center, Department of Biochemistry, Ohio State University, Columbus, OH.
Table 2.
Plasmids used in this study
| Plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| pCRBlunt II TOPO | Cloning vector; Kmr | Invitrogen |
| pUC18 | Cloning vector; Apr | 32 |
| pUC18_yjbIHSA | Apr | This study |
| pUC18_yjbIHSA spc | Apr Spr | This study |
| pDG1727 | Integration vector for B. subtilis; Spr Apr | 15 |
| pMAD | Temperature-sensitive S. aureus shuttle vector with the bgaB gene encoding a β-galactosidase; Apr Err | 1 |
| pMAD_yjbIHSA spc | Apr Err Spr | This study |
| pRMC2 | Apr Cmr | 4 |
| pDG1661 | Cmr | 15 |
| pDR111 | Insertional vector for IPTG-inducible expression from B. subtilis amyE locus; lacI; Apr Spr; Phyperspank | D. Rudner |
| pRMC2_yjbHSA | Cmr | This study |
| pCL25 | Tcr | 23 |
| pCL25_yjbIHSA | Tcr | This study |
| pCW7 | Cmr | 35 |
| pCW7_yjbHSA | Cmr | 35 |
| pCW101 | Insertional vector for IPTG-inducible expression from B. subtilis amyE locus; lacI; Apr Cmr; Phyperspank | This study |
| pCW101_yjbHBS | Cmr | This study |
| pCW101_yjbHSA | Cmr | This study |
| pCW101_yjbHBS-Cys | Cmr | This study |
| pCW101_yjbHSA-Cys | Cmr | This study |
| pDR111_spxBS | Cmr | This study |
| pCW101_spxSA | Cmr | This study |
| pCm::Tc | Tcr | 38 |
Apr, Cmr, Emr, Kmr, Spr, and Tcr indicate resistance to chloramphenicol, erythromycin, kanamycin, spectinomycin, and tetracycline, respectively.
To alter the cellular thiol redox balance, S. aureus or B. subtilis cells were treated with diamide [diazenedicarboxylic acid bis(N,N-dimethylamide)(CH3)2NCON = NCON(CH3)2] (5 mM [S. aureus] or 1 mM [B. subtilis]) at mid-exponential growth phase, and cells were harvested immediately before and 20 min after addition, unless stated otherwise.
Expression from the tetracycline-inducible promoter in pRMC2 and derived plasmids was induced by the addition of between 100 and 500 ng ml−1 anhydrotetracycline (Atet).
DNA manipulations and transformations.
All molecular biology techniques, including E. coli transformations, were performed as described by Sambrook and Russell (36). Preparations of B. subtilis chromosomal DNA and B. subtilis transformations with plasmid or chromosomal DNA were performed as described by Hoch (16). All oligonucleotides used in this work are described in Table 3. All cloned PCR-generated fragments were verified by sequencing.
Table 3.
Oligonucleotide sequences
| Oligonucleotide | Sequence (5′→3′) |
|---|---|
| SA2 | CTGCACTACGCATAAGAGTTAAAG |
| SA3 | CCGGATCCACACTTCTATATGAATTATTATG |
| SASPX1 | CCTCTAGATGGTAACATTATTTACTTCACC |
| SASPX2 | CCGTCGACACATAGTTAAATGGTTATTAG |
| SAYJBH1 | GGGGATCCATTAACGCCAACTAGAATG |
| SAYJBH2 | CGCATGCTTAATCCTCCTCT |
| SAYJBH3 | TCTAGATGGCTGGAGAATTACGAATAATG |
| SAYJBH4 | GTCGACTTATTTTGATTTGATTTTAGGCAT |
| SAYJBH10 | GGTACCAAAGGAGGTAAAGATGTATGGCTGGAGAATTACGAATAATGG |
| SAYJBH11 | TACGTTCTAATAATTTAATGTTGC |
| SCV1 | GCAACACCACATAATGGTTCAC |
| SCV8 | GCACATAATTGCTCACAGCCA |
| SPC2 | CGTATGTATTCAAATATATCCTCC |
| SPC10 | GCCGTATGATTTTAACTATGGACAC |
| NW-spxA-RT-fw | AAATGACTGAAGACGGTACTGATG |
| NW-spxA-RT-rev | CGTTGTGCTTCTTGTAATTGG |
| NW-trxB-RT-fw | AAGACGGCAAAGTGGGTTC |
| NW-trxB-RT-rev | AACATCTCCTGCTGCAAAAATAC |
| NW-yjbI-RT-fw | GCAGAAACAAGTCGTAAACAAAAAC |
| NW-yjbI-RT-rev | CTCCAACACCTTGTGGAAAC |
| NW-yjbH-RT-fw | AAGCCCCTTCTCTCGTTTTC |
| NW-yjbH-RT-rev | TTTAAAAGTTTTTCTGGCCATTC |
Construction of plasmids.
