Abstract
The recently discovered CRISPR-Cas adaptive immune system is present in almost all archaea and many bacteria. It consists of cassettes of CRISPR repeats that incorporate spacers homologous to fragments of viral or plasmid genomes that are employed as guide RNAs in the immune response, along with numerous CRISPR-associated (cas) genes that encode proteins possessing diverse, only partially characterized activities required for the action of the system. Here, we investigate the evolution of the cas genes and show that they evolve under purifying selection that is typically much weaker than the median strength of purifying selection affecting genes in the respective genomes. The exceptions are the cas1 and cas2 genes that typically evolve at levels of purifying selection close to the genomic median. Thus, although these genes are implicated in the acquisition of spacers from alien genomes, they do not appear to be directly involved in an arms race between bacterial and archaeal hosts and infectious agents. These genes might possess functions distinct from and additional to their role in the CRISPR-Cas-mediated immune response. Taken together with evidence of the frequent horizontal transfer of cas genes reported previously and with the wide-spread microscale recombination within these genes detected in this work, these findings reveal the highly dynamic evolution of cas genes. This conclusion is in line with the involvement of CRISPR-Cas in antiviral immunity that is likely to entail a coevolutionary arms race with rapidly evolving viruses. However, we failed to detect evidence of strong positive selection in any of the cas genes.
INTRODUCTION
CRISPR-Cas is a recently discovered adaptive immune system that is present in almost all archaea and many bacteria (7, 10, 37). A striking feature of the CRISPR-Cas system is that it can “remember” the identity of infectious agents (such as viruses and plasmids) by incorporating DNA sequences derived from the genomes of such agents (and possibly alien DNA in general) into the genome of a prokaryotic host. The CRISPR-Cas system thus allows prokaryotic cells to acquire information about the external environment (or more precisely, alien DNA present in the environment), incorporate this information into the host genome, and thereby transmit it to the progeny. Thus, CRISPR-Cas clearly exemplifies the principle of Lamarckian inheritance (28).
The CRISPR-Cas module is in the genome of archaea and bacteria in two parts, namely, arrays of repeat sequences known as clustered, regularly interspaced, short palindromic repeats (CRISPRs) and genes encoding CRISPR-associated (Cas) proteins (1, 26, 38). The operation of the CRISPR-Cas immune system can be divided into three functionally distinct stages, namely, adaptation, expression, and interference, each carried out through interactions between CRISPRs, their transcripts, Cas proteins, and foreign DNA. At the adaptation stage, DNA sequences of about 30 bp (called spacers) that are homologous to certain regions (called protospacers) in the genomes of infectious agents are incorporated into a CRISPR locus (7). The incorporation of a spacer is accompanied by the duplication of a similarly sized, CRISPR-constitutive repeat sequence, which joins the incoming spacer to an existing spacer, thereby elongating the CRISPR cassette by one unit. At the expression stage, CRISPR loci are transcribed, and the transcripts are processed into small RNA molecules (called crRNAs), which bind to an enzymatic complex consisting of Cas proteins, known as CASCADE (10, 13, 24). At the interference stage, a crRNA directs the bound CASCADE complex along with an additional Cas protein (Cas3) to destroy the foreign DNA or in some cases RNA after the crRNA forms a duplex with the cognate protospacer sequence (10, 23, 27, 64).
The acquisition of spacers at the adaptation stage is the critical step at which the distinction between self and nonself is made (also see reference 40). Otherwise, autoimmunity would ensue through the incorporation of the host's own DNA into the CRISPR loci; such self targeting indeed has been detected but appears to be extremely rare (58). The selection of protospacers in foreign DNA sequences is nonrandom: protospacers often are located adjacent to short, conserved motifs called protospacer adjacent motifs (PAMs), which are implicated in the selection of protospacers (14, 45). Moreover, PAMs are necessary for the recognition of foreign DNA sequences during the interference stage (14).
The CRISPR-Cas systems show remarkable diversity of protein sequences and genomic organization of the cas operons. At least 45 distinct protein families have been identified in association with CRISPR loci in various bacterial and archaeal genomes (22). Further analyses involving more sensitive methods of sequence and structure comparison supplemented by the analysis of cas operon architectures have revealed distant homologous relationships between many Cas protein families (33, 34). The recently developed classification divides CRISPR-Cas systems into three distinct types (I, II, and III) (35). All of these systems contain two universal genes: cas1, a metal-dependent DNase that is implicated, with no sequence specificity, in the integration of protospacers into CRISPR cassettes (39, 65); and cas2, a metal-dependent endoribonuclease that also appears to be involved in the adaptation stage (8). Apart from the conservation of cas1 and cas2, the three types of CRISPR-Cas systems substantially differ in their sets of constituent genes, and each is characterized by a unique signature gene (35). The signature genes for the three types are cas3 (a superfamily 2 helicase containing an N-terminal HD superfamily nuclease domain) (57), cas9 (a large protein containing a predicted RuvC-like and HNH nuclease domains), and cas10 (a protein containing a domain homologous to the palm domain of nucleic acid polymerases and nucleotide cyclases), respectively (35). Within the three major types, CRISPR-Cas systems can be further classified into subtypes based on a number of criteria, which include distinct signature genes along with the phylogeny of the universal cas1 gene (35). The Cas proteins known as RAMPs (for repeat-associated mysterious proteins) are present in several copies in both type I and III systems. Some of the RAMP proteins have been shown to possess sequence- or structure-specific RNase activity that is involved in the processing of pre-crRNA transcripts (10, 11, 24). The crystal structures of several RAMPs have been solved and shown to contain one or two RNA recognition motif (RRM) domains that show substantial structural variations in different Cas proteins (24, 32, 34, 53, 63).
