Abstract
Spo0A∼P is the master regulator of sporulation in Bacillus subtilis. Activity of Spo0A is regulated by a phosphorelay integrating multiple positive and negative signals by the action of kinases and phosphatases. The phosphatase Spo0E specifically inactivates the response regulator Spo0A∼P by dephosphorylation. We identified a σB-type promoter adjacent to spo0E that is activated by the general stress response sigma factor σB and is responsible for spo0E induction in vivo. Ectopic expression of σB and subsequent induction of spo0E cause a σB-dependent block of sporulation-specific transcription of the spo0A and spoIIE genes and produces a sporulation-deficient phenotype. This effect could be erased by a deletion of the σB promoter of spo0E and thus solely addresses σB activity. Here, a molecular mechanism is shown that integrates σB activity into the decision-making process of sporulation and provides a link to interconnect these two dominant and probably mutually exclusive adaptive responses in the regulatory network of B. subtilis.
INTRODUCTION
Upon severe stress in exponentially growing as well as starving nongrowing cells, the general stress response is one of the most noticeable components of the adaptational gene expression network (24, 41). The general stress regulon comprising approximately 200 genes is under the control of the alternative sigma factor σB, whose activation is controlled by protein-protein interactions in a central partner switch module consisting of σB, its anti-sigma factor, RsbW, and the RsbW antagonist RsbV (41). At least three distinct pathways integrate a large set of diverse stimuli into the signaling cascade. The energy stress module senses starvation for glucose, phosphate, and oxygen, as well as a drop in the cellular ATP level that is caused by treatment with azide, mycophenolic acid, or carbonyl cyanide m-chlorophenylhydrazone (CCCP). The stressosome integrates environmental stress stimuli, such as low pH, high and low temperature, salt, ethanol, manganese, sodium nitroprusside (SNP) exposure to blue light, and cell wall stress caused by addition of antibiotics, such as bacitracin and vancomycin (23, 41). The response mediated by the first two signaling pathways is rapid and transient, in contrast to the third σB-activating pathway that is observed under continuous growth close to the minimal or maximal growth temperatures of Bacillus subtilis. Here, a persistent activation of σB is triggered that is independent of the phosphorylation state of the anti-anti-sigma factor RsbV (6, 26a). The main and characteristic function of the σB-induced general stress proteins is to provide the cell with a comprehensive cross-protective and preventive multiple stress resistance (15a, 26b, 60): (i) cross-protective, because the response to one inducing stimulus not only includes resistance to all other inducing stimuli but also comprises improved resistance to oxidative (2, 12) or alkaline stress (15a), which are not σB-inducing stimuli; and (ii) preventive, because a nongrowing vegetative and nonsporulating cell is equipped with the protective functions of the general stress proteins to cope with possible future stress (26b, 60). Despite its physiological importance and a considerable overlap with other stress-specific responses in the regulatory network (23), the general stress response remains somehow isolated from the important processes of the decision-making stationary-phase network.
Starving B. subtilis cells can use a variety of alternative survival strategies, including diauxic growth by the consumption of secondary metabolites, motility and chemotaxis to actively seek for new resources, secretion of enzymes to break down extracellular proteins, lipids and polysaccharides, the production of antimicrobial agents to kill and feed on competitors as well as siblings, or the development of competence to take up and integrate exogenous DNA (16, 19, 51, 58).
Finally, starvation forces a majority of B. subtilis cells to undergo one of the most dramatic changes in cellular differentiation, the formation of a dormant cell type—the endospore (50). Spores are able to resist environmental extremes, such as desiccation, heat, toxic compounds, or even radiation, and thus are best equipped for long-term survival (50). Converting a cell into a spore is an energy- and time-consuming process that, in contrast to other cellular responses, becomes irreversible about 2 h after initiation (11, 34). Thus, a sporulating cell is highly susceptible to the further impact of stress, and it is not able to quickly switch back to vegetative growth in case of a nutrient influx (39, 58). Although sporulation is induced by starvation, it is not initiated immediately. It appears that responses allowing continued growth as long as possible are the cell's favored and mutually exclusive alternative to sporulation. Therefore, commitment to spore formation is a so-called “last resort” adaptive response to starvation after alternative responses failed to efficiently cope with the situation. To reach such a level of informed commitment, the input signals are processed by the combined action of a highly complex and sophisticated regulatory network, providing multiple opportunities for signal integration necessary for a finely tuned target gene expression in the context of the single cells as well as the community state (49).
Spo0A, the master regulator of spore development (26), is a member of the response regulator family of transcription factors. Hence, its activity is regulated by reversible phosphorylation of a specific aspartate residue (7). Phosphorylation of Spo0A is governed by a multicomponent phosphorelay (7). Activating signals are sensed by five histidine sensor kinases (KinA to -E) that introduce phosphoryl groups into the relay by phosphorylating Spo0F (27). The phosphoryl group of Spo0F∼P is rapidly transferred via Spo0B to the final acceptor Spo0A, thereby activating the regulator (7, 27). Inactivating signals are introduced into the relay by dedicated response regulator aspartate phosphatases (Rap's) that remove phosphoryl groups from Spo0F∼P (18, 35) or directly from Spo0A∼P (Spo0E) (33, 35). Once phosphorylated, Spo0A∼P acts as both a transcriptional activator and repressor, directly controlling 121 genes (13, 32) in a concentration-dependent manner (14). Due to different affinities of Spo0A∼P to the operator regions of its target genes, their expression responds to low or high threshold levels of the response regulator (14). This mechanism was an important finding to explain the observation that Spo0A∼P is not only essential for the process of sporulation but is also necessary for other stationary-phase responses, such as competence (1), cannibalism (14, 16), or biofilm formation (49a).
Here we provide evidence for a molecular mechanism that is able to integrate σB-inducing stimuli into the decision-making process of spore development by induction of the well-known regulatory key component Spo0E. We show that ectopic induction of σB leads to inactivation of the sporulation master regulator Spo0A, supporting the idea that σB may play an important role in the decision process that determines the cell's fate under certain physiological conditions.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study are listed in Table 1. Growth for initial Northern blots and primer extension (PE) experiments was performed in synthetic medium (57) and was monitored by measuring the optical density at 500 nm (OD500). Growth for sporulation-specific Northern blot experiments was performed in Difco sporulation medium (DSM) (47) and was monitored by measuring the OD540. Prewarmed growth medium (80 to 120 ml) was inoculated with exponentially growing cells to obtain a starting OD500 or OD540 of 0.05, respectively. Cultures were routinely grown in 500-ml Erlenmeyer flasks in a shaking water bath at 180 rounds per minute and 37°C.