To create pCW101, a derivative of the Phyperspank plasmid pDR111, plasmid pDG1661, was digested with SphI and treated with exonuclease Bal-31 to remove the SphI and SalI restriction sites, and finally the plasmid was recircularized with T4 DNA ligase. After propagation in E. coli, the resulting plasmid was isolated, and a fragment containing the chloramphenicol resistance gene and amyE was excised from the plasmid using the restriction enzymes EcoRI and PstI and ligated into pDR111 digested with the same enzymes. In the resulting plasmid, pCW101, the spectinomycin resistance gene of pDR111 was replaced by the chloramphenicol resistance gene of pDG1661.
The spxA gene was amplified by PCR from chromosomal DNA from S. aureus 8325-4, using primers SASPX1 and SASPX2. The PCR product was cloned into pCR-BLUNT II-TOPO (Invitrogen). The sequence of the cloned fragment was verified by sequencing and confirmed to have 100% sequence identity to the expected sequence. The insert was excised using XbaI and SalI and cloned into pCW7. The resulting plasmid was named pCW7_spxSA.
To create pCW101_spxSA, the spxSA fragment including a B. subtilis ribosome-binding site from pCW7_spxSA was excised using HindIII and SalI and cloned into pCW101. Plasmid pCW7_yjbHSA was created in the same manner as pCW7_spxSA except that the primers SAYJBH3 and SAYJBH4 were used for the PCR amplification. Plasmid pCW101_YjbHBS was created by excising the yjbH fragment from pCW7_YjbHBS with XbaI and HindIII and ligating it into pCW101 cut with the same enzymes.
Genes encoding cysteine-free variants of yjbH (yjbHSA-Cys and yjbHBS-Cys) were synthesized by Eurofins MWG Operon, and the fragments were cloned into pCW101 using HindIII, creating pCW101_yjbHSA-Cys and pCW101_yjbHBS-Cys, respectively. For complementation in S. aureus, the S. aureus yjbH gene (SAOUHSC_00938) was amplified by PCR from S. aureus 8325-4 chromosomal DNA using primers SAYJBH10 and SAYJBH11. The resulting fragment containing the yjbH gene and a ribosome binding site was isolated, cut (KpnI-EcoRI), and cloned into the KpnI-EcoRI sites of the expression vector pRMC2. This plasmid allows tetracycline-inducible expression of genes that are cloned downstream of a TetR-controlled promoter (4). The plasmid (pRMC2_yjbHSA) was amplified and purified from E. coli. The plasmid was then transformed into the restriction-negative strain S. aureus RN4220, isolated, and subsequently transformed into S. aureus Newman by electroporation.
Construction of B. subtilis strains.
Strain LUW428 was generated by transformation of LUW297 (spxΩery) with pCW101_spxSA and selection of transformants with chloramphenicol. The successful integration of the S. aureus spx gene into the amyE locus of the transformants was confirmed by the lack of halo formation upon growth on plates containing 1% starch with subsequent exposure of the plates to iodine.
Strain LUW421 was generated by transforming the wild-type strain 1A1 first with pCW101_yjbHBS and by subsequent transformation of the resulting strain with chromosomal DNA of strain LUW272 (ΔyjbH::spc). Confirmation of an AmyE-negative phenotype was done as described above. It should be noted that the deletion of yjbH severely affects competence development (21). Therefore, the introduction of the yjbH mutation was the final step in the strain constructions.
LUW448, LUW454, and LUW458 were created in the same manner as LUW421 with the exception that pCW101_yjbHSA, pCW101_yjbHSA-Cys, or pCW101_yjbHBS-Cys was used instead of pCW101_yjbHBS.
To create the B. subtilis strain LUW442, in which the yjbH and spx genes are replaced with their S. aureus counterparts, the marker for LUW428 was first switched from chloramphenicol to tetracycline by using plasmid pCm::Tc. The resulting strain was then transformed with pCW7_yjbHSA, selecting for chloramphenicol resistance. Finally, the yjbH mutation was introduced by transformation with chromosomal DNA from the ΔyjbH strain LUW272.
Diamide sensitivity assay.
Disc diffusion sensitivity assays were conducted by streaking a TBAB plate with a solution containing 1 × 108 cells/ml and then placing a 6-mm filter disc (AA discs; Whatman) containing 10 μl of 25 mM diamide onto it. Plates were incubated at 37°C for 18 h, and the zone of clearing around the discs was measured.
Pigment isolation.
Isolation and quantification of S. aureus carotenoid pigments were done according to Morikawa et al. (26).
Construction of an S. aureus yjbH deletion mutant.