The CRISPR-Cas modules could be expected to undergo rapid evolution in natural environments because of recurrent selection pressure exerted by coevolving viruses (20, 49). This expectation appears to be consistent with the extreme diversity of Cas protein sequences and structures. Moreover, in accord with the prediction of rapid evolution, it has been shown that the spacer composition of CRISPRs in biofilm-forming, acidophilic archaea evolves rapidly in a natural environment (62), turning over on a time scale of months (4). In addition, the evidence for the horizontal gene transfers (HGTs) of CRISPR-Cas modules has been accumulated by comparative sequence analyses focusing on various taxonomic ranks ranging from phyla to strains, indicating that the CRISPR-Cas modules undergo HGT on various evolutionary timescales (12, 19, 61, 62). However, in contrast to these observations, which are compatible with the rapid evolution of CRISPR-Cas modules, it has been shown that the spacer compositions of CRISPRs in Escherichia coli and Salmonella enterica evolve at a much slower rate, remaining unchanged for 103 to 105 years (60, 61). Because such slow evolution is at odds with the expectation for an active immune system interacting with evolving viruses, this finding led to the suggestion that, at least in some organisms, the CRISPR-Cas system could perform functions other than defense against infectious agents (60), a case in point being the reported involvement of Cas1 in DNA repair (6). Given these contrasting findings on the pace of CRISPR evolution, we sought to investigate the microevolution of the cas genes to gain further insights into tempo and mode in the evolution of different variants of the CRISPR-Cas system and potentially into the functions of cas genes.
In this work, we systematically examined the nature and intensity of selection pressure that affects different cas genes by estimating the ratio of nonsynonymous to synonymous substitutions (dN/dS), the generally accepted gauge of the type and strength of selection in the evolution of genes and individual amino acid sites. The results indicate that cas genes generally are subject to purifying selection, the intensity of which, however, varies greatly depending on the gene family and significantly differs between the stages of CRISPR immunity in which the genes are involved. Most of the cas genes evolve under much weaker selection pressure than the average selection pressure exerted on genes in the respective bacterial and archaeal genomes. However, we did not detect evidence of strong positive selection in any of the cas genes.
MATERIALS AND METHODS
Genomic data.
The completely sequenced genomes of 1,164 bacteria and archaea were downloaded from the NCBI FTP site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria) in August 2010. The profiles of 52 Cas proteins (or domains) reported by Makarova et al. (35) (ftp://ftp.ncbi.nih.gov/pub/wolf/_suppl/CRISPRclass/index.html) were obtained from Pfams (17) and TIGRFAMs (22, 55).
Gene sequences.
The Cas profiles were searched against the genomes using PSI-BLAST (3) (E value, 10−6), with the consensus sequence of each profile used as the master sequence. To remove false positives, the hits were searched against the Conserved Domains Database (CDD) using RPS-BLAST (36) (E value, 10−6) with hits to the NCBI Protein Clusters Database discarded. A PSI-BLAST hit was considered a bona fide Cas protein if a set of nonoverlapping, best-match profiles obtained as RPS-BLAST hits included (i) one of the profiles used in the previous PSI-BLAST search and/or (ii) the profile of a Cluster of Orthologous Groups of protein (COG) (59) describing this Cas protein (35). To increase the sample size, the obtained Cas sequences were searched against the genomes using BLASTP (2) with a stringent E value cutoff of 10−10, and the hits were considered additional true Cas sequences. All Cas sequences were pooled to remove redundancy and clustered with the BLASTClust program (ftp://ftp.ncbi.nlm.nih.gov) (similarity, >70%; bidirectional coverage, >90%). The clusters were categorized into groups, each representing a single Cas protein or a concatenation of multiple Cas proteins (domains), as follows. Every sequence of a cluster was searched against the profiles used in the previous PSI-BLAST search and those of COGs describing a single Cas protein with RPS-BLAST (E value, 10−1). If the union of nonoverlapping, best-match profiles consisted of multiple profiles, the cluster was considered to belong to a group representing the concatenation of the respective proteins (domains); if it consisted of a single profile, the assignment was done straightforwardly.
Orthology assignment.
For each cluster, the protein sequences were aligned with the MUSCLE program (16). DNA sequences were aligned based on the protein sequence alignments with the tranalign program from EMBOSS (52). The DNA sequences containing frame shifts were discarded together with the respective protein sequences. Protein sequences that were identical to each other were removed, except for one sequence, together with the respective DNA sequences. Phylogenetic trees were estimated from the protein alignments with the PhyML program (21). The trees were approximately rooted by the least-square distance method of Wolf et al. (66). For each tree, every monophyletic group of genes that reside in an identical genome was collapsed into one operational taxonomic unit (OTU) (inparalogs). Subsequently, every monophyletic group of genes each of which resides in a distinct genome belonging to an identical genus was considered a group of orthologous genes. To ensure a uniform level of sequence divergence within every orthologous group, genomes belonging to an identical Alignable Tight Genomic Cluster (AGTC) (46) were considered to belong to an identical genus (in the current data set, Salmonella, Citrobacter, Shigella, and Escherichia belonged to a single ATGC, and so did Nostoc and Anabaena). Otherwise, taxonomic classification was obtained from the NCBI Taxonomy Database (54). The procedure of orthology assignment described above was based on the assumption that the divergence time between genomes was too short to allow duplication followed by differential loss below the level of a genus (i.e., HGTs within a genus were ignored).
Detection of recombination events.
The aligned DNA sequences were examined for recombination signals with the RDP3 software (42). Recombination signals were accepted if at least 5 different methods detected statistically significant (P < 0.05) evidence of recombination (9, 18, 41, 43, 44, 48, 51). Based on the description in RDP3, MaxChi and Chimaera were considered the same method, and so were Chimaera and 3SEQ. MaxChi and 3SEQ were, however, considered different methods (no transitivity assumed). If a recombination signal was detected, all sequences involved in it (i.e., potential parental sequences and recombinants) were removed from a cluster, and the remaining sequences were examined with RDP3 again. A cycle of recombination detection and sequence removal was repeated until no recombination signals were detected in every alignment consisting of 3 or more DNA sequences (RDP3 cannot examine alignments consisting of 2 sequences because it is difficult, if not impossible, to detect recombination from 2 sequences). The sequences with recombination signals were discarded to improve the quality of the dN/dS ratio estimation (5, 56).
Estimation of the dN/dS ratios of cas genes.
