Abstract
Paralogous transcriptional regulators MarA, Rob, and SoxS act individually and together to control expression of more than 80 Escherichia coli genes. Deletion of marA, rob, and soxS from an E. coli clinical isolate prevents persistence beyond 2 days postinfection in a mouse model of pyelonephritis. We used microarray analysis to identify 242 genes differentially expressed between the triple deletion mutant and its parent strain at 2 days postinfection in the kidney. One of these, znuC of the zinc transport system ZnuACB, displayed decreased expression in the triple mutant compared to that in the parental strain, and deletion of znuC from the parental strain reduced persistence. The marA rob soxS triple deletion mutant was less viable in vitro under limited-Zn and Zn-depleted conditions, while disruption of znuC caused a reduction in the growth rates for the parental and triple mutant strains to equally low levels under limited-Zn or Zn-depleted conditions. Complementation of the triple mutant with soxS, but not marA or rob, restored the parental growth rate in Zn-depleted medium, while deletion of only soxS from the parental strain led to low growth in Zn-depleted medium. Both results suggested that SoxS is a major regulator responsible for growth under Zn-depleted conditions. Gel shift experiments failed to show direct binding of SoxS to the znuCB promoter, thus suggesting indirect control of znuCB expression by SoxS. While SoxS expression in the triple mutant fully restored persistence, increased expression of znuACB via a plasmid in this mutant only partially restored wild-type levels of persistence in the kidney. This work implicates SoxS control of znuCB expression as a key factor in persistence of E. coli in murine pyelonephritis.
INTRODUCTION
Escherichia coli is a highly adaptive bacterium that can inhabit a variety of niches. The ability of E. coli to colonize the human colon as well as lead to urinary tract disease is of particular interest for disease prevention, as E. coli is the leading cause of urinary tract infections (17, 18) and can produce bloody diarrhea and hemolytic uremic syndrome (21). The increasing prevalence of antibiotic resistance in clinical isolates of E. coli has led to the pursuit of an understanding of the association of antibiotic resistance with bacterial pathogenicity. Of interest are the overlapping regulons of the MarA, Rob, and SoxS transcriptional regulators, which control many functions, including antibiotic resistance (10).
MarA, produced from the marRAB operon, affects a variety of genes associated with antibiotic resistance (11) and virulence (10). A mutation in MarR, an autorepressor of this operon, leads to an antibiotic resistance phenotype (47). Rob increases antibiotic and organic solvent tolerance through induction of the mar operon (20) and is posttranscriptionally activated by such host physiological substances as bile salts (42) and antimicrobial peptides (50). The soxRS operon is induced by oxidative stress which, via SoxS, leads to direct activation of antibiotic resistance factors, including the marRAB operon (29). All three regulators affect expression of their own set of specific genes as well as an overlapping regulon of at least 80 genes (4, 39). Also, the three regulators are implicated in persistent kidney infection in the mouse (10). When all three factors are deleted in clinically isolated antibiotic-resistant E. coli strain KM-D, containing an inactivating mutation in marR, initial colonization of the mouse kidney is unaffected for 2 days but is greatly reduced over longer periods. Persistence is restored with the readdition of any one of the three transcription factors (10).
We utilized microarray technology to identify genetic loci which differed in expression based on the presence or absence of the MarA, Rob, and SoxS regulators in response to conditions within the host. We analyzed the transcriptional profiles of a marA rob soxS triple deletion mutant (PC1012) and its parent strain (KM-D) in mouse kidneys just before the triple deletion mutant was cleared from the kidney. Among other changes, we saw increased expression of the znuC gene in the parent strain. While our study was in progress, groups showed that the znuACB system is important for pathogenic fitness in uropathogenic E. coli (UPEC) (16, 43). Zinc uptake is similarly important for pathogens Proteus mirabilis (32), Brucella abortus (52), and Salmonella enterica (9). Therefore, we chose to pursue znuACB and its relation to MarA, Rob, and SoxS.
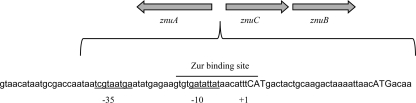
The znuACB operon encodes a three-component high-affinity Zn transport system used by the bacterium to import Zn2+ ions into the cell for use in many vital biological processes. The operon consists of znuA followed by a 24-bp intragenic sequence and the divergently transcribed znuCB genes (38) (Fig. 1). The znuACB operon is repressed by the Zur repressor protein, which binds to the intragenic region in times of sufficient Zn concentrations and slows transcription of the operon (37). This study identifies SoxS as a regulator of this operon in response to host factors in a mouse model of pyelonephritis.
Fig 1.
The znuACB locus features a 24-bp intragenic region between the divergently transcribed znuA and znuCB. The start codons are capitalized, and the +1, −10, and −35 sites for znuCB are noted (34). Additionally, the binding site of the Zur repressor (37) is noted.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Parental strain KM-D is a clinical isolate from an intestinal fistula containing a 43-amino-acid deletion within MarR, yielding a nonfunctional repressor protein and, consequently, MarA-mediated multidrug resistance (24). As shown previously, strain KM-D lacks many prominent uropathogenic factors (10). All strains were cultured on LB agar plates or in LB broth as described previously (50) unless otherwise noted. The antibiotic concentrations used were as follows: chloramphenicol (Cm), 20 μg/ml; kanamycin (Km), 50 μg/ml; and ampicillin (Ap), 100 μg/ml. When indicated, strains were grown in M9 minimal broth or agar (28) or dilute tryptone (DT) broth or agar (4 g/liter Bacto tryptone [Difco], 0.25% NaCl, 0.4% glucose, 1 μg/ml thiamine, 22 μg/ml histidine, and tryptophan, phenylalanine, and tyrosine [20 μg/ml each]) (44). Strains SIP468 and SIP775 and their descendants are offspring of the parent strain MC4100 and were P1 transduced with mutations from strain JW 4023-5 or JTG1078 as described previously (31) or transformed via chemical transformation. All strains are listed in Table 1.
Table 1.