The yjbIH region was amplified by PCR from S. aureus 8325-4 chromosomal DNA using primers SA2 and SA3 (Table 3). The fragment was cloned into pCR-BLUNT II-TOPO (Invitrogen). Transformants were selected for on kanamycin-containing plates. The resulting plasmid was cut with SalI and BamHI, and the 2,373-bp fragment containing the yjbIH region was ligated into pUC18 cut with the same enzymes and transformed into E. coli selecting for ampicillin resistance. The resulting plasmid was named pUC18_yjbIHSA (see Fig. S2 in the supplemental material). The spectinomycin resistance gene of pDG1727 (see Fig. S2 in the supplemental material) was excised with PvuII and EcoRV and was ligated into pUC18_yjbIHSA cut with MscI and EcoRV and transformed into E. coli selecting for spectinomycin and ampicillin resistance. The resulting plasmid, pUC18_yjbHSA_spc, was cut with SalI and BamHI, and the 3,117-bp fragment containing the S. aureus yjbIH region where the yjbH gene is partially replaced by the spectinomycin resistance gene was ligated into pMAD, which contains a temperature-sensitive S. aureus origin of replication, cut with SalI and BamHI (see Fig. S2 in the supplemental material). The ligate was used to transform E. coli, selecting for ampicillin resistance. The resulting plasmid was named pMAD_yjbIHSA_spc and was used to transform the restriction-deficient S. aureus strain RN4220 grown at 30°C to erythromycin resistance, selecting for blue colonies on plates containing X-Gal. The plasmid was then extracted and used to transform S. aureus NCTC 8325-4 grown at 30°C to erythromycin resistance, again selecting for blue colonies on plates containing X-Gal. Transformants were grown in TSB supplemented with erythromycin at 30°C for 2 h, after which they were harvested and resuspended in TSB without antibiotics. This culture was grown at 42°C for 6 h in order to allow for a double-crossover recombination event in the chromosomal yjbH locus. Cells were plated on TSA supplemented with spectinomycin and X-Gal and grown at 42°C, selecting for white colonies. The double-crossover recombination event was confirmed by PCR using primer pairs SPC10/SAYJBH1 and SAYJBH2/SPC2 (see Fig. S3 in the supplemental material). The yjbH mutant strain in the strain 8325-4 background, denoted LUSA1, did not show any apparent phenotype separating it from the wild type. One possible reason for this might be that strain 8325-4 lacks a functional copy of rsbU, which impairs signaling by the alternative sigma factor SigB that is involved in the general stress response (11). Because of this, the mutation was moved to S. aureus strain Newman by transduction as described by McNamara and Iandolo (25). The presence of the yjbH::spc mutation in strain Newman was confirmed by PCR as described for strain 8325-4 above.
Complementation of the S. aureus yjbH deletion mutant.
The ΔyjbH mutation was complemented with a chromosomally integrated copy of yjbH expressed from its own promoter (PyjbIH) using the single-copy integration plasmid pCL25 (24). The yjbH gene was excised from pUC18_yjbIHSA and cloned into pCL25 using HindIII and XbaI. The cloned fragment in the resulting plasmid was sequenced to confirm that the sequence was correct. The plasmid was transformed into RN4220 containing the pYL11219 plasmid encoding the integrase from the phage L54a that catalyzes site-specific integration of the plasmid (pCL25_yjbIH) into the attB site located within the geh gene (22). The integrated copy of pCL25_yjbIH was then transduced into the Newman strain using the lysogenic phage ϕ11. The ΔyjbH mutation in strain LUSA1 was then transduced into the strain containing the integrated copy of pCL25_yjbIH. PCR, using the primers SCV1 and SCV8, was used to confirm that the strain contained both the yjbH deletion and the intact copy of yjbH inserted into the geh gene. The strain was named LUSA8. A control strain with pCL25 instead of pCL25_yjbIH was created in the same way and denoted LUSA7.
Immunoblot analysis.
Soluble extracts were prepared by harvesting 25 ml of a culture of exponentially growing S. aureus or B. subtilis cells and dissolving the pellet in 100 mM Tris-HCl and 5 mM Na-EDTA (pH 8.0), supplemented with one tablet of Complete EDTA-free protease inhibitor (Roche). To lyse S. aureus cells, lysostaphin was added to a final concentration of 25 μg ml−1, followed by incubation at 37°C for 30 min, while B. subtilis cells were lysed by sonication. To remove cell debris, the lysates were centrifuged at 20,000 × g for 45 min at 4°C. The protein concentration was determined using the bicinchoninic acid method (Thermo Scientific) with bovine serum albumin as the standard. A total of 20 μg of the extracts was then separated by Laemmli sodium dodecyl sulfate-polyacrylamide gel electrophoresis (18%) and transferred to Hybond P membranes (GE Healthcare). Spx, YjbHBS, and YjbHSA were detected using an antiserum from a rabbit that had been immunized with a synthetic peptide (NHCOCH3)CGYNEDEIRRFLPRKVR(CONH2), (NHCOCH3)MLGDEPKPSETPPLC(CONH2), or (NHCOCH3)CGDFWKSKMPKIKSK(CONH2), respectively, conjugated at terminal cysteines to Keyhole limpet hemocyanin (KLH). The used peptides corresponded to sequence Gly-104 to Arg-119 of Spx, Met-221 to Leu-234 of YjbHBS, and Gly-248 to Lys-261 of YjbHSA. Spx antisera were used at a 1,250-fold dilution, and the YjbHBS and YjbHSA antisera were used at a 5,000-fold dilution. The secondary antibodies (ECL anti-rabbit IgG, horseradish peroxidase-linked species-specific whole antibody from donkey [GE Healthcare]) were used at a 5,000-fold dilution. Immunodetection was carried out by chemiluminescence using the Super Signal West Pico system (Thermo Scientific). Quantification was done using a Kodak image station 440CF.
RNA isolation and cDNA synthesis.