The dN/dS ratios were estimated with the maximum-likelihood method implemented in the PAML program (67) by comparing all possible pairs of DNA sequences within each cluster. Estimations yielding the number of synonymous substitutions per synonymous site (dS) that fell outside the range of 0.25 ≤ dS ≤ 1.5 were discarded to improve the quality of estimation (when the value of dS was too small, the dN/dS ratio seemed to be overestimated because of the inflation of a quotient by a small denominator; conversely, when the value of dS was too large, the estimation of dN/dS is not reliable because of saturation in synonymous sites). To compare the dN/dS ratios for different classes of genes, three statistical methods were applied: Mann-Whitney U test with the Holm-Bonferroni correction (25), t test with the Holm-Bonferroni correction, and the Dunnett T3 procedure of the Tukey-Kramer test (15).
Estimation of the genomic distributions of dN/dS ratios.
To map the dN/dS ratios of cas genes onto the genomic distributions of dN/dS ratios, dN/dS ratios were estimated for each gene from the respective pairs of genomes. The pairs of genomes were initially selected such that at least one of the pairs contained the cas genes for which the dN/dS ratios were estimated in the previous step. For each pair of genomes, a reciprocal BLASTP (2) search was done, and orthology was assigned to genes according to the bidirectional best-hit criterion (29). The dN/dS ratios of the orthologous genes were estimated with PAML (67) as described above; in this case, a search for recombination events was not performed because all subsequent analysis used only the median of the genomic distributions that would not be substantially affected by a small fraction of genes with detectable recombination (the median, in general, is robust to extreme values). If the genomic median of the dS values fell outside the range of 0.25 ≤ dS ≤ 1.5, such a pair of genomes was discarded to improve the quality of the estimation. Because many pairs fell outside this range, the scope of selections was extended to genomes belonging to the same genus (54) as that of the discarded genomes, assuming that organisms within the same genus have similar genomic distributions of dN/dS ratios (at least with respect to median values). Consequently, 39 genomic distributions of dN/dS ratios were obtained with the following characteristics. The median ranged from 0.025 to 0.11 with a mean (± standard deviation [SD]) value of 0.065 ± 0.021. The scaled median absolute deviation (MAD) ranged from 0.023 to 0.062 with a mean (±SD) value of 0.044 ± 0.011 (a scaling factor of 1.48 was used, because 1.48 MAD ≈ SD if the population distribution is Gaussian). The medians of the dN/dS ratio distributions were used to scale the dN/dS ratios of cas genes.
Estimation of the site-specific dN/dS ratios in cas genes.
Site-specific dN/dS ratios were estimated for the clusters consisting of at least 4 sequences with PAML (67) using 4 models, namely, M1a, M2a, M7, and M8, and unrooted phylogenetic trees estimated in the previous step. The likelihood ratio test was done between M1a and M2a and between M7 and M8 to detect evidence of positive selection. Sites were considered positively selected if the posterior probability for a site to be under positive selection was above 0.95 (with no correction for multiple comparisons).
RESULTS AND DISCUSSION
Selection pressure on different families of cas genes as measured by dN/dS ratios.
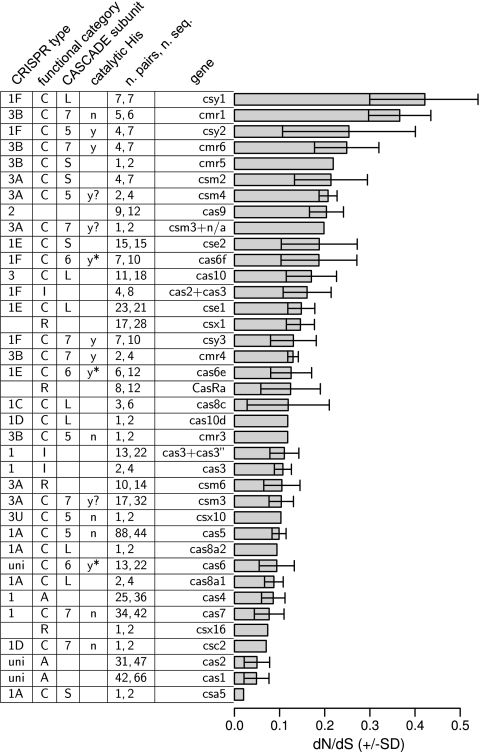
The dN/dS ratio is a gauge of selection affecting proteins under the assumption that synonymous sites in protein-coding sequences evolve neutrally (31, 68). The dN/dS ratios of all examined cas genes were less than 1 (Fig. 1), indicating that the cas genes generally evolve under purifying selection. However, the ratios varied greatly among the cas genes, covering a range between 0.05 and 0.3. This range spanned roughly 4 to 10 times the scaled median absolute deviation (MAD) of the genomic (gene by gene) distribution of dN/dS ratios: in the genomic distributions estimated from each of the 39 analyzed pairs of genomes, the scaled MAD ranged from 0.023 to 0.062 with a mean (±SD) value of 0.044 ± 0.011 (see Materials and Methods). This large variability of the dN/dS ratio among the cas genes likely reflects their diverse functions during different stages of CRISPR-Cas immune response and possibly roles beyond the immune response, such as the apparent involvement of Cas1 in DNA repair (6).
Fig 1.
dN/dS ratios of various cas genes. The table next to the bar plot shows various types of information about the genes. The first column (CRISPR type) describes the CRISPR-Cas type and subtypes in which the respective cas genes are represented. uni refers to the genes present in all types of CRISPR-Cas systems. The second column (functional category) describes the functional categories to which the respective cas genes belong: adaptation (A), CASCADE subunits (C), interference (I), and regulation (R). The third column (CASCADE subunit) describes the groups of CASCADE genes to which the respective cas genes belong: the large subunit (L), small subunit (S), cas5 (5), cas6 (6), and cas7 (7). The fourth column (catalytic His) describes the presence or absence of predicted catalytic histidine: y* (a site is present and a gene has been demonstrated as a nuclease), y (a site is present but a gene has not been demonstrated as an enzyme), y? (a site was detected to be present by Makarova et al. [35] but was not detected in the current data set), and n (a site is absent). The fifth column (n. pairs, n. seq) shows the number of estimated dN/dS ratios and the total number of sequences from which the ratios were estimated. The sixth column (gene) shows the names of the cas genes according to Makarova et al. (35). Plus signs indicate concatenated genes.