Strains and plasmids used in this study
| E. coli strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| KM-D | Clinical E. coli isolate from an intestinal fistula, marR | 24 |
| PC1012 | KM-D ΔsoxS Δrob ΔmarA | 10 |
| DH5α | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Invitrogen |
| S17 λpir | lamB F−supE44 thi-1 thr-1 leuB6 lacY1 tonA21 hsdR hsdM recA pro (RP4 2-Tc::Mu::Km::Tn7) λpir | 49 |
| KM-DznuC | KM-D znuC | This study |
| PC1012znuC | PC1012 znuC::Cm | This study |
| PC1037 | PC1012::soxS (soxS restored to chromosome) | 10 |
| PC1038 | PC1012::rob | 10 |
| PC1033 | PC1012::marA | 10 |
| PC1005 | KM-D ΔsoxS | 10 |
| SIP468 | MC4100 aroB znuA::MudX | 37 |
| SIP775 | MC4100 aroB znuC::MudX | 37 |
| JW4023-5 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ−rph-1 Δ(rhaD-rhaB)568 ΔsoxS756::kan hsdR514 | 3 |
| JTG1078 | soxR105 zjc-2204::Tn10kan; overexpression of soxS | 14 |
| SIP468soxS | SIP468 transduced with ΔsoxS756::kan from JW4023-5 | This study |
| SIP775soxS | SIP775 transduced with ΔsoxS756::kan from JW4023-5 | This study |
| SIP468soxR105 | SIP468 transduced with soxR105 zjc-2204::kan from JTG1078 | This study |
| SIP775soxR105 | SIP775 transduced with soxR105 zjc-2204::kan from JTG1078 | This study |
| Plasmids | ||
| pGEM | High-copy-no. cloning vector (Apr) | Promega |
| pSR47s | R6KoriV RP4oriT sacB (Kmr) | 22 |
| pKD3 | Source of cat cassette (Cmr) | 13 |
| pNTR-SD | Ptac lacIq; low copy no. (Apr) | 45 |
| pZnuC | Intact znuC gene with promoter cloned into A/T site in pGEM (Apr) | This study |
| pZnuC::Cm | znuC disrupted with the cat cassette; pGEM(Apr Chlr) | This study |
| pSRznuC::Cm | NotI fragment from pZnuC::Cm cloned into NotI site of pSR47s (Cmr Kmr) | This study |
| pZnuACB | Intact znuACB locus with promoter cloned into A/T site in pGEM (Apr) | This study |
| pNTznuACB | NotI fragment of pZnuACB cloned into NotI site of pNTR-SD (Apr) | This study |
| pSXS-Cm | IPTG-inducible soxS gene | 1 |
IPTG, isopropyl-β-d-thiogalactopyranoside.
Mouse infection and RNA extraction.
Groups of 6- to 8-week-old female CD-1 mice (Charles River) were diuresed with 5% glucose and limited food for 3 days preinfection in order to visualize the bladder during subsequent intrabladder injection of the inoculum. Bacterial strains, grown for 18 h shaking at 37°C in LB broth the night before infection, were pelleted and washed three times with phosphate-buffered saline (PBS) and resuspended at a final concentration of ∼109 CFU/ml. A 25-μl sample of the inoculum (∼2.5 × 107 CFU) was then injected into the bladders of ketamine-anesthetized mice as described previously (10). Inoculum titers were determined by culturing a sample on LB agar plates. Mice were sacrificed at 2 days postinfection, and kidneys were removed, weighed, and homogenized in 2 ml of the bacteriostatic RNAlater solution (Ambion). Fifty-microliter samples of homogenized kidney solutions were then used to determine bacterial loads for each mouse, and the remainder of the kidney solution was stored overnight at −20°C to maintain RNA integrity. Subsequent competitive and single-strain infections, from which RNA isolation was unnecessary, utilized PBS for sample collection.
Competitive infections in mice were performed by inoculating 8 to 10 mice with a mixture containing equal amounts of two strains, utilizing the appropriate antibiotic resistance markers to calculate mutant-parent strain ratios at the start of the experiment and at days 1 and 4 postinfection. Data from day 1 postinfection were used to evaluate ascension to the kidney and initial colonization, while data from day 4 postinfection were used to determine persistence. Each infection was performed at least twice. Titers of mutant strains were determined by culturing on medium with chloramphenicol, and numbers of CFU of the KM-D strain were calculated from the total number of CFU on plates without antibiotics by subtracting the mutant strain (Cm-resistant) titers. Strains were also tested in vitro for general growth differences by coculturing the strains in LB broth and determining the titers of the bacteria at different time points over 24 h to observe whether the competitive index (CI) values (ratio of mutant to wild type) remained close to the inoculated value, generally 1.0. All strain mixtures tested displayed CI values in a range of 0.9 to 1.1 (data not shown).
Single-strain infections used to evaluate the bacterial load of individual strains were also performed at least twice on 8 to 10 mice per experimental group as described previously (10). Data collected from these infections are shown as CFU/gram of kidney homogenate. In all cases, the bladder was not cultured, as previous findings showed no differences in persistence or colonization between strains KM-D and PC1012 at this body site (10).
Animal experiments were performed in the laboratory animal facility at Tufts University, an institution accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care, under protocol 5807, approved by the Tufts Institutional Animal Care and Use Committee. Buprenorphine (0.05 mg/kg of body weight) was used to relieve any postsurgical discomfort by subcutaneous injection of 0.05 ml of appropriately diluted analgesic. All euthanizing was performed via CO2 asphyxiation and cervical dislocation.
RNA isolation and microarray analysis.
Infected kidney suspensions were subjected to homogenization with a Tissue Tearor (Invitrogen) in RNAlater (Ambion). Homogenates were centrifuged at 5,000 rpm for 10 min to separate bacteria (pellet) from kidney (surface of RNAlater solution) cells. The kidney cells were evacuated from the top of the solution, and the remaining bacteria were centrifuged once more at 12,000 rpm for 10 min, followed by disposal of the supernatant. Bacterial cells were immediately resuspended in ice-cold Tris-EDTA (TE) buffer and treated with lysozyme (Sigma) at a 1 mg/ml final concentration for 5 min, and samples of 3 to 5 infected mice were pooled together for each infection group. Nucleic acids were extracted via the RNA extraction protocol using the RNeasy kit (Qiagen). After column purification, RNA samples were treated with 2 U of Turbo DNA-free (Ambion) for 30 min at 37°C, followed by purification and quantification of RNA at 260 nm. The remaining polyadenylated eukaryotic RNA was depleted from the samples with MICROBEnrich (Ambion) and resuspended in 20 μl of TE buffer. RNA was quantified spectrophotometrically, and equal amounts (∼300 to 500 ng) of RNA from KM-D (parental strain) and PC1012 (triple mutant) were subjected to the MessageAmp (Ambion) RNA amplification protocol. The resultant amplified RNA was quantified, and 10 μg was used in the Affymetrix protocol for prokaryotic cDNA synthesis and biotin labeling.