For isolation of total RNA, 10 ml of an exponentially growing S. aureus culture was added to a 50-ml centrifuge tube containing 10 g of crushed ice. The sample was centrifuged immediately at 17,000 × g for 5 min at 4°C, and the pellet was used for extraction of total RNA using the Qiagen RNeasy minikit according to the manufacturer's protocol. The integrity of the RNA was checked by electrophoresis on a 2% agarose gel containing ethidium bromide, and the RNA was quantified spectrophotometrically. A total of 2 μg RNA was treated with RNase-free DNase I (Fermentas) and was then used for cDNA synthesis with the RevertAid H Minus First-Strand cDNA synthesis kit (Fermentas) and random hexamer primers according to the manufacturer's protocol. A control without reverse transcriptase (NoRT) was made in parallel to ensure that no chromosomal DNA gave a false-positive signal in downstream applications.
qRT-PCR.
Quantitative real-time PCR (qRT-PCR) was performed in an Mx3000P qPCR system (Stratagene). Reaction mixtures (25 μl) were made using the Maxima SYBR green/ROX qPCR Master Mix (2×) (Fermentas) according to the manufacturer's protocol. Primers used for qRT-PCR are listed in Table 3. The cDNA and NoRT controls were diluted 5-fold in 10 mM Tris-HCl (pH 8.0), and 2 μl of these dilutions was used as templates in qRT-PCR. A total of 2 μl 10 mM Tris-HCl (pH 8.0) was added to the no-template controls. At least three biological replicates were made for each strain and condition, and each biological replicate was run in triplicate in the qRT-PCR. For each primer pair, a control sample containing the gene of interest at an appropriate concentration was included in all runs and used as the calibrator. Data were analyzed using the comparative quantitation feature of the MxPro software (Stratagene). A two-way Student t test assuming equal variances was used to compare data.
RESULTS AND DISCUSSION
Influence of diamide on cell growth and Spx concentration.
The azo compound diamide is a thiol-specific oxidant used to induce disulfide stress. In B. subtilis, the addition of 1 mM diamide to exponentially growing cells leads to a rapid increase in the intracellular level of Spx (Fig. 2A), thereby inducing the disulfide stress response. To investigate the effect of diamide on S. aureus, a series of different diamide concentrations were added to exponentially growing wild-type S. aureus cells in a broth medium, and growth was monitored for 60 min. The addition of ≥5 mM diamide led to growth inhibition, and the addition of ≥10 mM diamide completely inhibited growth (data not shown).
Fig 2.
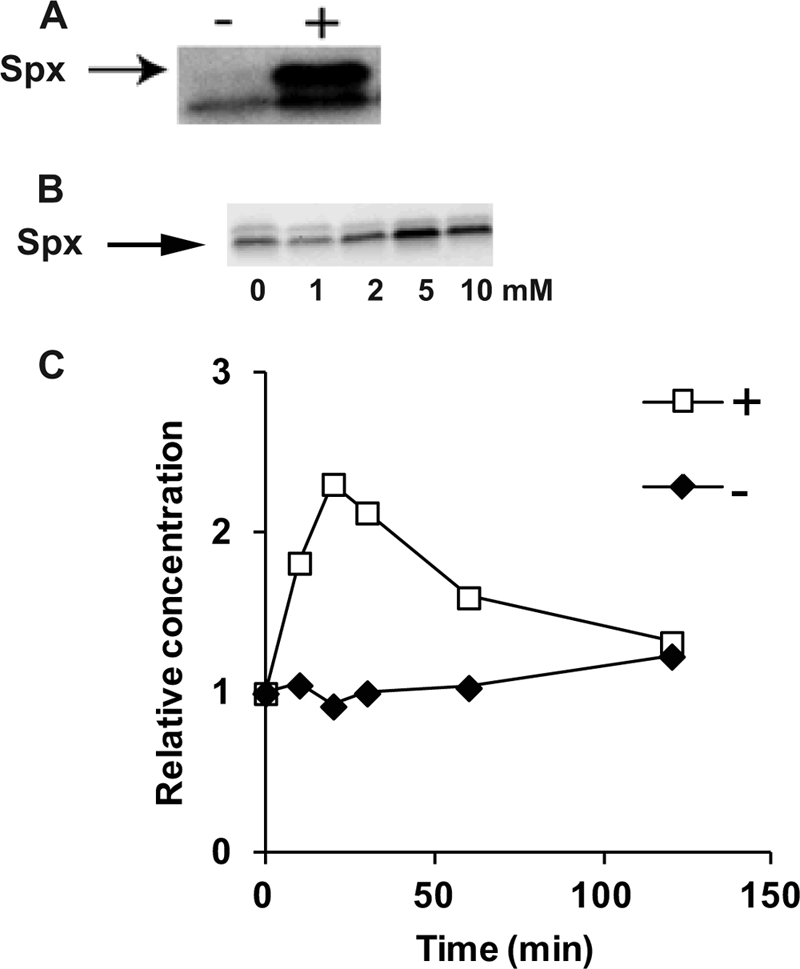
Immunoblot analysis of Spx content before and after the addition of 1 mM diamide to B. subtilis 1A1 (A) and 1 to 10 mM diamide to S. aureus Newman (B). The relative Spx content as a function of time after no addition (−) or addition (+) of 5 mM diamide to S. aureus Newman is shown in panel C.