Table 1.
B. subtilis strains and plasmids used in this study
| Strain or plasmid | Relevant feature(s) | Reference |
|---|---|---|
| Strains | ||
| 168 | trpC2 | 8 |
| BAR 17 | trpC2 ΔrsbVW-sigB::Tet | This study |
| BAR 18 | trpC2 ΔrsbVW-sigB::Tet amyE::PXyl-sigB::Cm(pX1) | This study |
| BAR 19 | trpC2 ΔrsbVW-sigB::Tet amyE::PXyl-sigB::Cm(pX1) ΔPσB of spo0E::Km | This study |
| BAR 20 | trpC2 ΔrsbVW-sigB::Tet amyE::PXyl-sigB::Cm(pX1) ΔabrB::Km | This study |
| Plasmids | ||
| pBR322 | Tetracycline resistance marker | 62 |
| pX1 | amyE integration vector; PxylA cat bla xylR | 28 |
| pDG148 | Kanamycin resistance marker | 52 |
Construction of mutant strains.
All gene deletions were created by insertions of resistance markers into the chromosome that erased the respective genes but left all regulatory regions unaffected. A modified two-step fusion PCR protocol (61) was used to generate a linear DNA fragment carrying a central resistance marker flanked by homologous sequences representing the chromosomal up- and downstream regions of the respective genes. The PσB promoter deletion of spo0E from the chromosome was achieved by the fusion of 4 fragments. The upstream fragment carried the PσΒ deletion of 28 nucleotides. The resistance marker with its own Shine-Dalgarno sequence was inserted into the chromosome right between the spo0E locus and its terminator, creating a transcriptional fusion. All mutants were selected on LB agar plates containing tetracycline (17 μg/ml), kanamycin (5 μg/ml), chloramphenicol (5 μg/ml), or a combination of these. A copy of the sigB gene was inserted into pX1 (28) through the BamHI sites and was finally integrated into the chromosomal amyE locus of B. subtilis. Chromosomal DNA of each mutant was sequenced to verify the correct mutation.
Xylose induction and cell sampling.
Cells were subjected to a final concentration of 0.3% (wt/vol) xylose when grown in synthetic medium or 0.1% (wt/vol) xylose when grown in DSM. Samples were taken under control conditions from untreated cultures right before and after addition of xylose at the time points indicated for the respective experiments. Cell samples for RNA extraction were mixed with 0.5 volume of ice-cold killing buffer (20 mM Tris-HCl, 5 mM MgCl, 20 mM NaN3). All samples were immediately cooled down to 0°C in liquid nitrogen, spun down at 10,000 × g for 8 min at 4°C, and stored in liquid nitrogen until further preparation.
RNA isolation.
Cells were mechanically disrupted as previously described by Hauser et al. (20). Total RNA was isolated according to the acid phenol method of Majumdar et al. (31). RNA samples were frozen and thawed three times (2 min on liquid nitrogen and 2 min at 40°C) to achieve properly dissolved RNA.
Northern blot analyses.
Northern blots were performed according to the method of Wetzstein et al. (63). RNA blots were methylene blue stained to check for RNA quality and equally loaded amounts. RNA probes for sigB, spo0E, spo0A, and spoIIE were digoxigenin labeled by in vitro transcription with T7 RNA polymerase from gene-specific PCR products fused to a T7 promoter. The primers used for generation of RNA probes are listed in Table 2.
Table 2.
Primers used in this study
| Purpose | Name | Sequence (5′→3′)a |
|---|---|---|
| Fusion PCR and gene deletion | abrB_up_for | GCCGTTTTTCTGTCGTGCGG |
| abrB_up_rev | GGTCCATTCACTATTCTCATTCTCCTCCCAAGAGATACTTA | |
| Km_for | ATGAGAATAGTGAATGGACC | |
| Km_rev | GATTAACAATTATTAGAGGTC | |
| abrB_do_for | GACCTCTAATAATTGTTAATCCCAGCTTCAAAACCTTAAATA | |
| abrB_do_rev | GCGATCGACACGCTGAAATC | |
| rsbV_up_for | AGCAGACGCTTCTCGGAACTA | |
| rsbV_up_rev | ATTGTGAATAGGATGTATTCATTCGTATCACCTCAAATTTTCCT | |
| tet_for | ATGAAATCTAACAATGCGCTCATCGT | |
| tet_rev | TCAGGTCGAGGTGGCCCGGCTCCATG | |
| sigB_do_for | AACATTCTCAAAGGGATTTCTAACCCTCGATGGAGTTAATGTAA | |
| sigB_do_rev | ACAGTTCCAGGTTTTCCCAC | |
| ΔPσB_far_up_for | AGAGTGCTTGAAGCGTATGAA | |
| ΔPσB_up_rev | TAGAAGAACCGCCGCCAGGCAGGAGGTAT | |
| ΔPσB_do_for | CTGGCGGCGGTTCTTCTAATCCTATCAAT | |
| ΔPσB_do_rev | TCTCATTTGATATGCCTCCTCTATTTATTTGCATCATATG | |
| km+SD_for | AGGAGGCATATCAAATGAGAATAGTGAATGGACC | |
| km_rev | GAGGTCATCGTTCAAAATGG | |
| spo0E_do_for | CCATTTTGAACGATGACCTCCGGCCTATCAGATGCATATA | |
| spo0E_do_rev | GAAAAAGGAACCGGTCTCGG | |
| Plasmid cloning | sigB_BamHI_pX1_for | TGGGATCCATGATCATGACACAACCATCA |
| sigB_BamHI_pX1_rev | TGGGATCCTTACATTAACTCCATCGAGGG | |
| Primer extension | spo0E_-400_for | CGGCATGACGATATACAGGA |
| -60b_PσB_rev | ATTACAGATTATACTTTATTT | |
| -7b_spo0E_ingene_rev | TCTTTCTTGTTCAGAAGAACC | |
| Northern blotting | sigB_Nor_for | ATCTGGTTGACATGCTTGCG |
| sigB_T7_rev | CTAATACGACTCACTATAGGGAGAATCGTGACAGTGCTTCCGTC | |
| spo0A_Nor_for | TCCCGATGTGCTCGTATTAG | |
| spo0A_T7_rev | CTAATACGACTCACTATAGGGAGATGGCGGATCGCTCTTTCTAC | |
| spo0E_Nor_for | ATGGGCGGTTCTTCTGAAC | |
| spo0E_T7_rev | CTAATACGACTCACTATAGGGAGATTTATTTGCATCATATGCTGG | |
| spoIIE_Nor_for | GTGCTTGCAGGTGCGCTGAC | |
| spoIIE_T7_rev | CTAATACGACTCACTATAGGGAGACACAACATAACGAGCCAATAT |
The underlined portions of the sequences represent T7 promoter sequences.
