Abstract
Escherichia coli dinJ-yafQ operon codes for a functional toxin-antitoxin (TA) system. YafQ toxin is an RNase which, upon overproduction, specifically inhibits the translation process by cleaving cellular mRNA at specific sequences. DinJ is an antitoxin and counteracts YafQ-mediated toxicity by forming a strong protein complex. In the present study we used site-directed mutagenesis of YafQ to determine the amino acids important for its catalytic activity. His50Ala, His63Ala, Asp67Ala, Trp68Ala, Trp68Phe, Arg83Ala, His87Ala, and Phe91Ala substitutions of the predicted active-site residues of YafQ abolished mRNA cleavage in vivo, whereas Asp61Ala and Phe91Tyr mutations inhibited YafQ RNase activity only moderately. We show that YafQ, upon overexpression, cleaved mRNAs preferably 5′ to A between the second and third nucleotides in the codon in vivo. YafQ also showed RNase activity against mRNA, tRNA, and 5S rRNA molecules in vitro, albeit with no strong specificity. The endoribonuclease activity of YafQ was inhibited in the complex with DinJ antitoxin in vitro. DinJ-YafQ protein complex and DinJ antitoxin alone selectively bind to one of the two palindromic sequences present in the intergenic region upstream of the dinJ-yafQ operon, suggesting the autoregulation mode of this TA system.
INTRODUCTION
Bacterial and archaeal type II toxin-antitoxin (TA) systems are pairs of genes coding for proteins, one a stable toxin and the other a proteolytically labile antitoxin (6, 10, 15). They are either encoded by low-copy-number plasmids or reside in the bacterial chromosome. Plasmid-borne TA systems ensure that only cells harboring plasmid survive upon segregation, whereas plasmid-free cells are postsegregationally killed by toxins when inherited labile antitoxin is degraded (21). The physiological role of chromosomally encoded TA is much less well defined. Normally, the cell produces equal amounts of both proteins, which bind into a stable complex where the activity of the toxin is inhibited. However, under certain circumstances, labile antitoxin is degraded by cellular proteases, and more stabile toxin inhibits essential cellular processes, causing growth inhibition or even cell death (6, 10, 14, 15). A growing number of TA systems observed more frequently in free-living microorganisms than in intracellular obligate microbes raised possibility that TA systems might mediate response to environmental stress by modulating the global level of biological processes such as translation and DNA synthesis (6, 10, 15, 40). In support of this hypothesis, numerous studies have demonstrated that certain E. coli TA systems are involved in the cell response to various stress conditions such as starvation, heat, oxidative shock, DNA damage, antibiotic treatment leading to bacteriostasis or cell death (7, 9, 26). Further insights into biological role of chromosomal TA systems showed their involvement in persistence, biofilm formation and multidrug tolerance (20, 27, 30, 47, 53, 55). Escherichia coli K-12 chromosome codes for at least 19 type II TA systems (57), including mazEF, relBE, yefM-yoeB, dinJ-yafQ, prlF-yhaV, chpBI-chpBK, hicAB, yafNO, hipBA, mqsAR, ygjNM, and ygiUT (3, 9, 15, 16, 23, 31, 33, 46). Nearly all E. coli chromosome-borne type II TA system toxins investigated to date inhibit translation. Most of them are endoribonucleases that, when expressed ectopically in bacterial cells, cleave mRNA with different specificity. RelE, YoeB, YafO, YafQ, and YhaV proteins exhibit translation-dependent RNase activity, whereas MazF, ChpBK, HicA, and MqsR toxins appear to act independently of the ribosomes (9, 24, 41, 42, 45, 58, 61, 62). HipA toxin inhibits translation by EF-Tu phosphorylation (48), while the recently identified RatA toxin prevents association of ribosome subunits into initiation complex (60). Some type II TA system toxins are known to inhibit replication by interacting with DNA gyrase (6, 15) and to affect peptidoglycan biosynthesis (34) and cell division (51).
We have shown earlier that E. coli YafQ toxin exhibits high structural similarity to RelE family of toxin RNases such as E. coli YoeB and Pyrococcus horikoshii RelE (33). More recently, Prysak et al. (42) demonstrated that YafQ specifically cleaves E. coli mRNA's exclusively at AAA (Lys) codons in the ribosome. These researchers showed that the DinJ-YafQ protein complex binds a palindrome which overlaps with the putative LexA binding site in the promoter DNA of the dinJ-yafQ operon.