The cas genes were classified into various groups on the basis of functional as well as structural features (Fig. 1) (33). The genes first were divided into four groups: (i) genes involved in the adaptation stage, (ii) genes involved in the interference stage, (iii) genes encoding CASCADE subunits, and (iv) genes encoding predicted transcription factors (regulation genes). The CASCADE gene group, the largest of the four groups, was further classified in two ways. The first classification included five gene groups: (i) genes for the large subunit of the CASCADE complex (also known as the CRISPR polymerase), (ii) genes for the small subunit of the CASCADE complex, and (iii to v) three groups of RAMP proteins, namely, the cas5, cas6, and cas7 families. In the second classification, the CASCADE subunits were partitioned into genes from type I CRISPR-Cas systems and genes from type III CRISPR-Cas systems (35). Finally, the RAMP genes (cas5, cas6, and cas7 groups) were reclassified according to the presence and absence of demonstrated or predicted enzymatic activity: demonstrated nucleases with a catalytic His (RAMP w/H*), predicted nucleases with a highly conserved His (RAMP w/H), and proteins without conserved His (RAMP w/o H). The cas9 gene was not included in any of these groups, because it shows no detectable similarity to any other cas gene (33).
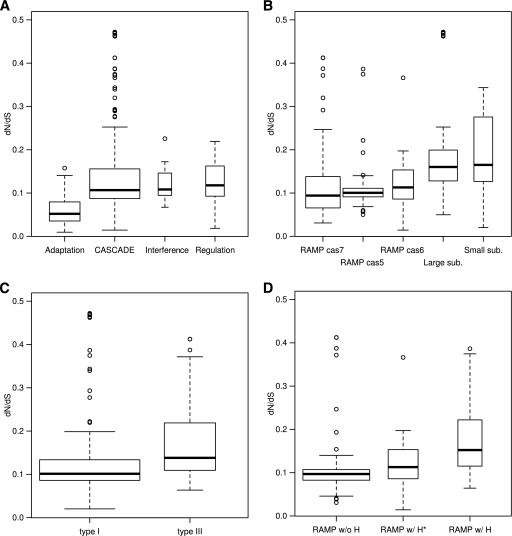
These different groups of cas genes displayed significant variations in dN/dS ratios (Fig. 2). The adaptation genes, especially cas1 and cas2, had significantly lower dN/dS ratios than the genes in all other groups (Fig. 2A) (P < 1.4 × 10−7 with the Mann-Whitney U test, P < 1.9 × 10−4 with the t test, and P = 1.2 × 10−6 with the Dunnett T3 test) (see Materials and Methods). These low dN/dS ratios indicate the slower evolution of protein sequences and accordingly stronger purifying selection (assuming that the effect of positive selection is negligible when considered on a whole-gene level). Therefore, this result indicates that cas1, cas2, and cas4 are under the strongest purifying selection of all cas genes. Although cas1, cas2, and cas4 appear to be involved in the adaptation stage of the CRISPR-Cas immune response, the finding of relatively strong purifying selection is poorly compatible with the hypothesis that these genes are directly involved in coevolution with infectious agents. Rather, there is a parallel between the universality of cas1 and cas2 in various types of CRISPR-Cas and stronger purifying selection exerted on them, in that both features, albeit from distinct angles and on different evolutionary scales, point to the strong evolutionary conservation of these genes (see the discussion of recombination below). This finding is compatible with the general trend of positive correlation between genes' propensity for loss and rate of sequence evolution (30).
Fig 2.
dN/dS ratios of the cas genes classified in four different manners (the classification is described in Fig. 1). The widths of bars are proportional to the square roots of the sample sizes (i.e., the number of estimates). (A) The genes were classified into four functional categories: genes involved in the adaptation stage of the CRISPR-Cas immune processes, those involved in the interference stage, predicted transcription regulators (denoted Regulation), and genes encoding the subunits of the CASCADE complex. (B) The CASCADE group of the genes was further divided into five groups: the cas5, cas6, and cas7 families of RAMPs and the large and small subunits of the CASCADE complex. (C) The CASCADE group of genes was classified according to the CRISPR-Cas types in which the genes were represented. The cas6 family of RAMPs (namely, cas6, cas6e, and cas6f) was excluded because a member of this family (cas6) is represented in both type I and type III systems. (D) The RAMP genes were classified according to the presence and absence of a predicted catalytic histidine: the genes that have a predicted catalytic histidine site and encode an experimentally characterized nuclease (RAMP w/H*), those that had a conserved histidine site but have not been demonstrated to possess nuclease activity (RAMP w/H), and those that do not have a predicted catalytic histidine (RAMP w/o H) (csm3 and csm4 were excluded).
The genes encoding the small subunit of the CASCADE complex had significantly greater dN/dS ratios than the cas5 and cas7 groups of the RAMP genes (P < 1.1 × 10−3 with the Mann-Whitney U test, P < 8.8 × 10−3 with the t test, and P = 0.044 with the Dunnett T3 test) and had the greatest median value among the five groups of the CASCADE group of cas genes (Fig. 2B). The elevated gene-wide dN/dS ratio might reflect positive selection on a subset of amino acid sites in the protein, and this interpretation seems to be compatible with the prediction that the small subunit recognizes PAMs in the foreign elements during the interference stage (33). However, given that this protein is small and mostly alpha-helical (33), an alternative and perhaps more plausible explanation is that the elevated dN/dS ratio reflects relaxed structural constraints (and hence weak purifying selection) on this protein. In addition, the genes encoding the large subunit of the CASCADE complex also had significantly greater dN/dS ratios than the cas5 and cas7 groups (P < 2.8 × 10−5 with the Mann-Whitney U test, P < 1.5 × 10−4 with the t test, and P = 0.021 with the Dunnett T3 test) (Fig. 2B). After removing the outliers with dN/dS ratios greater than 0.4, which all corresponded to csy1, the difference remained significant between the large subunit and the cas5 group (P = 1.1 × 10−5 with the Mann-Whitney U test, P = 0.013 with the t test, and P = 9.2 × 10−4 with the Dunnett T3 test). Taken together, these results suggest that the RAMP genes are under stronger gene-wide purifying selection than the non-RAMP components of the CASCADE complex (the small and large subunits).