Primer extension experiments.
The two oligonucleotide primers (-60b_PΒ_rev) and (-7b_spo0E_ingene_rev) (Table 2) were 5′ end labeled with [γ-32P]ATP and used for primer extension (PE) analyses as described previously (63). The spo0E promoter region was amplified by PCR using the primers (spo0E_-400_for) and (-7b_spo0E_ingene_rev), and sequencing was performed as described by Sanger et al. (46).
Sporulation assay.
To avoid premature sporulation or transfer of spores from precultures, we set up overnight cultures of the B. subtilis wild-type and strains BAR 17, BAR 18, and BAR 19 as dilution series in Luria broth (LB) first. Exponential-phase growing LB cultures were used the next morning to inoculate a 50-ml DSM preculture to a starting OD540 of 0.05. This preculture was grown to a final OD of 0.6 and used to inoculate the main cultures of 80 ml DSM to a starting OD of 0.05. Two control samples were taken from all cultures—the first during early exponential growth (OD, 0.4) and the second at the onset of the transient phase (OD, 2.0). An OD of 2.0 was set as time zero (t0) for all sporulation experiments. All experiments were grown in triplicate without and with the addition of 0.1% (wt/vol) xylose. Xylose was added immediately after t0 samples were taken. Growth was monitored every hour. The number of viable cells per milliliter of culture was determined as the total number of CFU on LB plates. The number of spores (spore-forming units [SFU]) per milliliter of culture was determined as the number of CFU after heat treatment at 80°C for 20 min. Samples for CFU and SFU determination were taken in 3-h intervals for a period of 24 h as well as after 48 h. CFU and SFU were determined by plating appropriate dilution series on LB agar plates. The sporulation frequency is the ratio of SFU to viable cells' CFU per milliliter.
RESULTS
Chromosomal organization and transcriptional regulation of the spo0E locus.
Transcription of spo0E has previously been described to occur from two distinct transcriptional start sites that are separated by three bases only (36). The first promoter, P1, produces a minor transcript starting at position −45 upstream of the spo0E start codon and seems to be constantly active throughout growth and remains unaffected by mutations of the spo0A or abrB genes (36). In contrast, the second promoter, P2, initiates transcription just three bases upstream from the P1 start site (Fig. 1A) and seems to be recognized by σA-containing RNA polymerase (36). P2 produced a predominant transcript in an abrB mutant strain that was about 4-fold stronger than the wild type; thus, it was shown that AbrB inhibits transcription of spo0E only from P2 (36).
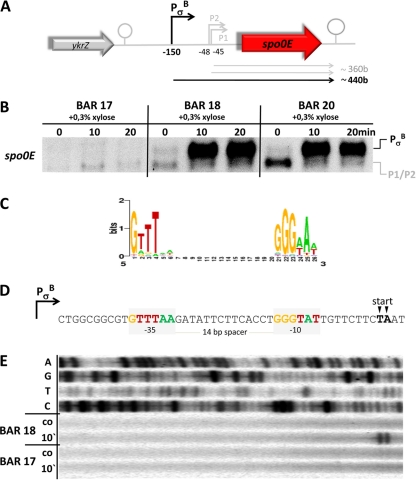
Fig 1.
Regulation of spo0E. (A) Schematic representation of the chromosomal spo0E region. (B) Northern blot experiment of the expression profiles of spo0E in response to addition of 0.3% (wt/vol) xylose to exponentially growing cultures of strains BAR 17 (ΔrsbVW-sigB), BAR 18 (ΔrsbVW-sigB amyE::PxylA-sigB), and BAR 20 (ΔrsbVW-sigB amyE::PxylA-sigB ΔabrB). Cells were grown in minimal media, and 0.3% xylose was added at t0 = OD500 of 0.4. Samples for RNA preparation were taken under control conditions before (0 min) as well as 10 and 20 min after addition of xylose. (C) Sequence logo of the σB consensus binding sites (http://dbtbs.hgc.jp/). The height of the letters in bits is proportional to their frequency (48). (D) Sequence of the σB promoter PσB of spo0E. The core promoter regions are marked with light gray boxes and bases matching the consensus were color coded accordingly. Transcriptional start points determined by the primer extension (PE) experiment are marked with arrows. (E) PE experiment to determine the 5′ end of the xylose-induced message observed in the Northern blot experiment. The upper 4 lines show dideoxy-sequencing ladders that were terminated with ddATP (line 1), ddGTP (line 2), ddTTP (line 3), and ddCTP (line 4). The lower 4 lines show reactions with RNA isolated from cells of the BAR 18 and BAR 17 strains under control conditions (co) before and 10 min (10′) after addition of 0.3% xylose.
We focused on spo0E regulation because it is one of many candidate σB regulon members that are preceded by σB-type promoter sequences but failed the criteria of σB-regulated genes in all previous studies (25, 38, 42). The core promoter sequences of PσB can be found about −150 bases upstream from the start codon of spo0E (Fig. 1A and D). PσB is not only perfectly conserved in every position known to be crucial for recognition by σB, but the −35 and −10 core regions are also perfectly separated by a 14-nucleotide spacer (Fig. 1C and D). To test whether the σB-type promoter is functional in vivo, we performed Northern blot experiments first (Fig. 1B). Due to the previous inability of the global approaches to identify spo0E as being a member of the general stress regulon in response to σB-inducing stress stimuli, we decided to create a mutant strain for stress-free ectopic expression of active σB. The first strain, BAR 17, carried a deletion of the σB-autoregulated genes rsbVW-sigB to avoid negative feedback regulation acting on ectopically expressed σB by autoinduction of the anti-anti-sigma factor RsbW. This strain served as a recipient to integrate a chromosomal copy of sigB under transcriptional control of PXylA into the amyE locus of B. subtilis, resulting in strain BAR 18. To further rule out any possible effects caused by AbrB binding to the downstream σA promoter (P2) of spo0E (36), we also introduced a ΔabrB mutation into the BAR 18 background resulting in strain BAR 20. The results of the Northern blots for spo0E transcription in strains BAR 17, BAR 18, and BAR 20 in response to addition of 0.3% xylose are shown in Fig. 1B. One short transcript with a length of approximately 360 bases only could be detected under control conditions as well as 10 or 20 min after addition of xylose in the ΔsigB strain (BAR 17).