In S. aureus, the basal level of Spx under nonstress conditions appears to be higher than that of B. subtilis. Using the same antibodies, raised against an Spx peptide, and comparable growth conditions, very low Spx-specific signal can be detected in cell extracts from nonstressed B. subtilis cells (Fig. 2A), while a clear band corresponding to Spx can be seen in immunoblots with cell extracts from nonstressed S. aureus cells (Fig. 2B).
The Spx content in extracts from cultures of S. aureus to which different concentrations of diamide had been added was analyzed. Concentrations up to 2 mM diamide could be added without any detected increase in the Spx level. However, increasing the added diamide concentration to 5 mM or more gave a significant increase in Spx concentration (Fig. 2B). Interestingly, previous studies have shown that the expression of the trxB gene is induced by as little as 0.5 mM diamide in S. aureus NCTC 8325-4 cells grown in the same growth medium as the one used in the present study (34). This induction of trxB is not seen in an spx mutant (34). This suggests that no increase above the basal concentration of Spx observed in nonstressed cells is required to induce trxB expression. It seems that the concentration of diamide where the level of Spx increases corresponds to the concentration where growth defects are seen, suggesting that this is a response to severe stress.
Heterologous complementation of a B. subtilis yjbH-null mutation.
To test whether S. aureus yjbH could complement the B. subtilis yjbH deletion, we constructed a strain with an IPTG-inducible S. aureus yjbH gene integrated in a single copy at the amyE locus on the chromosome of a B. subtilis yjbH-null mutant. Immunoblot analysis confirmed that the product of the exogenous yjbH gene was produced in an IPTG-dependent manner (see Fig. S7 in the supplemental material). B. subtilis yjbH mutant strains grow poorly and accumulate high levels of Spx compared with the wild-type strain (21). Expression of S. aureus yjbH alleviated the growth phenotype present in the mutant strain (data not shown), suggesting functional complementation by S. aureus yjbH. Moreover, the complementation with S. aureus yjbH restored the Spx level to wild-type levels as determined by immunoblotting (Fig. 3). The concentration of Spx was inversely dependent on the concentration of YjbH, which was shown by varying the amount of the inducer IPTG (see Fig. S4 in the supplemental material).
Fig 3.
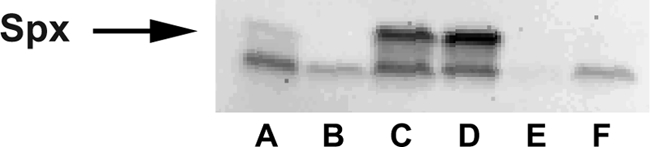
Immunoblot analysis of the Spx content in unstressed cells of different B. subtilis strains. Lane A, WT (1A1); lane B, spxΩpMUTIN2 (LUW297); lane C, ΔyjbH::spc (LUW272); lane D, ΔyjbH::spc (LUW400); lane E, ΔyjbH::spc amyE::yjbHBS (LUW421); lane F, ΔyjbH::spc amyE::yjbHSA (LUW448).
The strain complemented with S. aureus yjbH responded to the addition of 1 mM diamide by increasing the level of Spx in the same fashion as the strain complemented with B. subtilis yjbH and wild-type cells (data not shown). This shows that the ability of YjbHSA to facilitate the proteolysis of Spx is inhibited by diamide.
To further establish the functional conservation of the Spx/YjbH system, a complementation of the B. subtilis spx mutant with the spx gene from S. aureus was made. B. subtilis spx mutant strains are hypersensitive to diamide (29). Therefore, sensitivity to diamide was used to establish functional production of S. aureus Spx in B. subtilis. The complementation alleviated the sensitivity of the mutant to diamide as effectively as a complementation with spx from B. subtilis (Fig. 4). The concentration of S. aureus Spx was also shown to be regulated in a similar manner to B. subtilis Spx in response to diamide exposure, with a low level in the unexposed cells and a high level in exposed cells (Fig. 5). A B. subtilis strain where both yjbH and spx were deleted and complemented with their S. aureus counterparts was created (LUW442). This strain exhibited the same diamide-induced pattern of Spx regulation as the other complemented strains and showed similar resistance to diamide (Fig. 4 and 5). In conclusion, the experiments in this section show that despite the low sequence conservation between the B. subtilis and S. aureus YjbH proteins, they are similar in all functional aspects tested.
Fig 4.
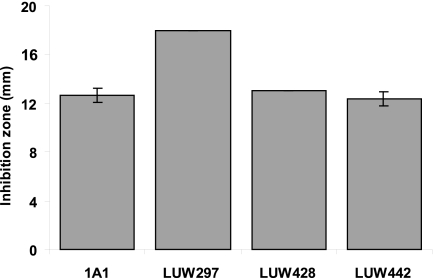
The diagram shows the diameter of the clearing zone of B. subtilis 1A1 (WT), LUW297 (spxΩpMUTIN2), LUW428 (spxΩpMUTIN2 amyE::spxSA), and LUW442 (spxΩpMUTIN2 ΔyjbH amyE::spxSA/pCW7_yjbHSA) in the presence of diamide. The error bars represent the standard deviations of the diameters of three separate clearing zones.