The genes encoding the CASCADE subunits from type III CRISPR-Cas systems had significantly greater dN/dS ratios than the CASCADE genes from type I CRISPR-Cas systems (P < 3.9 × 10−6 with the Mann-Whitney U test, and P < 1.8 × 10−4 with the t test; the T3 procedure was not applicable) (Fig. 2C), suggesting that the CASCADE subunits are subject to significantly stronger purifying selection in type I systems than in type III systems. The biological underpinning of this difference is unclear. It might be relevant that type III CRISPR-Cas modules often co-occur with CRISPR-Cas modules of other types in prokaryotic genomes, and those that do co-occur in some cases lack cas1 and cas2, the two genes that are otherwise present in all CRISPR-Cas systems (33, 35). Moreover, in the phylogenetic tree of Cas1 proteins, type III systems appear as polyphyletic groups, in contrast to type I and type II systems, which appear as monophyletic groups (35). These findings suggest that type III systems often are horizontally transferred to genomes that already encode other types of CRISPR-Cas systems. Thus, there is a parallel between this enhanced mobility and the faster evolution of the CASCADE subunits in the type III CRISPR-Cas due to relaxed purifying selection.
The RAMP genes that encode predicted nucleases with a highly conserved histidine had significantly greater dN/dS ratios than the genes encoding presumably noncatalytic RAMPs that lack a conserved histidine (Fig. 2D) (P = 4.8 × 10−6 with the Mann-Whitney U test, P = 4 × 10−6 with the t test, and P = 5.6 × 10−3 with the Dunnett T3 test). This difference persisted when the comparison was done between the noncatalytic RAMPs and experimentally characterized RAMP nucleases, although in this case the difference was only marginally significant (P = 0.05 with the Mann-Whitney U test, P = 0.10 with the t test, and P = 0.25 with the Dunnett T3 test). This finding appears counterintuitive, because in general enzymes would be expected to evolve under stronger purifying selection than homologous but inactive proteins. A possible interpretation that does not contradict this expectation is that the (predicted) catalytically active RAMPs are subject to positive selection in a small subset of its amino acid sites (see, however, the estimation of site-specific dN/dS ratios below).
Mapping the dN/dS ratios of cas genes onto the genomic distributions of dN/dS ratios.
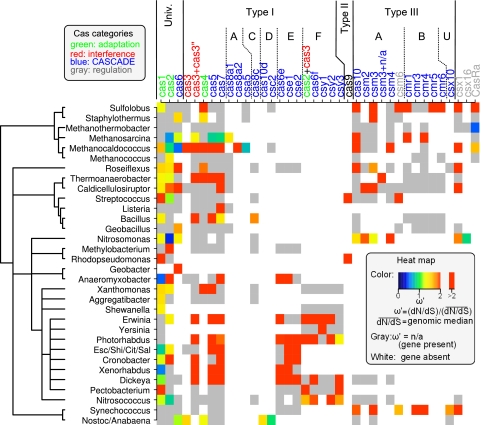
The intensity of selection pressure exerted on the genomes of prokaryotes is highly variable among different groups of bacteria and archaea, as indicated by at least an order-of-magnitude variation in the medians of the genomic (gene by gene) distributions of the dN/dS ratios (47). This variability is thought to reflect the diverse environments prokaryotes inhabit and their variable effective population sizes. To reduce the variation in the dN/dS ratios attributable to such taxon-dependent biases, the dN/dS ratios of cas genes were scaled by the median of the dN/dS ratios of the respective genomes (see Materials and Methods). This scaling showed that the dN/dS ratios of cas genes generally were greater than the genomic median in the respective genera, with several notable exceptions of cas1 and cas2, the two most slowly evolving cas genes (Fig. 3). Thus, most cas genes evolve under relatively relaxed purifying selection and/or relatively intensified positive selection compared to that of the genomic average. These two causes for the elevated levels of dN/dS ratios are difficult to discriminate, because dN/dS ratios reflect the superposition of both effects. However, it is relevant to note that among the genes that display clear evidence of positive selection (i.e., dN/dS > 1), the majority are involved in immune processes or in evasion thereof (68). Thus, it is tempting to draw a parallel between the high dN/dS ratios that we detected for the cas genes and this general trend. Among the cas genes, the relatively strong purifying selection on cas1 and cas2 remains apparent even after removing the taxonomic biases, which is in agreement with the findings described above.
Fig 3.
dN/dS ratios of cas genes scaled by the median dN/dS ratio of the respective genomes. The genes are categorized into four functional groups and also are grouped by the types of the CRISPR-Cas systems in which the genes are represented (the classification is described in Fig. 1). The genomes were grouped into genera according to the NCBI Taxonomy Database (54). The heat map indicates the scaled dN/dS ratios (the scale is shown in the inset). Gray indicates that the dN/dS ratios were unavailable because the dS values fell outside the acceptable range of 0.25 to 1.5. White indicates the absence of the respective gene in a given genome. Univ., universal; dN/dS, median of the genomic dN/dS ratio distribution of the respective genome; ω′, dN/dS ratio of a cas gene divided by the value of dN/dS in the respective genome.
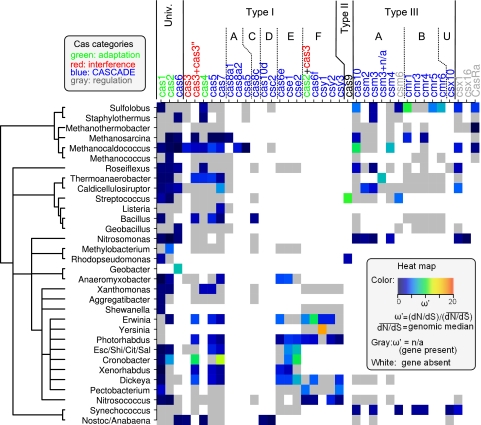
The scaled dN/dS ratios of cas genes did not show substantial variations among the taxonomic groups of bacteria and archaea (Fig. 4), indicating that the cas genes occupy similar places on the genomic spectrum of selection pressure, largely independent of the genomes to which the genes belong. Of particular note is the apparent absence of atypical values in the taxonomic group, including E. coli and S. enterica. The spacer composition of CRISPRs in these species evolves slowly, remaining unchanged for 103 to 105 years, suggesting that in these bacteria CRISPR-Cas members do not function as a typical immune system (60, 61). Although not in direct conflict with this hypothesis, the absence of atypical dN/dS values in the cas genes from these species reported here appears to be compatible neither with the interpretation that these genes are on the verge of degradation nor with the possibility that their functions are completely different from those of other CRISPR-Cas systems.