The same short transcripts of ∼360 bases were observed at all time points tested for the strains BAR 18 and BAR 20, whereas a second transcript of approximately 440 bases could be detected with the spo0E-specific probe 10 and 20 min after xylose-dependent induction of σB. Furthermore, the signals of the σB-dependent transcript obtained from the ΔabrB mutant strain BAR 20 did not differ from strain BAR 18, thus excluding a possible negative regulatory effect of AbrB on PσB. Nevertheless, an increased transcription rate could be observed for BAR 20 under control conditions compared to BAR 17 and BAR 18, consistent with the observations made previously by Perego and coworkers (36) that transcription originating from P2 is upregulated in a ΔabrB strain.
The starting points of all transcripts observed in the Northern blots were further analyzed and mapped by primer extension (PE) experiments. The transcript(s) of approximately 360 bases length that could be observed for all three strains in the Northern blot experiments clearly corresponded to the two starting points described earlier by Perego and coworkers (36) (data not shown). Furthermore, the PE experiments clearly associated the 5′ end of the xylose-induced transcript with the PσB promoter sequence preceding the spo0E locus (Fig. 1E). These data demonstrate that PσB of spo0E is responsive to active σB in vivo.
Ectopic expression of σB impairs sporulation-specific transcription by induction of spo0E.
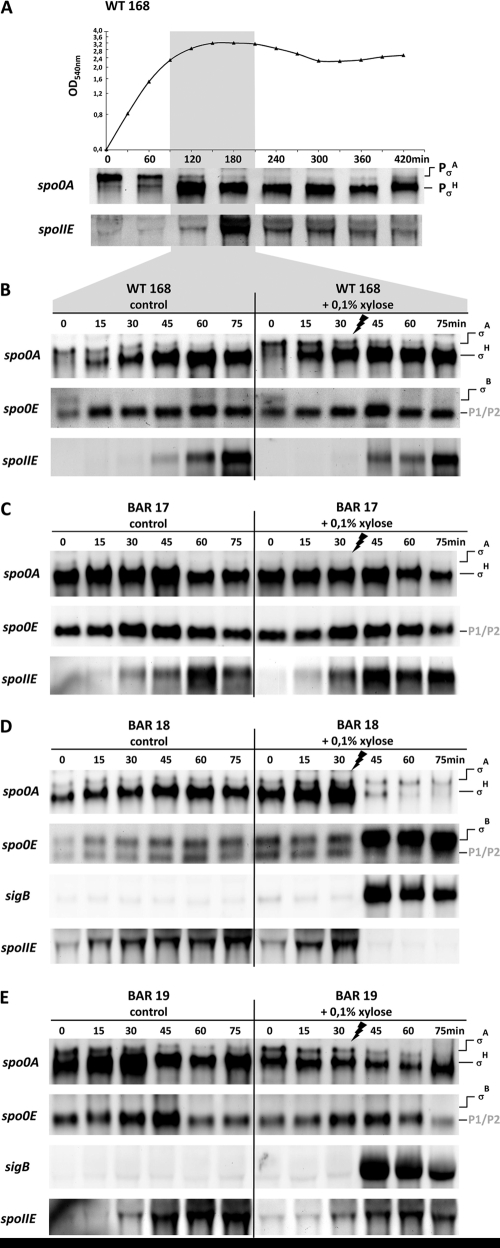
To test whether a negative effect on sporulation initiation is exerted via the σB-mediated expression of spo0E, we followed the transcription kinetics of spo0A, spoIIE, spo0E, and sigB in Northern blot experiments (Fig. 2). Thus, to set up a time frame for our experimental design, we first followed the induction kinetics of spo0A and spoIIE throughout the growth phases of the B. subtilis wild-type strain, 168, grown in Difco sporulation medium (DSM) (Fig. 2A). Transcription of spo0A is driven by two distinct promoters: an upstream σA promoter that is active during vegetative growth (9, 56) and a second sporulation-specific downstream promoter that is recognized by σH-containing RNA polymerase (40, 56) (Fig. 2A and see Fig. 4). The activity of the PσH promoter of spo0A can be taken as both a direct and indirect measurement of Spo0A activity because already low levels of Spo0A∼P (14) stimulate the synthesis of the alternative sigma factor σH (53) by inhibition and inactivation of its transcriptional repressor, AbrB (37, 55) (Fig. 4). Furthermore, spo0A transcription from PσH is also under direct positive and negative control by Spo0A∼P in a concentration-dependent manner (56). Low levels of Spo0A∼P activate and high levels limit spo0A transcription to a constant upper or maximal level (15, 26, 56). The switch from vegetative growth transcription originating from PσA to the sporulation-specific transcript induced from PσH of spo0A can be followed in Fig. 2A. The expression of the second gene, spoIIE, monitored by the Northern blot experiments (Fig. 2A) reflects the presence of high threshold levels of Spo0A∼P (14). In the case of spoIIE transcription, Spo0A∼P acts as a direct activator in conjunction with σA-containing RNA polymerase (39). The kinetics of spoIIE transcription and the temporal shift compared to initiation of spo0A transcription from PσH are depicted in Fig. 2A. In the left panel of Fig. 2B are shown more detailed kinetics with higher time resolution of spo0A, spo0E, and spoIIE transcription of the B. subtilis wild type in the chosen time window after entry into stationary phase. The kinetics most likely reflect that Spo0A∼P levels are low at the beginning, sufficient to activate the positive autoregulatory loop at PσH of spo0A, and continuously increase until high levels of Spo0A∼P are reached about 45 min later, which are necessary for spoIIE induction. To eliminate secondary regulatory mechanisms potentially perturbing the effect caused by σB on sporulation initiation and to clearly address the σB effect on its specific activity at the PσB promoter of spo0E, the stress-free expression system was used again, and one additional mutant strain was created. Thus, besides the B. subtilis wild-type strain 168, the experimental setup included the ΔrsbVW-sigB strain (BAR 17), the strain for ectopic expression of σB (BAR 18), and a strain isogenic to BAR 18, carrying a chromosomal deletion of just 28 nucleotides that erased the PσB core promoter elements but left the remaining regulatory regions P1 and P2 as well as the spo0E locus intact (BAR 19). Transcription of spo0A, spo0E, spoIIE, and sigB was monitored in detailed kinetics every 15 min for a total period of 75 min in stationary-phase cells of the B. subtilis wild type and the three mutant strains under control conditions (Fig. 2B to E, left panels) as well as upon addition of 0.1% xylose 30 min after cells reached t0 (Fig. 2B to E, right panels). No differences could be observed for the control and the xylose-treated cultures of the wild type (Fig. 2B) as well as the ΔsigB operon strain BAR 17 (Fig. 2C). These two control experiments clearly demonstrate that the addition of xylose to