Fig 5.
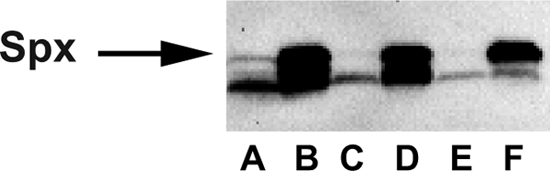
Immunoblot analysis of the heterologous S. aureus Spx content in B. subtilis. Lanes A and B, LUW362 (spxΩpMUTIN2 amyE::spxBS); lanes C and D, LUW428 (spxΩpMUTIN2 amyE::spxSA); lanes E and F, LUW442 (spxΩpMUTIN2 ΔyjbH amyE::spxSA/pCW7_yjbHSA) before (lanes A, C, E) and after (lanes B, D, F) diamide treatment.
Role of cysteine residues in YjbH.
Cysteines have the ability to be reversibly oxidized and covalently modified and are often involved in redox sensing. B. subtilis YjbH contains an N-terminal CXXC motif (two cysteines separated by two other residues) at residues 31 to 34 and five additional cysteines (Fig. 1). In thioredoxins and related proteins, the CXXC motif is directly involved in redox reactions, suggesting that this motif could have a similar role in YjbH. However, in S. aureus YjbH, the cysteine corresponding to C31 is replaced by a serine. Moreover, it has been suggested that YjbH binds zink via a His-Cys-rich domain and that redox-induced inactivation of Zn-binding leads to Zn2+ release and liberation of Spx from YjbH (9). In order to investigate the role of the cysteine residues in YjbHBS, a mutant where all seven cysteine residues (C13, C31, C34, C89, C175, C236, C297) were exchanged for serines was constructed. Similarly, a mutant of YjbHSA where all four cysteines (C37, C71, C121, C123) were exchanged for serines was also constructed. These cysteine-free YjbH variants were used to complement the B. subtilis yjbH-null mutant in the same way as described in the previous section. Immunoblot analysis showed that both of the cysteine-free YjbH variants were produced at similar levels as the wild-type protein and that they were able to facilitate the efficient proteolysis of Spx (data not shown and Fig. 6). It is concluded that the cysteine residues are dispensable for the function of the protein and that the activity of the adaptor is not activated or deactivated via redox-active cysteines. When our investigation was completed, Göhring et al. (12) reported a study on the role of the cysteine residues in S. aureus YjbH using a complementary approach. The cysteine residues were replaced with glycine residues, and plasmid-encoded cysteine replacement mutants of YjbH were tested for their ability to complement an S. aureus yjbH mutant strain. In contrast to our results, Göhring et al. (12) suggest that the cysteine residues indeed are important for the role of YjbH in disulfide stress management. A possible difference underlying the obtained results is the stability of the mutated YjbH proteins. The difference in helix propensities between cysteine (intermediate), serine (intermediate), and glycine (very low) might affect the stability and folding of YjbH.
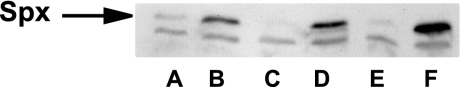
Fig 6.
Immunoblot analysis of the Spx content before (lanes A, C, and E) and after (lanes B, D, and F) the addition of diamide to B. subtilis WT (lanes A and B) and ΔyjbH mutant cells complemented with YjbHBS without cysteines (lanes C and D) or YjbHSA without cysteines (lanes E and F).
Pleiotropic properties of an S. aureus yjbH mutant.
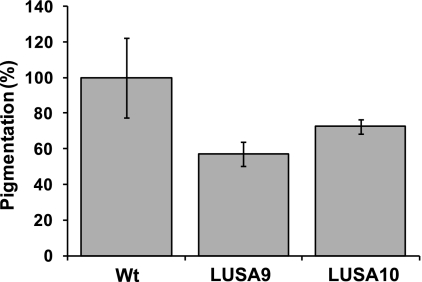
To investigate the possible role of YjbH in the regulation of Spx in S. aureus, an allelic replacement was used to exchange yjbH with a spectinomycin resistance cassette (for details, see Materials and Methods). We compared the growth of the yjbH deletion strain with that of the wild type (Newman) and found that the mutation caused a decreased growth rate in rich medium. This is similar to a B. subtilis yjbH mutant, which displays growth defects both on plates and in broth medium (21). Loss of YjbH also resulted in decreased carotenoid pigment production by S. aureus (Fig. 7). The pigment protects S. aureus against oxidative stress due to its ability to absorb excess energy from reactive oxygen species and is linked to virulence (23). We do not know if the reduced pigmentation is due to lack of YjbH or increased levels of Spx. However, it has recently been shown that pigment production is linked to several different metabolic processes in the cell (20). To confirm that the phenotype was caused by the deletion of yjbH, we performed complementation analysis by inserting the yjbH gene in the pRMC2 plasmid (4) to generate the plasmid pRMC2_yjbHSA. The complementation restored the pigment production; how-ever, the growth rate phenotype was not restored in the strain complemented with pRMC2_yjbHSA. This observation suggests that timing or level of yjbH expression is important. To better mimic the wild-type situation, an additional complementation construct, pCL25_yjbIHSA, containing the yjbIH operon under the control of its native promoter integrated into the chromosome, was constructed. The resulting strain, LUSA10, alleviated the growth phenotype seen in the mutant and also complemented the pigmentation phenotype (see Fig. S5 in the supplemental material). In B. subtilis, the disruption of yjbH results in an increased resistance to diamide due to increased levels of thioredoxin (21). In contrast to this, the S. aureus ΔyjbH mutant showed decreased resistance to diamide compared to that of the wild type and the complemented strain (see Fig. S6 in the supplemental material).