Two micrograms of biotin end-labeled cDNA from three independent infection groups were individually hybridized to GeneChip E. coli Genome 2.0 arrays (Affymetrix). Chips were then blocked, washed, and labeled with fluorescent streptavidin conjugates using a microfluidics 450 station, following the protocol provided by Affymetrix. Hybridized chips were read with a GeneChip Scanner 3000 (Affymetrix), and fluorescence was compared to the E. coli array information obtained from Affymetrix (www.affymetrix.com). Fluorescence data were preprocessed and normalized by mas5 median normalization to account for background fluorescence and to equalize all chip expression levels to one another. Then all 6 chips (3 independent replicates of strain KM-D and 3 of PC1012) were compared to each other using GeneChip Operating Software (GCOS) (Affymetrix). Normalized expression levels were compared between the two strains, and fold changes, standard errors, and significance levels were determined by Student's t tests for each gene. Fold changes with P values of >0.05 were considered nonsignificant.
Quantitative real-time PCR.
One hundred nanograms of RNA from bacteria grown in rich medium or minimal medium or of nonamplified RNA isolated from mice was converted into cDNA using SuperScript II reverse transcriptase (Invitrogen). Briefly, RNA was added to random hexamers (250 ng) and deoxynucleoside triphosphates (dNTPs) (10 mM each) before heating at 65°C for 10 min and then transferred to an ice bath. Next, 5× First-Strand buffer, 0.1 M dithiothreitol (DTT), and RNaseOUT were added to the solutions, followed by incubation at 25°C for 2 min. Each sample was treated in duplicate, with one treated with 200 U of reverse transcriptase and the other acting as a nontranscriptase control. Samples were incubated at 25°C for 10 min and then at 42°C for 50 min, with inactivation at 70°C for 15 min. Samples were then treated with 2 U of RNase H (Invitrogen) at 37°C for 20 min. The final cDNA product was then used as a template for real-time PCRs using the primers listed in Table S1 in the supplemental material and SYBR green master mix (provided by Tufts Expression Array Core). All reactions were performed in duplicate with a no-template control and a no-reverse-transcriptase control. Expression levels were determined using the comparative method of calculation; gapA served as the housekeeping gene.
β-Galactosidase assays.
β-Galactosidase assays were performed as described previously (28) to compare levels of induction of the znuA and znuC promoters in the MC4100 background of strains SIP468 and SIP775 in the presence of wild-type soxS, mutated soxS, or overexpressed soxS via the soxR105 mutation or the plasmid pSXS-Cm. Data shown are from three separate experiments in which β-galactosidase expression levels of the indicated mutants of strains SIP468 and SIP775 were normalized to the expression levels of the wild-type soxS parent strains.
Zinc sensitivity assays.
Growth differences under Zn-depleted conditions were obtained by monitoring growth in dilute tryptone broth or plates and in M9 minimal medium broth or plates supplemented with 0.4% glucose, with the addition of the divalent ion chelator EGTA (Sigma) (2.0 or 5.0 mM in agar or broth, respectively) and the addition of divalent cations in the forms of ZnSO4, FeSO4, MnSO4, CaCl2, and MgSO4 at a final concentration of 500 μM when indicated. Growth curves in broth were performed by diluting cultures grown overnight to a starting optical density at 600 nm (OD600) of 0.025 and incubating them with shaking at 37°C for 24 h, taking OD600 readings at the indicated time points. Growth on plates was examined by spotting 5 μl of dilutions of mid-log-phase bacteria onto the indicated agar and incubating for 24 h at 37°C. The results shown are from three separate experiments.
Electromobility shift assays.
The 6His-SoxS purified protein, kindly provided by Paratek Pharmaceuticals, Boston, MA, or purified as described previously (36), was produced by cloning the soxS gene into the pET-15b expression plasmid and subsequently purifying the His-tagged protein. DNA containing the fumC, gnd, or znuC promoters was amplified by PCR using the primer pairs fumCF1/fumCR1, gndF/gndR, and znuC-proF/znuC-proR; the products were gel purified. The fumC and gnd promoter PCR products were kindly provided by Laura McMurry (our laboratory). The 283-bp fumC promoter fragment (−102 to +181 bp, with respect to the transcriptional start site) has a well-characterized marbox (27), while the 167-bp gnd promoter fragment (−112 to +55) contains no such sequence (46). A 271-bp znuC promoter PCR fragment (−229 to +22) containing a putative mar/soxbox just upstream of the −35 motif was amplified. A shorter 171-bp promoter fragment (−149 to +22) was also amplified using primer pair znuC-proF2/znuC-proR to isolate the putative forward soxbox just upstream of the −35 motif. The SoxS protein and the PCR products containing the various promoters were mixed in reaction buffer (20 mM NaCl, 50 mM KCl, 20 mM HEPES [pH 7.9], 0.4 mg/ml of bovine serum albumin, 10% glycerol) to final concentrations of 0 to 1,000 nM SoxS and 10 nM DNA in a total volume of 20 μl. Protein-DNA mixtures were incubated for 20 min at 4°C in the presence of salmon sperm DNA at a final concentration of 5 μg/ml before the addition of loading buffer. The samples were electrophoresed in 6% acrylamide gel run in Tris-borate-EDTA (TBE) buffer (90 mM Tris base, 90 mM boric acid, 2 mM EDTA), and double-stranded DNA was stained in TBE buffer with a 1:10,000 dilution of SYBR green reagent (Applied Biosystems). Fluorescence of the stained gels was photographed using the Bio-Rad Gel Imager 1000.
Genetic constructs.
Disruption of znuC was undertaken in strains KM-D and PC1012 using the suicide vector pSR47s (22). Briefly, znuC with 500 bp of flanking DNA was amplified with primers znuC-F5 and znuC-R4 from strain KM-D by PCR and cloned into plasmid pGEM. This construct (pZnuC) was transformed into DH5α, and Apr transformants were confirmed to have the plasmid by plasmid purification (Qiagen) and PCR screening. Plasmid pZnuC was digested with restriction enzyme MscI and ligated to a blunt-ended cat cassette amplified by PCR from plasmid pKD3 (13), yielding plasmid pZnuC::Cm. This new plasmid was selected for its Apr Cmr phenotype, and the increased size of the znuC gene was confirmed with PCR using the original cloning primers. The disrupted gene was then removed from the pGEM vector by digestion with NotI, gel purified, and ligated into the NotI site of the suicide vector pSR47s, yielding plasmid pSRznuC::Cm, which was selected in strain S17λpir on LB agar plates containing Cm and Km. Cmr Kmr transformants were screened with PCR for the disrupted znuC::Cm gene, and PCR products were sequenced at the Tufts Core Facility.