stationary-phase cells had no effect on transcription of spo0A, spoIIE, or spo0E. As expected, a different picture emerged for strain BAR 18 carrying the xylose-inducible copy of sigB (Fig. 2D). The Northern blot data show a weak basal-level expression under control conditions due to residual activity of PXylA and a strong but transient induction of sigB upon addition of xylose (Fig. 2D). The transcription pattern of spo0E is consistent with the observed profile for sigB expression. A basal activity for PσB of spo0E could be detected under control conditions followed by a massive increase in transcription rate in correlation with sigB induction (Fig. 2D). With the induction of sigB and spo0E, transcription of spo0A from the sporulation-specific PσH promoter as well as Spo0A∼P-dependent transcription of spoIIE is completely abolished. Finally, the data observed for strain BAR 19 (Fig. 2E) clearly address the negative effect of sigB induction on sporulation-specific transcription to the presence of PσB and the expression of spo0E. Despite the xylose-dependent induction of sigB, no expression of spo0E was induced due to the lack of the PσB sequence, and neither the sporulation-specific transcription of spo0A nor spoIIE was affected, as observed for strain BAR 18 (Fig. 2D). Apart from a slight decrease in the signals detected for spo0A and spoIIE, these data point to the pivotal role of spo0E induction in the σB-mediated mechanism of Spo0A inactivation.
Fig 2.
Northern blot analysis. (A) Growth curve of the B. subtilis wild-type strain 168 in Difco sporulation medium (DSM) and corresponding Northern blot experiments showing the expression profiles of spo0A and spoIIE at the time points t0 (OD540 of 0.4), 60, 120, 180, 240, 300, 360, and 420 min. The two promoters used for differential expression of spo0A are indicated on the right-hand side. The time window of sporulation initiation is boxed (light gray) and was analyzed with more detailed kinetics as shown in panels B to E. (B to E) Northern blot experiments showing detailed time-resolved kinetics of the expression profiles of spo0A, spo0E, spoIIE, and sigB under control conditions (left panels) and in response to addition of 0.1% (wt/vol) xylose (right panels) to stationary-phase cultures of the B. subtilis wild-type strain, 168 (B), as well as its isogenic ΔrsbVW-sigB strain (BAR 17) (C), (D) a ΔrsbVW-sigB strain carrying an amyE::PXylA-sigB integration (BAR 18) (D), and a ΔrsbVW-sigB amyE::PXylA-sigB strain carrying a deletion of the core promoter elements of PσB of spo0E (BARf 19) (E). All strains were grown in DSM, and the respective starting points (t0) for cell sampling were reached with an OD540 of 2.3 for the wild type and BAR 18 as well as an OD540 of 1.9 for the BAR 17 and BAR 19 strains, respectively. Samples for RNA preparation were taken every 15 min for a total period of 75 min, and 0.1% xylose was added to one-half of the cultures 30 min after cells reached t0. The promoters used for differential expression of spo0A and spo0E are indicated on the right side.
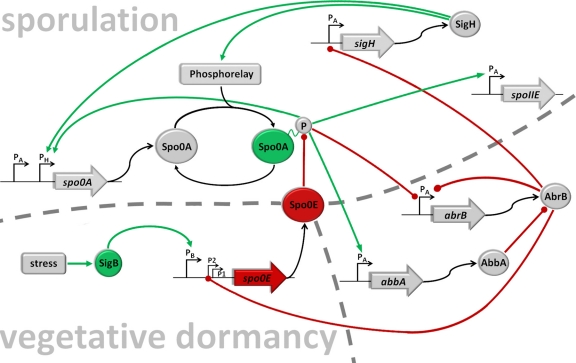
Fig 4.
The regulatory network. Shown is a simplified schematic representation of the core components relevant for the integration of the σB input into the regulatory network that governs initiation of sporulation. Lines ending with a dot (red) represent negative actions, and arrows (green) represent positive actions. To be active, Spo0A needs to be phosphorylated by a multicomponent phosphorelay (7) that integrates positive signals (27) and negative signals (18, 35). The spo0A gene is transcribed from a vegetative σA promoter and a sporulation-specific σH promoter (9, 40). Low levels of Spo0A∼P set in motion two parallel pathways of repression and antirepression that relieve AbrB-mediated repression of sigH (54), by directly repressing transcription of the abrB gene (15, 37, 53) and the induction of abbA encoding an antirepressor of the AbrB protein (3, 32). Derepression of sigH encoding the alternative sigma factor σH results in a positive autoregulatory loop by induction of the spo0A gene itself (15, 40) as well as the phosphorelay components KinA and Spo0F (15, 40). Furthermore, spo0A transcription from PσH is also under direct positive and negative control by Spo0A∼P in a concentration-dependent manner (56). Low levels of Spo0A∼P activate and high levels limit spo0A transcription to a constant upper or maximal level (15, 56). AbrB is a repressor for a wide variety of stationary-phase genes as well as its own synthesis (54). Inactivation of AbrB also causes a negative-feedback loop by derepression of spo0E from P2 (36) encoding a phosphatase Spo0E that directly removes phosphoryl groups from Spo0A∼P (33), converting it from an active to an inactive form; P1 of spo0E is constitutively active at low rates (36), and the third upstream promoter is recognized and induced by σB-containing RNA polymerase (this work). The spoIIE gene is directly activated by high threshold levels of Spo0A∼P from a σA-dependent promoter (10, 14) encoding a protein phosphatase that participates in σF activation.
Ectopic expression of σB produces a sporulation-deficient phenotype by induction of spo0E.