Fig 7.
Pigment formation in S. aureus. The bars represent the relative amounts isolated from 24-h culture by methanol extraction. The error bars represent the standard deviations from three experiments. Wt, Strain Newman; LUSA9, ΔyjbH strain; LUSA10, ΔyjbH + yjbIH strain.
YjbH influences the level of Spx in S. aureus.
In B. subtilis, a yjbH-null mutation leads to the accumulation of Spx (21), which in turn is responsible for the observed pleiotropic phenotype. To investigate if YjbH is required for efficient proteolytic degradation of Spx also in S. aureus, the amounts of Spx in the wild type and the yjbH mutant strain were analyzed by immunoblotting. A band corresponding to the expected size of Spx was detected at elevated levels in the yjbH mutant strain (Fig. 8). In both the complemented strains, the level of Spx returned to a low level similar to that of the wild-type strain. For the pRMC2_yjbHSA construct, the complementation was seen without adding the inducer (Atet) for YjbHSA production. This implies that residual expression yielding very small quantities of YjbHSA are enough to control the level of Spx. Larger amounts of YjbHSA were achieved by adding increasing concentrations of the inducer (see Fig. S8 in the supplemental material). This resulted in Spx concentrations below wild-type levels and a decreased growth rate. It was previously shown (34) that the spx mutant has a severe growth defect, so the depletion of Spx seen under YjbHSA overexpression is most likely the cause of the observed growth defect.
Fig 8.

Immunoblot analysis of the Spx content of different S. aureus strains. Lane A, Newman (wild type); lane B, LUSA2 (Newman ΔyjbH); lane C, 8325-4 Δspx mutant; lane D, LUSA2/pRMC2; lane E, LUSA2/pRMC2-yjbHSA.
Transcriptional analysis.
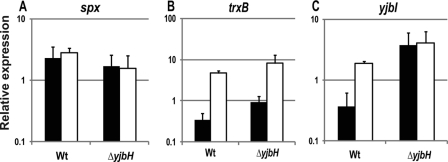
To determine whether the effect on Spx seen at the protein level was due to regulation at the transcriptional or the posttranscriptional level, qRT-PCR was used to quantify the amount of spx mRNA (Fig. 9A). No significant difference in the expression of spx before or after the addition of 5 mM diamide was found, nor was any significant difference seen between the expression of spx in the wild type and the yjbH mutant. This suggests that the increased level of Spx is due to changes at the posttranscriptional level that lead to a decreased degradation of Spx.
Fig 9.
Transcript levels of spx (A), trxB (B), and yjbI (C) and in wild-type (Newman) and ΔyjbH (LUSA2) strains before (black columns) and after (white columns) addition of diamide (5 mM). The error bars represent the standard deviations from three experiments.
Previous studies identified trxB expression in S. aureus as an important target of Spx regulation (34). As expected, the expression of trxB was increased after the addition of diamide in the wild-type strain (Fig. 9B). The yjbH mutant showed a significantly higher expression of trxB in the unstressed cells but also an increase after the addition of diamide, indicating that the amount of Spx and the oxidation state of the cell effect trxB expression.
In B. subtilis, YjbH has a negative effect on its own expression (21), which is likely mediated through Spx. This ensures that there will be YjbH present in the cell to mediate the breakdown of Spx once the stress is gone. The expression of the S. aureus yjbIH operon was investigated. In the wild type, the addition of diamide leads to an induction of the expression of both yjbI and yjbH. In the yjbH mutant, only the level of yjbI could be measured and was assumed to be representative of yjbH. In the mutant, the level of yjbI was elevated compared to that of the unstressed wild type both before and after the addition of diamide (Fig. 9C). The transcriptional data presented here suggest that, when the level of Spx is high in the cell (stressed cells and ΔyjbH mutant) the level of expression from the yjbIH operon is elevated.
In conclusion, we here demonstrate that despite the low sequence similarity (approximately 30% identity) between YjbH in B. subtilis and S. aureus, the S. aureus protein can functionally complement its counterpart in B. subtilis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter Kjelgaard for technical assistance and David Rudner for the plasmid pDR111.
This work was supported by Crafoordska stiftelsen, Carl Tryggers Stiftelse, and Magnus Bergvalls Stiftelse.