Strain S17λpir harboring pSRznuC::Cm was then used as the donor in conjugations with the desired recipient strains. Conjugations were performed on LB agar in which the bacterial pairs were allowed to grow for 8 h at 37°C. Bacteria were then swabbed off the plates and transferred to M9 agar supplemented with glucose (0.4%), Km, and Cm which selected against the donor strain (leucine, threonine, and proline auxotroph) and allowed growth of the single crossover recipient strain. Kmr Cmr strains were then transferred to sucrose to select against the sacB gene on the pSR47s plasmid. Resultant colonies were screened for double crossover by transferring them to LB agar plates supplemented with Km or Cm. Cmr Kms colonies were then screened by PCR for the disrupted gene.
Primers znuA-revC and znuB-revC were used to amplify the wild-type znuACB operon, including its native promoter, which was then cloned into pGEM, yielding plasmid pZnuACB. This plasmid was transformed into DH5α via electroporation with selection on Ap. The znuACB operon was removed from pGEM with enzyme NotI and cloned into the low-copy-number plasmid pNTR-SD (45). This plasmid (pNTznuACB) was used for complementations and to overexpress the znuACB operon.
Microarray data accession number.
Microarray data were deposited into the Genome Expression Omnibus (MIAME compliant) under accession number GSE23417.
RESULTS
Microarray studies.
The previous finding that deletion of marA, rob, and soxS causes severe attenuation in the bacterial load at day 3 postinfection in a mouse model of pyelonephritis (10) led us to investigate the in vivo transcriptional differences between the parental strain (KM-D) and the triple mutant (PC1012). We chose to extract RNA at day 2 postinfection because attenuation 3 days postinfection would likely be mediated by changes in transcription on day 2. Moreover, at the time, there were bacterial numbers sufficient to isolate ∼300 to 500 ng of bacterial mRNA from pooled kidney samples of mice infected with either strain. Bacterial mRNA was prepared for microarray analysis as described in Materials and Methods. Genes with significant differences in mRNA levels are shown in Tables S2 and S3 in the supplemental material.
As expected, marA and rob were detected in KM-D but not in PC1012 (see Table S2 in the supplemental material). These findings and the lack of marA, rob, and soxS in strain PC1012 were further confirmed with quantitative real-time PCR (qRT-PCR) using mouse-derived cDNA as template (data not shown). We analyzed the list of differentially regulated genes, searching for multiple genes in a pathway. A total of 50 of the 112 transcripts found to be higher and 50 of the 130 transcripts found to be lower in strain KM-D than in strain PC1012 (see Tables S2 and S3 in the supplemental material) were hypothetical genes and were not studied further. Of the genes and pathways which may play a role in persistence, we found factors, such as alr, an alanine racemase, which are known to downregulate expression of proinflammatory cytokines by urothelial cells in vitro (7) (expressed 10-fold more in strain KM-D than in PC1012). Additionally, several genes important for survival in both anaerobic acidic environments (amtB, citA, hyaD, gadY, rffH, atpC, sufC, ybeL, gatR, and asr) (2, 19) and DNA-damaging conditions (recA and dnaT) (25, 33), which are experienced in the kidney, were differently transcribed in strains KM-D and PC1012 (see Tables S2 and S3 in the supplemental material). Recent studies concerning the role of zinc uptake in infection led us to further investigate the relationships between the Zn transporter locus znuACB and the mar-rob-sox regulon.
Effect of triple deletion of mar-rob-sox on znuACB expression.
The mouse-derived microarray data revealed that znuC, a gene in the znuACB locus (Fig. 1) which encodes a high-affinity Zn transporter, was expressed 8-fold more in KM-D than in strain PC1012. qRT-PCR found a 10-fold increase in mice and a 2.5-fold increase with mid-log-phase strains grown in vitro (Table 2). Of note, in vivo and in vitro expression data showed little if any effect of the triple deletion on transcription of the divergently transcribed znuA gene (1.2- to 1.5-fold) or of the zinc uptake regulator zur (0.83- to 1.2-fold) (Table 2).
Table 2.
Differences in gene expression between strains KM-D and PC1012
| Gene | Test and RNA sourcea |
||
|---|---|---|---|
| Microarray analysis with murine kidneys | qRT-PCR |
||
| Murine kidneys | In vitro | ||
| znuC | 18.9 ± 8.1 | 9.9 ± 0.8 | 2.5 ± 0.5 |
| znuA | 0.42 ± 0.5 | 1.2 ± 0.2 | 1.4 ± 0.3 |
| zur | 1.03 ± 3.0 | 1.1 ± 0.1 | 1.2 ± 0.3 |
All mouse-derived cDNA was synthesized from RNA extracted from the infected kidneys of mice at day 2 postinfection. All in vitro results are from tests performed on cDNA synthesized from RNA isolated from bacteria grown to mid-log phase in M9 minimal medium.
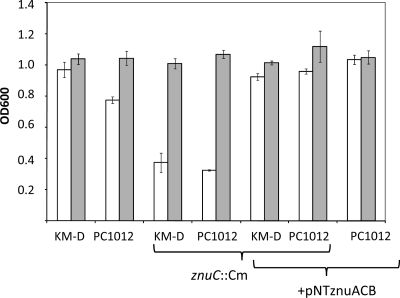
Further testing demonstrated that strains KM-D and PC1012 could be distinguished by growth under Zn-depleted conditions. Expression levels of znuACB can be observed phenotypically by monitoring the growth of bacteria under low-Zn conditions, such as those found in M9 minimal medium, in which, over an 18-h growth period, KM-D grew to a higher density than PC1012. This growth difference was negated by the addition of Zn2+ (Fig. 2). ZnuACB is the major transporter of Zn2+ in low-Zn conditions; however, other low-affinity transporters which act to equilibrate M9 growth with the addition of Zn2+ exist (5, 6). The effects seen here were attributed to znuACB expression differences, since inactivation of znuC equalized growth for strains KM-D and PC1012 in low-Zn2+ conditions, and expression of the znu operon from a plasmid (pNT-znuACB) caused the growth of strain PC1012 to equal that of strain KM-D (Fig. 2). Addition of other divalent cations (Fe, Ca, Mn, and Mg) did not cause these effects (data not shown).
Fig 2.