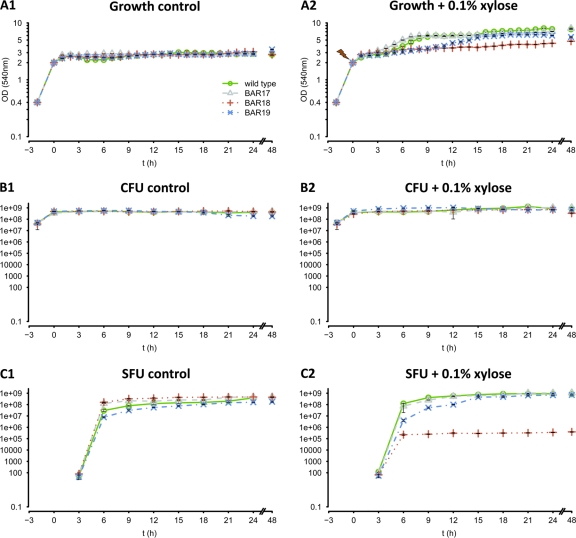
To test whether the observed decrease of spo0A and spoIIE transcription by σB-dependent induction of spo0E also results in a diminished-sporulation phenotype, we monitored sporulation frequencies in all four strains used before in the Northern blot experiments. We monitored growth, the total number of CFU, as well as the total number of spores (SFU) that were formed under control conditions without and with addition of 0.1% xylose (Fig. 3). Two control samples were taken from all cultures: the first during early exponential growth (OD, 0.4) and the second at the onset of the transient phase, when cultures reached an OD of 2.0, which was also set as t0 for all sporulation experiments (Fig. 3). Xylose was added immediately after t0 samples were taken. Growth was monitored every hour, and samples for CFU and SFU determination were taken in 3-h intervals for a period of 24 h, as well as a final sample taken after 48 h. The growth curves of the control experiments (Fig. 3A1) show that no differences could be observed for all four strains tested. All strains entered stationary phase after t0 and retained an OD between 2.2 and 2.8. The same was true for the determined number of CFU (Fig. 3B1). All strains reached a constant upper level between 4 × 108 and 6 × 108 CFU during stationary phase. No spores could be detected from exponentially growing (OD, 0.4) or transient-phase (t0) cells (Fig. 3C1). After 3 h at stationary phase, the first few spores emerged, followed by a drastic increase after 6 and 9 h. All strains reached sporulation frequencies of approximately 100% between 12 and 15 h after entry into stationary phase. Apart from some marginal differences in SFU observed for the 6- and 9-h samples, sporulation kinetics of all strains were strikingly similar under control conditions.
Fig 3.
Sporulation assay. Combination of all relevant data of a sporulation assay for the B. subtilis wild-type strain (green lines and open circles), the rsbVW-sigB mutant strain BAR 17 (gray lines and open triangles), strain BAR 18 carrying the xylose-inducible sigB construct in amyE (red dotted lines and crosses), and strain BAR 19 carrying the sigB promoter deletion ΔPB of spo0E (blue dashed and dotted lines and crosses). The symbol legend is shown in panel A1. The left column shows the control experiments (A1, growth; B1, CFU; C1, spore-forming units [SFU]) for all strains grown in DSM without the addition of xylose. The right column (A2, B2, and C2) shows the corresponding data for all strains grown in DSM with the addition of 0.1% xylose when cultures reached an OD540 of 2.0. A control sample was taken for all experiments at an OD540 of 0.4, and t0 was set when cultures reached an OD540 of 2.0. After this, growth was monitored hourly, and samples for CFU and SFU determination were taken every 3 h for 24 h, and a final sample was taken after 48 h. All experiments were performed in triplicate, and error bars are depicted.
Different results could be observed for the xylose-treated cultures. After addition of 0.1% xylose, all strains seemed to slowly continue growth after a short lag phase of approximately 2 h (Fig. 3A2). The ΔrsbVW-sigB mutant (BAR 17) and the wild-type strain reached their ultimate stationary phase approximately 8 h after addition of xylose, with final ODs between 7 and 7.5. The promoter deletion strain ΔPB (BAR 19) reached its stationary phase approximately 15 h after addition of xylose, with final ODs between 5.8 and 6.1, whereas the ODs of strain BAR 18 carrying the xylose-inducible construct of sigB continuously increased throughout the whole period observed but never exceeded values above 4.7 (Fig. 3A2). Surprisingly, despite the fact that different ODs could be observed for the four strains after addition of xylose, no such effect appeared for the determined CFU (Fig. 3B2). Unless the CFU of the xylose-treated cultures (Fig. 3B2) were consistently higher than those of the control cultures (Fig. 3B1), indicating slow continued growth, they did not differ significantly among each other. Thus, the observed differences in the ODs between the four strains after addition of xylose cannot be simply explained by growth phenomena or different cell densities. The most obvious explanation for this effect is probably the different sporulation kinetics of the four strains observed after addition of xylose, which seems to correlate with the respective ODs (Fig. 3A2 and C2). Consistent with the results of the control cultures, no spores could be detected from exponentially growing (OD, 0.4) or transient-phase cells (t0) (Fig. 3C2). After 3 h, similar amounts of the first few spores could be detected for all strains. After this, the ΔrsbVW-sigB mutant (BAR 17) and the wild type show slightly increased sporulation frequencies after 6 and 9 h compared to the control cultures (Fig. 3C1) as well as the ΔPB strain, BAR 19 (Fig. 3C2). Whereas strain BAR 19 converges to wild-type levels within the next 9 h, sporulation frequencies in strain BAR 18 are drastically and constantly decreased by approximately 3 log levels, or 99.9%, compared to the wild type throughout the whole period of 48 h observed. Thus, the σB-dependent induction of spo0E and the decrease of sporulation-specific spo0A and spoIIE transcription indeed cause a clear sporulation-deficient phenotype. With respect to this, it is interesting to note that the CFU level of strain BAR 18 remains approximately stable throughout the observed time course of 48 h, indicating that the large amount of remaining vegetative cells (∼3 × 108 to 6 × 108 per ml) do not increasingly die or lyse, but seem to continue in a nongrowing vegetative dormant state.
DISCUSSION
According to our results, the alternative sigma factor σB is able to induce the spo0E gene encoding a phosphatase specifically inactivating the sporulation master regulator Spo0A∼P (33) (Fig. 4). We could show that ectopic induction of σB activity significantly decreased sporulation-specific transcription of spo0A and spoIIE and produces a clear sporulation-deficient phenotype. Furthermore, a deletion of the σB-dependent promoter restored spo0A and spoIIE transcription as well as sporulation frequencies to wild-type levels.