Fig 4.
Heat map shown in Fig. 3 rescaled to facilitate comparison between different genomes. The scale is shown in the inset.
Given the high dN/dS ratios of the cas genes relative to that of the genomic median, we searched for evidence of positive selection in these genes through the estimation of site-specific dN/dS ratios (see Materials and Methods). The results, however, did not reveal any consistent evidence of positive selection (although statistically significant evidence of positive selection was detected in two clusters of cas9, the sites predicted to be under positive selection did not overlap between the two clusters; moreover, an additional analysis using Datamonkey [50] did not detect any statistically significant evidence either). These observations suggest that, the immune functions of CRISPR-Cas notwithstanding, there are no sites under strong positive selection in cas genes. This result, however, should be taken with caution, because the size of the samples generally was small (e.g., the number of sequences in the two cas9 clusters was effectively four, the bare minimum required for statistical significance according to PAML). Thus, the further analysis of larger data sets is required to characterize potential effects of positive selection in cas genes.
Recombination within cas genes.
Recombination signals were detected in 22 of the 4,130 clusters from 15 cas genes that belonged to various groups defined above: adaptation genes, interference genes, RAMPs, the large and small subunits of the CASCADE complex, and cas9 (Table 1). Notably, recombination was not predicted for cas1, although this ubiquitous gene represents the largest fraction of all cas genes in the analyzed data set (689 of 6,079).
Table 1.
Recombination signals in the cas genes detected with RDP3
| Gene(s) |
P value by statistical method |
Gene(s) involved in recombination (GI) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RDP | Gncnva | Btscnb | MaxChi | Chmrc | SiScan | 3Seq | Recombinant sequence(s) | Parent sequenced |
||
| Major | Minor | |||||||||
| cas6 | 5.9e−08 | 7.8e−08 | 2.7e−07 | 2.0e−05 | 2.7e−06 | 1.6e−11 | 4.8e−08 | 297545575 | 167036552 | 20809011 |
| cas6 | 1.5e−06 | 6.2e−03 | 1.7e−06 | 1.0e−03 | 2.9e−03 | NS | 1.9e−02 | 257388871 | 257052531 | 55376272 |
| cas3-cas3″ | 5.2e−07 | 1.6e−08 | 2.6e−08 | 9.8e−13 | 1.8e−08 | 1.4e−16 | 8.7e−09 | 170018993, 16130668, 89109548, 170082336, 238901898 | 157162211 | (218547719) |
| cas3-cas3″ | 1.0e−07 | 2.8e−06 | 1.0e−07 | 1.3e−05 | 6.8e−05 | 1.1e−08 | 4.7e−06 | 218547719 | 157162211 | 170018993 |
| cas3-cas3″ | NSe | 1.8e−04 | 1.5e−04 | 1.8e−04 | NS | 3.3e−02 | 2.4e−06 | 170018993 | 238901898 | (218547719) |
| cas3-cas3″ | 2.3e−03 | 1.3e−02 | 1.1e−02 | 4.3e−02 | NS | NS | 8.6e−03 | 157162211 | 218547719 | (238901898) |
| cas3-cas3″ | 7.1e−11 | 3.7e−09 | NS | 4.1e−07 | 2.4e−07 | 2.3e−06 | 2.7e−12 | 74313331, 15803278, 209398890, 291284088, 15832869, 254794701 | 260869440 | (260845407) |
| cas3-cas3″ | 7.9e−04 | 1.7e−05 | 6.8e−08 | 2.3e−05 | 5.0e−04 | 1.6e−13 | NS | 260845407, 218696359 | (218706256) | 74313331 |
| cas3-cas3″ | 7.2e−03 | 4.0e−04 | 1.2e−03 | 1.4e−03 | 1.4e−02 | 8.6e−07 | NS | 170680605 | (291284088) | 218706256 |
| cas3-cas3″ | 1.7e−02 | 4.8e−02 | 1.1e−02 | NS | NS | 1.6e−06 | 2.4e−14 | 218706256 | 170680605 | 291284088 |
| cas4 | 1.2e−03 | NS | 4.2e−03 | 2.1e−02 | 3.6e−03 | 1.0e−10 | 8.3e−03 | 146297393, 222528175 | (302870840) | 146295385 |
| cas7 | 8.0e−03 | NS | 1.4e−03 | 4.1e−06 | 3.5e−05 | 5.1e−03 | 2.6e−04 | 229584436, 227827215, 238619365 | 227829662 | (15898280) |
| cas7 | 5.4e−07 | NS | 5.0e−05 | 2.3e−06 | 1.8e−08 | 7.3e−05 | 4.5e−08 | 285019649, 21244564, 34496682, 58580492, 84622452, 188578571 | 226945225 | 251792128 |
| cas7 | NS | 3.1e−02 | 3.2e−06 | 4.9e−07 | 9.8e−07 | 1.4e−11 | 1.1e−18 | 194451697, 16766247 | 224584720 | 207858201 |
| cas7 | 2.1e−04 | NS | 2.1e−04 | 1.5e−04 | 9.9e−07 | 3.9e−02 | 3.1e−11 | 207858201, 16766247, 194451697, 205353882 | 198244668 | 197248466 |
| cas8c | 1.2e−05 | 1.5e−04 | 1.2e−05 | 7.1e−04 | 2.2e−04 | 1.3e−03 | 3.5e−06 | 94994792 | 94990879 | (209559722) |
| cse1 | 2.8e−05 | 1.8e−03 | 2.8e−05 | 3.9e−06 | 2.3e−07 | 2.2e−08 | 3.6e−08 | 238901897, 16130667, 89109547, 170082335 | 170018994 | (157162209) |
| cse1 | 3.1e−06 | NS | 3.2e−04 | 1.9e−13 | 2.6e−14 | 2.4e−10 | 9.1e−17 | 205353884, 16766249, 194447974, 198243398, 207858203 | 224584722 | 283786686 |
| cse1 | 3.9e−09 | 3.2e−03 | 6.4e−04 | 3.8e−15 | 1.1e−02 | NS | 6.3e−03 | 260845406, 82545167 | 218555307 | (283786686) |
| cse1 | 1.3e−03 | 5.2e−03 | 6.8e−03 | 1.2e−03 | 5.7e−04 | 1.1e−04 | 8.4e−08 | 205353884, 198243398, 207858203 | (194447974) | 197251297 |
| cse2 | 1.6e−03 | 2.3e−03 | 7.2e−03 | 5.9e−07 | 1.1e−07 | 1.8e−13 | 5.1e−13 | 260869438, 209920205, 260856870 | 218696357 | 218706254 |
| cas2-cas3 | 1.2e−17 | 5.5e−15 | 2.5e−13 | 1.3e−06 | 8.6e−09 | 4.4e−05 | 6.7e−05 | 271501956 | 251788336 | 242238185 |
| csy1 | 1.1e−14 | 2.6e−14 | 1.8e−07 | 4.8e−10 | 2.2e−11 | NS | 1.3e−05 | 271501955, 251788337 | 242238184 | 261822893 |