Effect of disruption of znuC and overexpression of the znuACB locus on growth in minimal medium. Strains were grown overnight (18 h) in M9 minimal broth as described in Materials and Methods, and OD600 values were taken. Cultures were grown alone (open) or with ZnSO4 (500 μM) (shaded). These data are the means of 3 independent experiments. The error bars are the standard errors of the means.
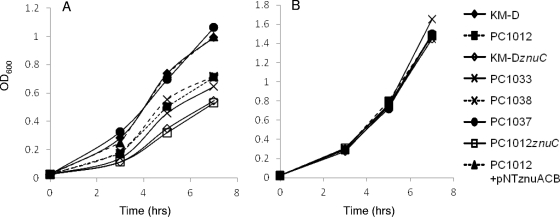
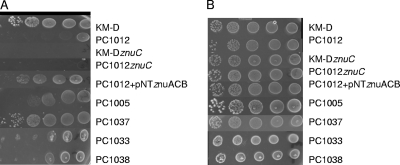
In order to compensate for any growth differences due to the lack of other factors in M9 minimal medium, we also assayed znu expression in more complex media by using chelation of available Zn2+. Dilute tryptone (DT) broth and agar growth assays were used with and without the metal chelator EGTA. We included experiments in which Zn2+ (ZnSO4) was added to ascertain if the chelator effects were due to the inability of strains to get Zn2+ from depleted medium. Strain KM-D grew better in complex medium under Zn-depleted conditions than did PC1012 (Fig. 3 and 4). The differences in growth under chelating conditions were eliminated when znuC mutations were introduced into the chromosomes of both KM-D and PC1012 (Fig. 3A and 4A). Complementation with plasmid pNT-znuACB caused both znuC::Cm mutants to display increased growth under Zn-chelated conditions. Additionally, pNT-znuACB produced increased growth of strain PC1012 in the presence of the chelator (Fig. 3 and 4).
Fig 3.
Growth under Zn-chelating conditions. (A) Strains were grown in DT broth supplemented with EGTA (5 mM) as described in Materials and Methods. (B) Growth in medium with EGTA and the addition of ZnSO4 (500 μM). Growth in DT broth without EGTA showed similar growth for all strains (data not shown).
Fig 4.
Growth on Zn-limited medium. Mid-log-phase strains grown in DT broth were serially diluted 10-fold and spotted on DT agar with 2.0 mM EGTA (A) or DT agar with EGTA and 500 μM ZnSO4 (B). Growth on EGTA plates supplemented with Mn, Mg, Fe, and Ca was no different from growth on EGTA alone for all strains (data not shown).
Single-deletion mutants of marA, rob, or soxS in strain KM-D revealed that only the lack of soxS (strain PC1005) produced results similar to those of PC1012 under low-Zn conditions. In accord, complementation of the triple mutant with marA (PC1033) or rob (PC1038) showed no growth differences from that of strain PC1012, while complementation with soxS (strain PC1037) showed near wild-type levels of growth under Zn-limiting conditions (Fig. 3 and 4A). These findings point to SoxS among the three transcriptional regulators deleted in strain PC1012 as the chief regulator of Zn2+ uptake.
Additional testing of lacZ fusion strains SIP468 and SIP775 (a kind gift from Silke Patzer) showed that the absence of soxS led to a 70% decrease in expression of znuC compared to that of the wild type, while znuA expression did not experience significant changes (Table 3). These findings were further verified by introducing the constitutively active soxR105 mutation into the lacZ fusion strains, revealing that high levels of soxS induction caused increased expression of znuC compared to that of the wild-type strain (SIP 775), with minimal effects on expression of znuA (SIP468). Additionally, introduction of an inducible soxS plasmid (pSXS-Cm) into strain SIP775 and SIP775soxS caused a 5.2- to 1.8-fold increase in expression of the znuC promoter (Table 3). These findings agree with the qRT-PCR findings (Table 2) that the control of znu by SoxS is unidirectional, only affecting znuCB. Importantly, these findings also show that this transcriptional relationship exists in a different bacterial background, as the SIP strains are from the parental laboratory strain MC4100. In accord with these findings, expression of SoxS and known members of its regulon is reportedly increased under Zn-depleted conditions in a microarray study comparing E. coli grown in rich medium to E. coli grown in rich medium treated with a Zn chelator (48).
Table 3.
Effects of removal or overexpression of soxS on znu expression in strain MC4100 as determined by β-galactosidase activity
| Strain | Ratio of LacZ activity ± SEMb |
|---|---|
| SIP468 (znuA::lacZ) soxS::Km | 0.96 ± 0.09 |
| SIP775 (znuC::lacZ) soxS::Km | 0.27 ± 0.02 |
| SIP468 (znuA::lacZ) soxR105 | 0.90 ± 0.05 |
| SIP775 (znuC::lacZ) soxR105 | 1.4 ± 0.2 |
| SIP775 (znuC::lacZ)/pSXS-Cma | 5.2 ± 0.5 |
| SIP775 (znuC::lacZ) soxS::Km/pSXS-Cma | 1.8 ± 0.4 |
Strains containing pSXS-Cm were grown in the presence of 0.5 mM IPTG and 30 μg/ml chloramphenicol.
β-Galactosidase activity values are expressed as a ratio of the LacZ activity of the soxS deletion mutants or overexpressers to that of the wild-type parent, with a value of 1.0 indicating no difference between the two strains. All experiments were performed in triplicate, and the error margins are the standard errors of the mean.
Regulation of znuCB by SoxS.
Regulation of znuCB by SoxS has not been reported. To deduce whether SoxS directly controls transcription of the znu operon, we examined the binding of His-tagged SoxS to the promoter of znuCB. Sequence analysis of the znuCB promoter revealed a mar/soxbox in the class II forward orientation just upstream of the −35 of the transcriptional start site. However, gel shift experiments using a 171- or 251-bp promoter fragment containing this mar/soxbox showed no shift of DNA by SoxS (Fig. 5). These findings strongly suggest that SoxS induces expression of the znuCB operon in an indirect manner.
Fig 5.