Notably, all previous global approaches failed to assign the spo0E gene to the σB regulon, including our detailed transcriptome and proteome analysis in response to ethanol stress (25, 38, 42, 44). These problems were most likely due to the complex control of spo0E from three different promoters and the resulting heterogeneous population of spo0E transcripts. Certainly the use of tiling arrays in future global approaches will circumvent the problem of signal attribution to distinct promoters, at least for a large portion of genes that are under complex control. Furthermore, Spo0E is a relatively small protein of 9.6 kDa and can only be found at the lower detection limit of two-dimensional (2D) gels, and thus it just cannot be visualized properly by gel-based global proteome approaches. The central question that arises from these observations is whether PσB of spo0E is responsive to physiological σB-inducing stimuli. Besides the clear activity of the σB-type promoter upon ectopic σB expression, a σB-dependent transcript of spo0E, although weak, can be observed at t0 of the Northern blot experiments for the wild type (Fig. 2B) and is lacking in the sigB mutant strain BAR 17 (Fig. 2C), as well as the ΔPσB mutant BAR 19 (Fig. 2E). The detected transcript corresponds to the onset of the transient phase, and induction is most likely caused by nutrient limitation activating σB. Thus, already indicating that σB-dependent induction of spo0E is given under physiological conditions, a comprehensive screening of PσB promoter activity under various σB-inducing stimuli will be part of detailed future studies. In this context, it will also be of great importance to investigate whether a physiological condition exists that produces a clear σB-dependent sporulation-deficient phenotype for which the ΔPσB promoter deletion mutant of spo0E will be a helpful tool to arrive at these findings. At least some σB-activating conditions, including ethanol (4), cold (6a, 31a), and salt (29, 45) stresses, were reported previously to cause sporulation defects. Unpublished data on ethanol stress experiments already point to a pivotal role of the newly identified regulatory pathway in blocking sporulation under stress conditions, but due to the complexity caused by multiple pleiotropic regulatory effects, these results will be published elsewhere (unpublished data). Nevertheless, new roles for Spo0A in stress adaptation and stationary-phase survival of B. subtilis have also been indicated under various stress conditions that were independent of Spo0A function in spore formation (31a).
Taken together, our data shown here represent another example that multiple opportunities for signal integration are provided in the regulatory network necessary for a finely tuned target gene expression in the context of decision-making processes. In this context, we suggest that sporulation and σB-dependent stress adaptation are interconnected pathways that allow an adequate response of B. subtilis to its fluctuating environment. In essence, for the first time a molecular mechanism is shown that sheds a new light on the discussion about the role of the general stress response for stationary-phase adaptation and long-term survival (Fig. 4). We propose that sporulation is the last resort adaptation for cells in response to starvation conditions, contrary to a “vegetative dormant” state that may be preferred under conditions of long periods of physical stress. The state of a vegetative dormant and stress-resistant cell could be an essential part of a survival strategy alternative to sporulation in B. subtilis (22, 41).
ACKNOWLEDGMENTS
We are grateful to R. Losick for helpful advice and D. Höper, S. Michalik, and A. Elsholz for great discussions on this work.
This work was supported by the BMBF (FK2:0313978A) and the DFG (HE 1887/7-4 and HE 1887/8-1).
A.R. designed and performed experiments, analyzed data, and wrote the paper. U.G. and M.H. supervised the project. All authors discussed the results and implications and commented on the manuscript at all stages.
The authors declare they have no competing financial interests.
Footnotes
Published ahead of print 30 December 2011
REFERENCES
- 1. Albano M, Hahn J, Dubnau D. 1987. Expression of competence genes in Bacillus subtilis. J. Bacteriol. 169:3110–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antelmann H, Engelmann S, Schmid R, Hecker M. 1996. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J. Bacteriol. 178:6571–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Banse AV, Chastanet A, Rahn-Lee L, Hobbs EC, Losick R. 2008. Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 105:15547–15552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bohin JP, Rigomier D, Schaeffer P. 1976. Ethanol sensitivity of sporulation in Bacillus subtilis: a new tool for the analysis of the sporulation process. J. Bacteriol. 127:934–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reference deleted.
- 6. Brigulla M, et al. 2003. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185:4305–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a. Budde I, Steil L, Scharf C, Völker U, Bremer E. 2006. Adaption of Bacillus subtilis to growth at low temperature: a combined transcriptomic and proteomic appraisal. Microbiology 152:831–853 [DOI] [PubMed] [Google Scholar]
- 7. Burbulys D, Trach KA, Hoch JA. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552 [DOI] [PubMed] [Google Scholar]
- 8. Burkholder PR, Giles NH. 1947. Induced biochemical mutations in Bacillus subtilis. Am. J. Bot. 34:345–348 [PubMed] [Google Scholar]
- 9. Chibazakura T, Kawamura F, Takahashi H. 1991. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J. Bacteriol. 173:2625–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chung JD, Stephanopoulos G, Ireton K, Grossman AD. 1994. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J. Bacteriol. 176:1977–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dworkin J, Losick R. 2005. Developmental commitment in a bacterium. Cell 121:401–409 [DOI] [PubMed] [Google Scholar]
- 12. Engelmann S, Hecker M. 1996. Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol. Lett. 145:63–69 [DOI] [PubMed] [Google Scholar]
- 13. Fawcett P, Eichenberger P, Losick R, Youngman P. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 97:8063–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujita M, Gonzalez-Pastor JE, Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187:1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujita M, Sadaie Y. 1998. Feedback loops involving Spo0A and AbrB in in vitro transcription of the genes involved in the initiation of sporulation in Bacillus subtilis. J. Biochem. (Tokyo) 124:98–104 [DOI] [PubMed] [Google Scholar]
- 15a. Gaidenko TA, Price CW. 1998. General stress transcription factor sigma(B) and sporalation transcripton factor sigma(H) each contribute to survival of Bacillus subtilis under extreme growth conditions. J. Bacteriol. 180:3730–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gonzalez-Pastor JE, Hobbs EC, Losick R. 2003. Cannibalism by sporulating bacteria. Science 301:510–513 [DOI] [PubMed] [Google Scholar]
- 17. Reference deleted.
- 18. Grossman AD. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477–508 [DOI] [PubMed] [Google Scholar]
- 19. Hamoen LW, Venema G, Kuipers OP. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9–17 [DOI] [PubMed] [Google Scholar]
- 20. Hauser NC, et al. 1998. Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast 14:1209–1221 [DOI] [PubMed] [Google Scholar]
- 21. Reference deleted.