In the present study, we used site-specific mutagenesis to determine which amino acids are important for E. coli YafQ functional activity. We show that YafQ cleaves cellular mRNA at a lower stringency than was previously described (42), preferentially at 5′ to A between second and third nucleotides in the codon and that this activity is abolished in the putative active-site mutants of YafQ. The region upstream of the dinJ-yafQ operon contains two palindromes, including one previously unrecognized palindrome that specifically interacts with the DinJ-YafQ protein complex and antitoxin DinJ alone, indicating its involvement in the autoregulation of dinJ-yafQ transcription.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
The E. coli strains and plasmids used in the present study are listed in Table 1. Bacterial cultures were grown in liquid or solid Luria-Bertani (LB) medium at 37°C. When needed, 100 μg of ampicillin ml−1 was added. For growth inhibition analysis and total RNA extraction, E. coli BW25113 containing pBAD30 and pBADyafQ plasmids or pBADyafQ mutant variants (Table 1) was grown in liquid LB medium to an optical density at 600 nm (OD600) of 0.3. The culture was then diluted 100-fold, and bacteria were grown to an OD600 of 0.1. l-Arabinose was added to a concentration of 0.2% to induce the expression of cloned genes. Incubation was continued for 2 to 4 h. At the indicated time points, the OD600 was measured, or the cell suspension was serially diluted and plated onto LB agar plates containing ampicillin. The plates were incubated at 37°C for 16 h, and the colonies were counted. All experiments were repeated at least three times.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| BL21 | F−ompT (rB− mB−) | 49 |
| BW25113 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-4) lacIp-400(lacIq) λ−rpoS396(Am) rph-1Δ(rhaD-rhaB)568 rrnB-4 hsdR514 | 12 |
| Plasmids | ||
| pBAD30 | Expression plasmid | 17 |
| pBADyafQ | 376-bp fragment with yafQ gene cloned into pBAD30 | 32 |
| pBADyafQQ20A | pBADyafQ with Gln20Ala mutation | This study |
| pBADyafQH50A | pBADyafQ with His50Ala mutation | This study |
| pBADyafQD61A | pBADyafQ with Asp61Ala mutation | This study |
| pBADyafQH63A | pBADyafQ with His63Ala mutation | This study |
| pBADyafQD67A | pBADyafQ with Asp67Ala mutation | This study |
| pBADyafQW68A | pBADyafQ with Trp68Ala mutation | This study |
| pBADyafQW68F | pBADyafQ with Trp68Phe mutation | This study |
| pBADyafQR83A | pBADyafQ with Arg83Ala mutation | This study |
| pBADyafQH87A | pBADyafQ with His87Ala mutation | This study |
| pBADyafQF91A | pBADyafQ with Phe91Ala mutation | This study |
| pBADyafQF91Y | pBADyafQ with Phe91Tyr mutation | This study |
| pBluescript II KS | Expression plasmid | Stratagene |
| pBluescript-hns | 160-bp fragment of the 5′ part of hns gene cloned into pBluescript II (KS) expression vector | This study |
| pAS2 | 1.3-kb DNA fragment with asr gene cloned into pUC19 | 50 |
| pWSK29 | Expression plasmid | 54 |
| pWasr3 | 1.3-kb DNA fragment with asr gene from pAS2, cloned into pWSK29 | This study |
| pRSETdinJyafQ | 559-bp DNA fragment with dinJ and yafQ genes cloned into pRSETb (Invitrogen) | This study |
PCR reagents, restriction enzymes, other DNA- and RNA-modifying enzymes, and kits for the molecular biology experiments were from Fermentas. All enzymes and kits were used as recommended by the supplier. The oligonucleotide primers used in the study are listed in Table 2.