| csy1 | 3.0e−03 | 3.0e−02 | NS | 1.8e−02 | 3.9e−04 | 1.5e−30 | 3.3e−68 | 251788337, 271501955 | 261822893 | 242238184 |
| csy1 | 1.3e−02 | 4.2e−02 | NS | 4.4e−02 | 3.4e−03 | 1.0e−04 | 1.9e−03 | 271501955, 251788337 | 50122602 | 242238184 |
| csy2 | 6.3e−05 | 6.9e−06 | 6.2e−06 | 5.0e−03 | 1.3e−03 | NS | 2.0e−02 | 271501954 | 251788338 | 242238183 |
| cas9 | 1.4e−03 | 1.2e−02 | 8.4e−05 | 4.3e−05 | 1.3e−02 | NS | 9.9e−04 | 218563121 | 153952471 | 157415744 |
| cas9 | 9.6e−08 | 1.3e−08 | 2.7e−08 | 3.4e−05 | 8.3e−10 | NS | 3.8e−12 | 55822627 | 55820735 | 116627542 |
| cas9 | 5.1e−48 | 1.1e−48 | 4.8e−56 | 8.0e−36 | 2.4e−22 | 7.5e−92 | 3.0e−38 | 195978435 | 94990395 | (94994317) |
| cas9 | 1.0e−16 | 1.0e−14 | 4.0e−02 | 3.3e−05 | 3.2e−05 | 9.4e−06 | 1.2e−12 | 94994317 | 251782637 | 28896088 |
| cas9 | 1.6e−13 | 3.9e−12 | 6.9e−07 | 8.9e−04 | 5.5e−04 | 1.1e−45 | NS | 251782637, 94994317 | (28896088) | 15675041 |
| cas9 | 1.9e−02 | 2.6e−02 | 1.6e−02 | 2.0e−04 | 2.7e−04 | 6.6e−06 | NS | 209559356 | 28896088 | (15675041) |
| cas10 | NS | 4.3e−06 | 1.3e−07 | 8.3e−10 | 3.5e−09 | 8.2e−13 | 2.6e−13 | 148270227 | 170288804 | 281412438 |
| cmr6 | 3.0e−07 | 3.3e−07 | 1.4e−06 | 6.3e−09 | 1.5e−09 | 1.6e−09 | 1.2e−06 | 229578558 | 15898785 | 227829744 |
| cmr6 | 4.5e−02 | 3.1e−07 | 3.8e−03 | NS | 9.3e−07 | 2.8e−03 | 6.5e−08 | 229578429 | 229578558 | 227829744 |
| csm4 | 3.4e−02 | NS | 1.7e−02 | 5.2e−04 | 1.0e−02 | 1.5e−12 | 5.8e−04 | 148270224, 170288807 | 281412441 | 15644552 |
| csm5 | 4.7e−04 | NS | 4.5e−03 | 1.3e−02 | 5.9e−03 | 7.5e−05 | 1.4e−02 | 15644551 | 281412442 | 148270223 |
Gncnv, GENECONV method.
Btscn, Bootscan method.
Chmr, Chimaera method.
Numbers in parentheses indicate sequences used to infer unknown parents.
NS, not significant (P > 0.05).
Several studies have presented ample evidence of HGT in CRISPR-Cas modules, thus revealing the high evolutionary mobility of the cas genes (12, 19, 62). Moreover, there are indications that entire cas gene cassettes have been transferred between the identical loci associated with CRISPRs in the genomes of E. coli and S. enterica, suggesting that the genomic regions associated with CRISPRs are hot spots of recombination (61). Extending these results, our findings indicate that recombination also occurs in a wide variety of cas genes. The only major exception to this trend is the absence of evidence of microscale recombination in cas1. This finding parallels the other lines of evidence on the strong conservation of cas1 that is manifest both in the ubiquitous representation of this gene in CRISPR-Cas modules and in the low dN/dS ratios described above.
Conclusions.
We report here that cas genes evolve under purifying selection that is typically much weaker than the median strength of purifying selection affecting genes in the respective genomes. The exceptions are the cas1 and cas2 genes, which evolve at levels of purifying selection close to the genomic median. Taken together with the evidence of frequent HGT in the cas genes reported previously and wide-spread microscale recombination in the genes described here, these findings reveal the dynamic evolution of cas genes. This conclusion is in line with the involvement of CRISPR-Cas in antiviral immunity that is likely to entail a coevolutionary arms race with rapidly evolving viruses. However, we failed to detect evidence of strong positive selection in any of the cas genes.
Additionally, two notable observations were made regarding the biological correlates of the selection intensity estimated for different cas genes. The genes that are implicated in the adaptation stage of the CRISPR-Cas process (spacer acquisition), in particular cas1 and cas2, were found to be subject to the strongest purifying selection among all cas genes. This finding is compatible with the (near) ubiquity of these genes in CRISPR-Cas systems and, in the case of cas1, with the absence of evidence of microscale recombination within this gene. These results do not seem to support the possibility that cas1 and cas2 are directly engaged in the coevolutionary arms race, although they are likely to physically interact with foreign genetic elements. A potentially important factor underlying the relatively strong purifying selection that affects cas1 and cas2 could be the additional involvement of these genes in processes distinct from the CRISPR-Cas-mediated immunity, as suggested by experiments implicating cas1 in DNA repair functions (6). Another notable observation is that the RAMPs containing a predicted catalytic histidine had higher dN/dS ratios (weaker purifying selection) than those observed for predicted noncatalytic RAMPs. This result is unexpected, because within the same protein family stronger purifying selection generally would be predicted for enzymatically active proteins. One interpretation of this observation is that the (predicted) catalytic RAMPs experience positive selection in a small subset of amino acid sites. Although the estimation of site-specific dN/dS ratios did not reveal convincing evidence of positive selection in these genes, further analysis of expanded data sets is required to clarify this issue.