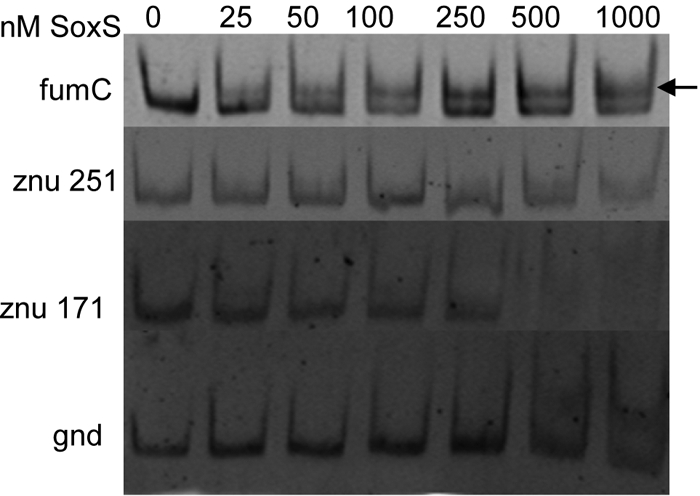
Electromobility shift assays with 6His-SoxS. Purified PCR fragments (10 nM final concentration) were incubated in increasing concentrations of SoxS-6His protein (0 to 1,000 nM) for 20 min at 4°C. The fumC promoter has an established mar/soxbox and acts as a positive control, while the gnd promoter has no such sequence and acts as a negative control. The znuCB upstream regions (−229 to +22 [marked znu 251] and −149 to +22 [marked znu 171]) were amplified as detailed in Materials and Methods. Protein-bound DNA is denoted by an arrow.
Role of znuACB in persistence in mouse kidneys.
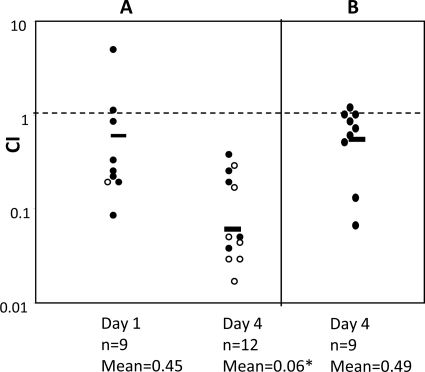
A recent study revealed that the zinc uptake system ZnuACB is important to the uropathogenic E. coli strain CFT073 in a mouse model of ascending urinary tract infection (43). Along with our own microarray results, these findings led us to ask if this system was a player in the persistence difference we observed between nonuropathogenic strains KM-D and PC1012. First, we performed a competitive infection study with strain KM-D and the isogenic znuC mutant (KM-DznuC). At day 1 postinfection, there was only a small difference in initial kidney colonization between the znuC mutant and the parent strain (geometric mean of competitive index [CI] = 0.47). By day 4 postinfection, however, there was a much greater decrease in CI (mean CI = 0.06) (Fig. 6A). Competitive growth of the wild-type and mutant strains in vitro in LB broth showed no differences in CI (data not shown), which indicated that the attenuation of znuC mutants in mice was due to conditions specific to the murine kidney. Additionally, complementation of the strain KM-DznuC with plasmid pNTznuACB caused a return to day 1 levels of competitive persistence on day 4 (mean CI = 0.49) (Fig. 6B).
Fig 6.
Competitive infection studies of zinc uptake mutants in mouse kidneys. Mice were infected (see Materials and Methods) with equal mixtures of either strains KM-D and KM-DznuC (A) or strains KM-D and KM-DznuC with pNT-znuACB (B). Experimental groups were sacrificed at day 1 and day 4 postinfection. Data are the competitive indices from at least two independent experiments, and the geometric means are noted with a bar. Results were calculated using the following equation: competitive index = (mutantoutput/wild typeoutput)/(mutantinput/wild typeinput). Open circles represent mice from which no znuC mutants were cultured, in which cases the limit of detection was used to calculate the competitive index. The significance level was determined by a Mann-Whitney test. The asterisk denotes a P value of 0.01 when comparing the day 1 and day 4 data for infections of KM-D and KM-DznuC.
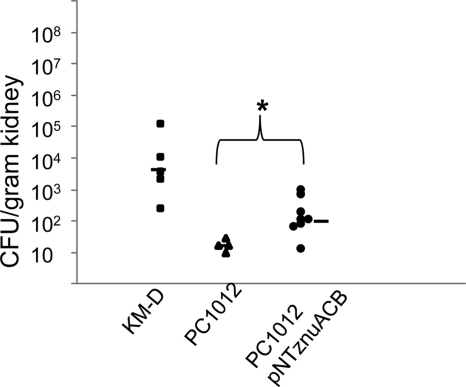
To assess the degree to which decreased Zn uptake played a role in decreased persistence of PC1012, we infected mice with strain PC1012 harboring pNTznuACB. The presence of pNTznuACB increased the median bacterial load of PC1012 10-fold at day 4 postinfection (Fig. 7). Still, survival of PC1012/pNTznuACB in the kidney did not reach levels seen in mice infected with strain KM-D alone. This finding presumably reflects the contributions of factors besides Zn uptake that are controlled by MarA, Rob, and/or SoxS.
Fig 7.
Impact of increased Zn uptake on persistence of strain PC1012 in a single-strain infection. Mice were inoculated with a single dose of the PC1012 (triangles), KM-D (squares), or PC1012/pNT-znuACB (circles) strain. Each data point represents the bacterial load of the indicated strain in kidneys from one mouse on day 4 postinfection. The median values are shown with bars. The significance level was determined by Student's t test. The asterisk denotes a P value of 0.03 when comparing mice infected with PC1012 to mice infected with PC1012 with pNT-znuACB.
DISCUSSION
Persistent kidney E. coli infection is affected by the regulators MarA, Rob, and SoxS (10). The use of a nonuropathogenic strain of E. coli allowed us to look beyond the dominant pathogenic factors already characterized for kidney infection and highlight other genes that are affected by marA, rob, and soxS which play a role in pyelonephritis (21).
The in vivo microarray findings mimic in vitro studies showing that overproduction of the SoxS protein upregulates fldA, gltA, yadM, astD, and malF and downregulates galF and gatR (8, 39). Additionally, a previous study of MarA overexpression in vitro shows upregulation of gltA and downregulation of ybfW (4), which we also found here in mice. While it was encouraging to observe similarities across studies, we also noted several genes or operons expressed oppositely in mice than in previous in vitro findings for soxS, namely, cysB, cysM, pdhR, and nikB for SoxS (12, 41) and yadH for MarA (11). These differences may relate to the unique environment of the murine kidney, to differences between findings of a single gene and those of our studies using the triple deletion mutant, and/or to our use of a clinical strain rather than a laboratory strain.