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| M_Q20A | AAAACTTGCAGCGAAGCGTCATAAGGAT |
| R_Q20A | ACATCCTTTGAATATTGTCCCGAG |
| M_H50A | TATAAAGACGCGCCGCTGCAAGGTT |
| R_H50A | AACAGCTGGAAGCGGTAAAGTATTA |
| M_D61A | GGTTATCGCGCGGTCCATGTCGAAC |
| R_D61A | TTTCCATGAACCTTGCAGCGGGTG |
| M_H63A | CGCGATGCTGCGGTCGAACCGGACT |
| R_H63A | ATAACCTTTCCATGAACCTTGCAGC |
| M_D67A | GTCGAACCGGCGTGGATCCTGAT |
| R_D67A | ATGAGCATCGCGATAACCTTTCCAT |
| M_W68A | GAACCGGACGCGATCCTGATTTACA |
| M_W68F | GAACCGGACTTTATCCTGATTTACA |
| R_W68X | GACATGAGCATCGCGATAACCTTTC |
| M_R83A | CGATTTGAGGCGACTGGAACTCAC |
| R_R83A | TAAAAGTTTATCGGTAAGTTTGT |
| M_H87A | ACTGGAACTGCGGCGGCGCTCTTTG |
| R_H87A | TCTCTCAAATCGTAAAAGTTTATCG |
| F_F91X | GGGTAAAGCCCGCTTCCGATGAAA |
| M_F91A | CGCGAGCGCCGCGTGAGTTCCAGT |
| M_F91Y | GTAGAGCGCCGCGTGAGTTCCAGT |
| R acpP | CTTCAACGAAAGAAGCATTGTTGG |
| R hns | CGTTCGTTAACAACAACTTC |
| R lpp2 | AGGATTACCGCGCCCAGTACCAG |
| dinJ2 | GTAGCTGAAAGAGATATGTG |
| dinJPrF | GCTGCCTGATTCTTCAGAT |
| dinJ_Pal_Z12 | CGCTGTTGCTCATTTGAGCTACAATT |
| dinJ_Pal_Z12_R | AATTGTAGCTCAAATGAGCAACAGCG |
| dinJ_Pal_Z23 | TTTGAGCTACAATTCAAGCTGAATAA |
| dinJ_Pal_Z23_R | TTATTCAGCTTGAATTGTAGCTCAAA |
| dinJ_Pal_Z34 | TCAAGCTGAATAAATATACAGCACAG |
| dinJ_Pal_Z34_R | CTGTGCTGTATATTTATTCAGCTTGA |
| random_26 | ACTAATTACATAGTACCTCCGGGACA |
| random_26_R | TGTCCCGGAGGTACTATGTAATTAGT |
Site-directed mutagenesis of yafQ.
The oligonucleotides for the construction of YafQ predicted active-site mutants (Gln20Ala, His50Ala, Asp61Ala, His63Ala, Asp67Ala, Trp68Ala, Trp68Phe, Arg83Ala, His87Ala, Phe91Ala, and Phe91Tyr) are listed in Table 2. Mutant DNA was amplified using primers containing the mutations of interest (M_Q20A, M_H50A, M_D61A, M_H63A, M_D67A, M_W68A, M_W68F, M_R83A, M_H87A, M_F91A, and M_F91Y), appropriate reverse primers, and pBADyafQ plasmid DNA as a template using a Long-Range PCR kit (Fermentas) according to the manufacturer's recommendations. After the PCR, template DNA was digested by DpnI restriction endonuclease, ligated, and transformed into BW25113 strain. The introduced mutations and the absence of secondary mutations were verified by sequencing of plasmid DNA.
Total RNA extraction.
For isolation of total RNA for primer extension analysis, E. coli BW25113 containing pBAD30 and pBADyafQ plasmids or pBADyafQ mutant variants (Table 1) were induced as described earlier. At 0, 30, 60, and 120 min after induction, samples were withdrawn, and the total RNA was isolated using hot phenol extraction (44).
Protein purification.
Purification of protein YafQ fused to His6 [YafQ(His)6] and DinJ(His)6, as well as the DinJ-YafQ(His)6 protein complex, was performed as described earlier (34). (His)6DinJ-YafQ protein complex was obtained after overexpression of pRSETdinJyafQ plasmid (Table 1) in BL21 strain, and (His)6DinJ protein was purified from (His)6DinJ-YafQ protein complex as described earlier (33).
RNA cleavage analysis in vitro.