ACKNOWLEDGMENTS
We thank David Kristensen for help with data analysis and Igor Rogozin for helpful discussions.
This research was supported by the Intramural Research Program of the NIH, National Library of Medicine.
Footnotes
Published ahead of print 16 December 2011
REFERENCES
- 1. Al-Attar S, Westra ER, van der Oost J, Brouns SJ. 2011. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol. Chem. 392:277–289 [DOI] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersson AF, Banfield JF. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047–1050 [DOI] [PubMed] [Google Scholar]
- 5. Anisimova M, Nielsen R, Yang Z. 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164:1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Babu M, et al. 2011. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 79:484–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrangou R, et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 8. Beloglazova N, et al. 2008. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 283:20361–20371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boni MF, Posada D, Feldman MW. 2007. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brouns SJ, et al. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carte J, Wang R, Li H, Terns RM, Terns MP. 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22:3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chakraborty S, et al. 2010. Comparative network clustering of direct repeats (DRs) and cas genes confirms the possibility of the horizontal transfer of CRISPR locus among bacteria. Mol. Phylogenet. Evol. 56:878–887 [DOI] [PubMed] [Google Scholar]
- 13. Deltcheva E, et al. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deveau H, et al. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dunnett CW. 1980. Pairwise multiple comparisons in the unequal variance case. J. Am. Stat. Assoc. 75:796–800 [Google Scholar]
- 16. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gibbs MJ, Armstrong JS, Gibbs AJ. 2000. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582 [DOI] [PubMed] [Google Scholar]
- 19. Godde JS, Bickerton A. 2006. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 62:718–729 [DOI] [PubMed] [Google Scholar]
- 20. Gomez P, Buckling A. 2011. Bacteria-phage antagonistic coevolution in soil. Science 332:106–109 [DOI] [PubMed] [Google Scholar]
- 21. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 22. Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS. Comput. Biol. 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hale CR, et al. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6:65–70 [Google Scholar]
- 26. Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170 [DOI] [PubMed] [Google Scholar]
- 27. Jore MM, et al. 2011. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 18:529–536 [DOI] [PubMed] [Google Scholar]
- 28. Koonin EV, Wolf YI. 2009. Is evolution Darwinian or/and Lamarckian? Biol. Direct 4:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kristensen DM, Wolf YI, Mushegian AR, Koonin EV. 2011. Computational methods for Gene Orthology inference. Brief Bioinform. 12:379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krylov DM, Wolf YI, Rogozin IB, Koonin EV. 2003. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 13:2229–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li W. -H. 1997. Molecular evolution. Sinauer Associates, Sunderland, MA [Google Scholar]
- 32. Lintner NG, et al. 2011. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J. Biol. Chem. 286:21643–21656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Makarova KS, Aravind L, Wolf YI, Koonin EV. 2011. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct. 6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Makarova KS, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11:181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marraffini LA, Sontheimer EJ. 2009. Invasive DNA, chopped and in the CRISPR. Structure 17:786–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marraffini LA, Sontheimer EJ. 2010. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463:568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563 [DOI] [PubMed] [Google Scholar]
- 42. Martin DP, et al. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martin DP, Posada D, Crandall KA, Williamson C. 2005. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retrovir. 21:98–102 [DOI] [PubMed] [Google Scholar]
- 44. Maynard Smith J. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126–129 [DOI] [PubMed] [Google Scholar]
- 45. Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740 [DOI] [PubMed] [Google Scholar]
- 46. Novichkov PS, Ratnere I, Wolf YI, Koonin EV, Dubchak I. 2009. ATGC: a database of orthologous genes from closely related prokaryotic genomes and a research platform for microevolution of prokaryotes. Nucleic Acids Res. 37:D448–D454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Novichkov PS, Wolf YI, Dubchak I, Koonin EV. 2009. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J. Bacteriol. 191:65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Padidam M, Sawyer S, Fauquet CM. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225 [DOI] [PubMed] [Google Scholar]
- 49. Paterson S, et al. 2010. Antagonistic coevolution accelerates molecular evolution. Nature 464:275–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pond SL, Frost SD. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533 [DOI] [PubMed] [Google Scholar]
- 51. Posada D, Crandall KA. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. U. S. A. 98:13757–13762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 53. Sakamoto K, et al. 2009. X-ray crystal structure of a CRISPR-associated RAMP Cmr5 protein from Thermus thermophilus HB8. Proteins 75:528–532 [DOI] [PubMed] [Google Scholar]
- 54. Sayers EW, et al. 2009. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 37:D5–D15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Selengut JD, et al. 2007. TIGRFAMs and genome properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 35:D260–D264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shriner D, Nickle DC, Jensen MA, Mullins JI. 2003. Potential impact of recombination on sitewise approaches for detecting positive natural selection. Genet. Res. 81:115–121 [DOI] [PubMed] [Google Scholar]
- 57. Sinkunas T, et al. 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 30:1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 26:335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tatusov RL, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Touchon M, et al. 2011. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J. Bacteriol. 193:2460–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Touchon M, Rocha EP. 2010. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One 5:e11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tyson GW, Banfield JF. 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 10:200–207 [DOI] [PubMed] [Google Scholar]
- 63. Wang R, Preamplume G, Terns MP, Terns RM, Li H. 2011. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure 19:257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wiedenheft B, et al. 2011. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477:486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wiedenheft B, et al. 2009. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure 17:904–912 [DOI] [PubMed] [Google Scholar]
- 66. Wolf YI, Aravind L, Grishin NV, Koonin EV. 1999. Evolution of aminoacyl-tRNA synthetases–analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 9:689–710 [PubMed] [Google Scholar]
- 67. Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591 [DOI] [PubMed] [Google Scholar]
- 68. Yang Z, Bielawski JP. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15:496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]