We chose to focus our investigation on the zinc uptake system ZnuACB. The marA rob soxS deletion mutant displayed a lower level of znuC mRNA than did the wild type, as observed with the microarray and qRT-PCR studies, both in vivo and in vitro. This feature was manifested phenotypically by decreased growth of strain PC1012 under Zn-depleted conditions compared to that of the parent strain. These differences were negated when both strains had a disruption of znuC. Moreover, overexpression of znuACB or complementation with soxS in strain PC1012 led to correction of the persistence/growth defect (Fig. 7). These findings show that znuCB expression is enhanced by SoxS. In this regard, SoxS has been shown to affect the expression of other zinc transport factors during cobalt stress (40). Our findings also agree with a study that showed increased expression of members of the soxS regulon in response to low extracellular Zn2+ concentrations in vitro (48). Gel shift experiments performed using SoxS and znuCB promoter regions did not identify a functional soxbox within the promoter or upstream regions of the operon (15, 26) (Fig. 5). These results are in line with the findings that MarA and Rob do not affect Zn uptake as might be expected if there were a mar/soxbox motif present in the znuCB promoter. Therefore, we favor the explanation that the regulation of znuCB expression by SoxS is indirect and unique, suggesting several possibilities about the regulation. For example, regulation could occur via a small RNA (sRNA) akin to the effect of SoxS on ompF through the sRNA micF (51). While the gel shift studies did not reveal binding of SoxS to the znuCB promoter, we cannot completely rule out a unique direct mechanism not detected by the gel shift experiments, especially since the phenotype is seen in the cell under zinc starvation, which may result in expression of other regulators which assist SoxS in binding to the znuCB promoter, an effect that would not be observed in the cell-free environment of the gel shift assay.
Zinc is an important cofactor for bacteria and is in short supply in the mammalian host due to host sequestration factors (23, 41). Hence, disruption of znuC in the wild-type strain caused decreased persistence within the mouse kidneys (Fig. 6). Our competitive index values were similar to those found in another study of znuACB in the uropathogenic E. coli strain CFT073 (43). It is important to note that the previous study of murine kidney infection with strains KM-D and PC1012 found that a deletion of soxS alone yielded decreased persistence of the strain (10). Loss of Zn uptake may have been responsible for some of this effect, although the previous study also showed that restored persistence was not specific to SoxS. The readdition of any of the three studied transcription factors (marA, rob, and soxS) to strain PC1012 restored persistence levels to those of the wild-type strain (10). Here we showed that a low-copy-number plasmid expressing znuACB in strain PC1012 increased the bacterial load of the strain at day 4 postinfection compared to that of strain PC1012 without the plasmid, though it was only a partial restoration of the persistence levels displayed by the KM-D strain. This partial restoration of persistence with increased znuACB expression, compared to the previously observed complete restoration of persistence with the readdition of soxS to strain PC1012 (10), suggests that other SoxS-controlled factors play a role in the persistence phenotype. Additionally, the observation that readdition of marA or rob also restores persistence of strain PC1012 (10) suggests that factors other than ZnuACB that are regulated by marA and rob contribute to persistence. Besides transcriptional differences, the attenuation of strain PC1012 may be partially due to differences in the ability of PC1012 to resist the host immune factors, a sensitivity which is affected by MarA, Rob, and SoxS (50). Additional studies are being performed with other genes uncovered in the microarray to identify and assess other persistence factors controlled by these proteins in a mammalian host.
The SoxRS system senses and responds to reactive oxygen species by controlling various factors important for survival in their presence. The primary cells responsible for increased reactive oxygen species in the host also release metal ion chelators, including calprotectin (30), which has been shown to chelate Zn, causing decreased bacterial growth at the site of infection (12). Therefore, it makes evolutionary sense for SoxS to also affect the expression of zinc uptake factors to increase survival in the presence of the producers of reactive oxygen species in the host.
The use of Zn as a healing agent has been proposed (35), and several studies show that it decreases bacterial loads of various pathogens. Our data demonstrate that SoxS, a controller of oxidative stress response and antibiotic resistance, also activates znuCB expression. These findings, in addition to the fact that members of the SoxS regulon are upregulated in times of Zn depletion (48), suggest that the addition of Zn to a bacterial infection could decrease (via lowered SoxS levels) the expression of antibiotic resistance induced by SoxS. Of note, homologous znuACB-based Zn uptake systems have been linked to motility, one of the dominant pathogenicity factors necessary for bladder infection, in several uropathogens (16, 32). Also, analysis of convalescent-phase serum from Streptococcus suis revealed its ZnuA homolog to be an immunodominant protein recognized by the host in an animal infection model (53). Our finding that ZnuACB is controlled by SoxS further defines control of this important pathogenic factor.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by United States Public Health Service grant AI56021 from the National Institutes of Health and NIH training grant T32 DK07542 (to D.M.W.).
We thank Chris Parkin and the Tufts University Expression Array Core for assistance in microarray and data analysis. We thank Laura McMurry for advice and critical reading of the manuscript. We also thank Silke Patzer of Universität Tübingen for the strains SIP448 and SIP775.