- 22. Hecker M, Pane-Farre J, Völker U. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61:215–236 [DOI] [PubMed] [Google Scholar]
- 23. Hecker M, Reder A, Fuchs S, Pagels M, Engelmann S. 2009. Physiological proteomics and stress/starvation responses in Bacillus subtilis and Staphylococcus aureus. Res. Microbiol. 160:245–258 [DOI] [PubMed] [Google Scholar]
- 24. Hecker M, Völker U. 1990. General stress proteins in Bacillus subtilis. FEMS Microbiol. Ecol. 74:197–213 [Google Scholar]
- 25. Helmann JD, et al. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318–7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoch JA. 1991. Spo0 genes, the phosphorelay, and the initiation of sporulation, p 747–755 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and other Gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC [Google Scholar]
- 26a. Holtmann G, et al. 2004. RsbV-independent induction of the SigB-dependent general stress regulon of Bacillus subtilis during growth at high temperature. J. Bacteriol. 186:6150–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26b. Höper D, Völker U, Hecker M. 2005. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis at low growth temperature. J. Bacteriol. 187:2810–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang M, Shao WL, Perego M, Hoch JA. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535–542 [DOI] [PubMed] [Google Scholar]
- 28. Kim L, Mogk A, Schumann W. 1996. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181:71–76 [DOI] [PubMed] [Google Scholar]
- 29. Kunst F, Rapoport G. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reference deleted.
- 31. Majumdar D, Avissar YJ, Wyche JH. 1991. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA—a new approach for isolating DNA. Biotechniques 11:94–101 [PubMed] [Google Scholar]
- 31a. Mendez MB, Orsaria LM, Philippe V, Pedrido ME, Grau RR. 2004. Novel roles of the master transcription factors SpoOA and sigma(B) for survival and sporulation of Bacillus subtilis at low growth temperature. J. Bacteriol. 186:989–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Molle V, et al. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683–1701 [DOI] [PubMed] [Google Scholar]
- 33. Ohlsen KL, Grimsley JK, Hoch JA. 1994. Deactivation of the sporulation transcription factor Spo0A by the Spo0E protein phosphatase. Proc. Natl. Acad. Sci. U. S. A. 91:1756–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parker GF, Daniel RA, Errington J. 1996. Timing and genetic regulation of commitment to sporulation in Bacillus subtilis. Microbiology 142:3445–3452 [DOI] [PubMed] [Google Scholar]
- 35. Perego M, Hoch JA. 2002. Two-component systems, phosphorelays, and regulation of their activities by phosphatases, p 473–481 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 36. Perego M, Hoch JA. 1991. Negative regulation of Bacillus subtilis sporulation by the spo0E gene product. J. Bacteriol. 173:2514–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perego M, Spiegelman GB, Hoch JA. 1988. Structure of the gene for the transition-state regulator, AbrB—regulator synthesis is controlled by the Spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689–699 [DOI] [PubMed] [Google Scholar]
- 38. Petersohn A, et al. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Piggot PJ, Losick R. 2002. Sporulation genes and intercompartmental regulation, p 483–517 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 40. Predich M, Nair G, Smith I. 1992. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing sigma H. J. Bacteriol. 174:2771–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Price CW. 2000. Protective function and regulation of general stress response in Bacillus subtilis and related Gram-positive bacteria, p 179–197 In Storz G, Hengge-Aronis R. (ed), Bacterial stress responses. ASM Press, Washington, DC [Google Scholar]
- 42. Price CW, et al. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757–774 [DOI] [PubMed] [Google Scholar]
- 43. Reference deleted.
- 44. Reder A, et al. 2008. The Spx paralogue MgsR (YqgZ) controls a subregulon within the general stress response of Bacillus subtilis. Mol. Microbiol. 69:1104–1120 [DOI] [PubMed] [Google Scholar]
- 45. Ruzal SM, López C, Rivas E, Sánchez-Rivas C. 1998. Osmotic strength blocks sporulation at stage II by impeding activation of early sigma factors in Bacillus subtilis. Curr. Microbiol. 36:75–79 [DOI] [PubMed] [Google Scholar]
- 46. Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schaeffer P, Millet J, Aubert JP. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 54:704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schultz D, Wolynes PG, Ben Jacob E, Onuchic JN. 2009. Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 106:21027–21034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a. Shafikhani SH, Mandic-Mulec I, Strauch MA, Smith I, Leighton T. 2002. Postexponential regulation of sin operon expression in Bacillus subtilis. J. Bacteriol. 184:564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stephens C. 1998. Bacterial sporulation: a question of commitment? Curr. Biol. 8:R45–R48 [DOI] [PubMed] [Google Scholar]
- 51. Stragier P. 2006. To kill but not be killed: a delicate balance. Cell 124:461–463 [DOI] [PubMed] [Google Scholar]
- 52. Stragier P, Bonamy C, Karmazyn-Campelli C. 1988. Processing of a sporulation sigma factor in Bacillus subtilis—how morphological structure could control gene expression. Cell 52:697–704 [DOI] [PubMed] [Google Scholar]
- 53. Strauch M, Webb V, Spiegelman G, Hoch JA. 1990. The Spo0A protein of Bacillus subtilis is a repressor of the AbrB gene. Proc. Natl. Acad. Sci. U. S. A. 87:1801–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Strauch MA. 1995. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promoter. J. Bacteriol. 177:6999–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Strauch MA, et al. 1989. The transition-state transcription regulator AbrB of Bacillus subtilis is a DNA-binding protein. EMBO J. 8:1615–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Strauch MA, Trach KA, Day J, Hoch JA. 1992. Spo0A activates and represses its own synthesis by binding at its dual promoters. Biochimie 74:619–626 [DOI] [PubMed] [Google Scholar]
- 57. Stülke J, Hanschke R, Hecker M. 1993. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139:2041–2045 [DOI] [PubMed] [Google Scholar]
- 58. Veening JW, et al. 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl. Acad. Sci. U. S. A. 105:4393–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reference deleted.
- 60. Völker U, Maul B, Hecker M. 1999. Expression of the sigmaB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259–265 [DOI] [PubMed] [Google Scholar]
- 62. Watson N. 1988. A new revision of the sequence of plasmid pBR322. Gene 70:399–403 [DOI] [PubMed] [Google Scholar]
- 63. Wetzstein M, et al. 1992. Cloning, sequencing, and molecular analysis of the DnaK locus from Bacillus subtilis. J. Bacteriol. 174:3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]