Footnotes
Published ahead of print 22 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Åslund F, Beckwith J. 1999. Bridge over troubled waters: sensing stress by disulfide bond formation. Cell 96:751–753 [DOI] [PubMed] [Google Scholar]
- 3. Baker TA, Sauer RT. 2006. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem. Sci. 31:647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Corrigan RM, Foster TJ. 2009. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61:126–129 [DOI] [PubMed] [Google Scholar]
- 5. Duthie ES, Lorenz LL. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95–107 [DOI] [PubMed] [Google Scholar]
- 6. Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671–683 [DOI] [PubMed] [Google Scholar]
- 7. Frees D, Qazi SN, Hill PJ, Ingmer H. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48:1565–1578 [DOI] [PubMed] [Google Scholar]
- 8. Frees D, Savijoki K, Varmanen P, Ingmer H. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63:1285–1295 [DOI] [PubMed] [Google Scholar]
- 9. Garg SK, Kommineni S, Henslee L, Zhang Y, Zuber P. 2009. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J. Bacteriol. 191:1268–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gerth U, Krüger E, Derre I, Msadek T, Hecker M. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787–802 [DOI] [PubMed] [Google Scholar]
- 11. Giachino P, Engelmann S, Bischoff M. 2001. Sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Göhring N, et al. 2011. New role of the disulfide stress effector YjbH in beta-lactam susceptibility of Staphylococcus aureus. Antimicrob. Agents Chemother. 55:5452–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gottesman S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19:565–587 [DOI] [PubMed] [Google Scholar]
- 14. Grundling A, Missiakas DM, Schneewind O. 2006. Staphylococcus aureus mutants with increased lysostaphin resistance. J. Bacteriol. 188:6286–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guerout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61 [DOI] [PubMed] [Google Scholar]
- 16. Hoch JA. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol. 204:305–320 [DOI] [PubMed] [Google Scholar]
- 17. Kajfasz JK, et al. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J. Bacteriol. 192:2546–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kommineni S, Garg SK, Chan CM, Zuber P. 2011. YjbH-enhanced proteolysis of Spx by ClpXP in Bacillus subtilis is inhibited by the small protein YirB (YuzO). J. Bacteriol. 193:2133–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krüger E, Witt E, Ohlmeier S, Hanschke R, Hecker M. 2000. The Clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J. Bacteriol. 182:3259–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lan L, Cheng A, Dunman PM, Missiakas D, He C. 2010. Golden pigment production and virulence gene expression are affected by metabolisms in Staphylococcus aureus. J. Bacteriol. 192:3068–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Larsson JT, Rogstam A, von Wachenfeldt C. 2007. YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis. Mol. Microbiol. 66:669–684 [DOI] [PubMed] [Google Scholar]
- 22. Lee CY, Buranen SL, Ye ZH. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105 [DOI] [PubMed] [Google Scholar]
- 23. Liu CI, et al. 2008. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 319:1391–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luong TT, Lee CY. 2007. Improved single-copy integration vectors for Staphylococcus aureus. J. Microbiol. Methods 70:186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McNamara PJ, Iandolo JJ. 1998. Genetic instability of the global regulator agr explains the phenotype of the xpr mutation in Staphylococcus aureus KSI9051. J. Bacteriol. 180:2609–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morikawa K, et al. 2001. Overexpression of sigma factor, sigma(B), urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactams. Biochem. Biophys. Res. Commun. 288:385–389 [DOI] [PubMed] [Google Scholar]
- 27. Nakano MM, Hajarizadeh F, Zhu Y, Zuber P. 2001. Loss-of-function mutations in yjbD result in ClpX-and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 42:383–394 [DOI] [PubMed] [Google Scholar]
- 28. Nakano S, Erwin KN, Ralle M, Zuber P. 2005. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 55:498–510 [DOI] [PubMed] [Google Scholar]
- 29. Nakano S, Küster-Schöck E, Grossman AD, Zuber P. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 100:13603–13608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakano S, Nakano MM, Zhang Y, Leelakriangsak M, Zuber P. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. U. S. A. 100:4233–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Newberry KJ, Nakano S, Zuber P, Brennan RG. 2005. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 102:15839–15844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Norrander J, Kempe T, Messing J. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101–106 [DOI] [PubMed] [Google Scholar]
- 33. Novick R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155–166 [DOI] [PubMed] [Google Scholar]
- 34. Pamp SJ, Frees D, Engelmann S, Hecker M, Ingmer H. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188:4861–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rogstam A, Larsson JT, Kjelgaard P, von Wachenfeldt C. 2007. Mechanisms of adaptation to nitrosative stress in Bacillus subtilis. J. Bacteriol. 189:3063–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 37. Shaw LN, et al. 2006. Investigations into sigmaB-modulated regulatory pathways governing extracellular virulence determinant production in Staphylococcus aureus. J. Bacteriol. 188:6070–6080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Steinmetz M, Richter R. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79–83 [DOI] [PubMed] [Google Scholar]
- 39. Veiga P, et al. 2007. SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J. Biol. Chem. 282:19342–19354 [DOI] [PubMed] [Google Scholar]
- 40. Winstedt L, Yoshida K, Fujita Y, von Wachenfeldt C. 1998. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J. Bacteriol. 180:6571–6580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zemansky J, et al. 2009. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J. Bacteriol. 191:3950–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zuber P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186:1911–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.