Footnotes
Published ahead of print 30 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Amabile-Cuevas CF, Demple B. 1991. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 19:4479–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Armalyte J, Seputiene V, Melefors O, Suziedeliene E. 2008. An Escherichia coli asr mutant has decreased fitness during colonization in a mouse model. Res. Microbiol. 159:486–493 [DOI] [PubMed] [Google Scholar]
- 3. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barbosa TM, Levy SB. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beard SJ, Hashim R, Membrillo-Hernandez J, Hughes MN, Poole RK. 1997. Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol. Microbiol. 25:883–891 [DOI] [PubMed] [Google Scholar]
- 6. Beard SJ, et al. 2000. Evidence for the transport of zinc(II) ions via the pit inorganic phosphate transport system in Escherichia coli. FEMS Microbiol. Lett. 184:231–235 [DOI] [PubMed] [Google Scholar]
- 7. Billips BK, Schaeffer AJ, Klumpp DJ. 2008. Molecular basis of uropathogenic Escherichia coli evasion of the innate immune response in the bladder. Infect. Immun. 76:3891–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blanchard JL, Wholey WY, Conlon EM, Pomposiello PJ. 2007. Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks. PLoS One 2:e1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campoy S, et al. 2002. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 70:4721–4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Casaz P, et al. 2006. MarA, SoxS and Rob function as virulence factors in an Escherichia coli murine model of ascending pyelonephritis. Microbiology 152:3643–3650 [DOI] [PubMed] [Google Scholar]
- 11. Cohen SP, Hachler H, Levy SB. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corbin BD, et al. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962–965 [DOI] [PubMed] [Google Scholar]
- 13. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greenberg JT, Chou JH, Monach PA, Demple B. 1991. Activation of oxidative stress genes by mutations at the soxQ/cfxB/marA locus of Escherichia coli. J. Bacteriol. 173:4433–4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Griffith KL, Wolf RE., Jr 2001. Systematic mutagenesis of the DNA binding sites for SoxS in the Escherichia coli zwf and fpr promoters: identifying nucleotides required for DNA binding and transcription activation. Mol. Microbiol. 40:1141–1154 [DOI] [PubMed] [Google Scholar]
- 16. Gunasekera TS, Herre AH, Crowder MW. 2009. Absence of ZnuABC-mediated zinc uptake affects virulence-associated phenotypes of uropathogenic Escherichia coli CFT073 under Zn(II)-depleted conditions. FEMS Microbiol. Lett. 300:36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gupta K, Hooton TM, Stamm WE. 2001. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann. Intern. Med. 135:41–50 [DOI] [PubMed] [Google Scholar]
- 18. Gupta K, Stamm WE. 1999. Pathogenesis and management of recurrent urinary tract infections in women. World J. Urol. 17:415–420 [DOI] [PubMed] [Google Scholar]
- 19. Hayes ET, et al. 2006. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 6:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jair KW, et al. 1996. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 178:2507–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 22. Kolter R, Helinski DR. 1978. Construction of plasmid R6K derivatives in vitro: characterization of the R6K replication region. Plasmid 1:571–580 [DOI] [PubMed] [Google Scholar]
- 23. Liuzzi JP, et al. 2005. Interleukin-6 regulates the zinc transporter Zip14 in liver and contributes to the hypozincemia of the acute-phase response. Proc. Natl. Acad. Sci. U. S. A. 102:6843–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maneewannakul K, Levy SB. 1996. Identification for mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40:1695–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marinus MG, Casadesus J. 2009. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 33:488–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin RG, Gillette WK, Rhee S, Rosner JL. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34:431–441 [DOI] [PubMed] [Google Scholar]
- 27. McMurry LM, Levy SB. 2010. Evidence that regulatory protein MarA of Escherichia coli represses rob by steric hindrance. J. Bacteriol. 192:3977–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 29. Miller PF, Gambino LF, Sulavik MC, Gracheck SJ. 1994. Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob. Agents Chemother. 38:1773–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Motley ST, et al. 2004. Simultaneous analysis of host and pathogen interactions during an in vivo infection reveals local induction of host acute phase response proteins, a novel bacterial stress response, and evidence of a host-imposed metal ion limited environment. Cell. Microbiol. 6:849–865 [DOI] [PubMed] [Google Scholar]
- 31. Nicoloff H, Perreten V, McMurry LM, Levy SB. 2006. Role for tandem duplication and lon protease in AcrAB-TolC-dependent multiple antibiotic resistance (Mar) in an Escherichia coli mutant without mutations in marRAB or acrRAB. J. Bacteriol. 188:4413–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nielubowicz GR, Smith SN, Mobley HL. 2010. Zinc uptake contributes to motility and provides a competitive advantage to Proteus mirabilis during experimental urinary tract infection. Infect. Immun. 78:2823–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oberto J, Nabti S, Jooste V, Mignot H, Rouviere-Yaniv J. 2009. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One 4:e4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Outten CE, O'Halloran TV. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488–2492 [DOI] [PubMed] [Google Scholar]
- 35. Overbeck S, Rink L, Haase H. 2008. Modulating the immune response by oral zinc supplementation: a single approach for multiple diseases. Arch. Immunol. Ther. Exp. (Warsz.) 56:15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paterson ES, Boucher SE, Lambert IB. 2002. Regulation of the nfsA gene in Escherichia coli by SoxS. J. Bacteriol. 184:51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Patzer SI, Hantke K. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275:24321–24332 [DOI] [PubMed] [Google Scholar]
- 38. Patzer SI, Hantke K. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199–1210 [DOI] [PubMed] [Google Scholar]
- 39. Pomposiello PJ, Bennik MH, Demple B. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Puskarova A, et al. 2002. Regulation of yodA encoding a novel cadmium-induced protein in Escherichia coli. Microbiology 148:3801–3811 [DOI] [PubMed] [Google Scholar]
- 41. Rink L, Haase H. 2007. Zinc homeostasis and immunity. Trends Immunol. 28:1–4 [DOI] [PubMed] [Google Scholar]
- 42. Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609–1619 [DOI] [PubMed] [Google Scholar]
- 43. Sabri M, Houle S, Dozois CM. 2009. Roles of the extraintestinal pathogenic Escherichia coli ZnuACB and ZupT zinc transporters during urinary tract infection. Infect. Immun. 77:1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sabri M, Leveille S, Dozois CM. 2006. A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology 152:745–758 [DOI] [PubMed] [Google Scholar]
- 45. Saka K, et al. 2005. A complete set of Escherichia coli open reading frames in mobile plasmids facilitating genetic studies. DNA Res. 12:63–68 [DOI] [PubMed] [Google Scholar]
- 46. Schneiders T, Barbosa TM, McMurry LM, Levy SB. 2004. The Escherichia coli transcriptional regulator MarA directly represses transcription of purA and hdeA. J. Biol. Chem. 279:9037–9042 [DOI] [PubMed] [Google Scholar]
- 47. Seoane AS, Levy SB. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177:3414–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sigdel TK, Easton JA, Crowder MW. 2006. Transcriptional response of Escherichia coli to TPEN. J. Bacteriol. 188:6709–6713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Simon R, Priefer U, Puehler A. 1983. A broad host range mobilizing system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784–791 [Google Scholar]
- 50. Warner DM, Levy SB. 2010. Different effects of transcriptional regulators MarA, SoxS and Rob on susceptibility of Escherichia coli to cationic antimicrobial peptides (CAMPs): Rob-dependent CAMP induction of the marRAB operon. Microbiology 156:570–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wood TI, et al. 1999. Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol. Microbiol. 34:414–430 [DOI] [PubMed] [Google Scholar]
- 52. Yang X, Becker T, Walters N, Pascual DW. 2006. Deletion of znuA virulence factor attenuates Brucella abortus and confers protection against wild-type challenge. Infect. Immun. 74:3874–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang A, et al. 2009. Identification of three novel in vivo-induced expressed antigens during infection with Streptococcus suis serotype 2. FEMS Microbiol. Lett. 295:17–22 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.