The asr and hns mRNAs were synthesized in vitro using T3 or T7 transcription kits (Fermentas) according the manufacturer's recommendations. Plasmid pWasr3 was created by cloning a 1.3-kb fragment encoding asr gene from plasmid pAS2 into pWSK29 through EcoRI and BamHI restriction sites. Plasmid pBluescript-hns was constructed by cloning 160-bp fragment of the 5′ part of hns gene into pBluescript IIKS expression vector (Table 1). Plasmids with cloned E. coli asr and hns genes under T3 or T7 promoters, respectively, were used as templates for in vitro transcription reactions. The asr and hns mRNAs, tRNA, and 5S rRNA were analyzed for YafQ-mediated cleavage in vitro. The digestion reaction mixtures (10 μl) contained 7.5 μg of the asr mRNA, different amounts (0.1, 0.25, and 0.5 μM) of DinJ(His)6, YafQ(His)6, or DinJ-YafQ(His)6 in 50 mM Tris-HCl (pH 7.0). hns mRNA digestion mixtures (5 μl) contained 0.2 μg of mRNA and 2 μM YafQ(His)6. When stable 5S rRNA and tRNA were used, 0.16 μg of RNA was incubated with 1 μM DinJ(His)6, YafQ(His)6, or DinJ-YafQ(His)6. The reaction mixtures were incubated at 37°C for different time intervals. Reactions were stopped by heat inactivation or by placing the samples in −20°C. asr mRNA, tRNA, and 5S rRNA digestion mixtures were loaded onto a 2% agarose gel containing 2 μg of ethidium bromide/ml with TAE buffer. hns mRNA digestion mixtures were used for primer extension reactions with hns-specific 5′-32P-labeled primer. Primer extension reactions were carried out as described below.
In vivo RNA cleavage analysis by primer extension.
For primer extension analysis, oligonucleotides complementary to different abundant E. coli mRNAs were used (Table 2). The 5′ ends of reverse primers (R acpP, R hns, and R lpp2) were labeled with [γ-32P]ATP using T4 polynucleotide kinase (Fermentas). Primer extension reactions were carried out with 5 μg of total RNA for in vivo and 0.05 μg of hns mRNA for in vitro cleavage analysis and with 1 pmol of 5′-32P-labeled primer, denaturing the mixture for 10 min in 70°C and annealing the primer by putting the sample on ice. The reverse transcription was carried out at 42°C for 1 h using Moloney murine leukemia virus reverse transcriptase (Fermentas) according to the manufacturer's recommendations in 5-μl samples. Reactions were stopped by adding 1.5 μl of formamide loading buffer (44). The extended products were incubated at 95°C for 4 min analyzed on the 6% polyacrylamide gel containing 7 M urea. The sequencing ladder was obtained by dideoxy DNA sequencing using the same primer.
EMSA.
The 376-bp DNA fragment corresponding to hypothetical dinJ-yafQ promoter/operator region was obtained by PCR using the primers dinJPrF and dinJ2 (Table 2). Reaction mixtures (10 μl) consisting of 8 pmol of PCR-amplified DNA and different amounts of DinJ-YafQ(His)6 (0, 0.5, 1, 2, and 4 pmol), DinJ(His)6 (10, 20, 40, and 80 pmol), or (His)6DinJ (10, 20, 40, and 80 pmol) in binding buffer (10 mM Tris-HCl, 50 mM NaCl, 1 mM dithiothreitol [DTT], 5 mM MgCl2, 2.5% glycerol [pH 7.5]) were incubated at 22°C temperature for 30 min. For the DinJ-YafQ(His)6 protein complex interaction with the palindromic sequences of dinJ-yafQ promoter region, the DNA fragments were prepared as follows. The dinJ_Pal_Z12, dinJ_Pal_Z23, dinJ_Pal_Z34, and random_26 oligonucleotides (Table 2) were incubated for 10 min at 95°C with appropriate reverse oligonucleotides and then left to cool to room temperature. Reaction mixtures for electromobility shift assay (EMSA; 10 μl) consisted of 12.5 pmol of each DNA fragment and 0, 8, 15, and 30 nmol of DinJ-YafQ(His)6 protein complex in binding buffer. After incubation, the samples were mixed with loading dye (50% glycerol, 10 mM Tris-HCl [pH 8.0], 0.1% Nonidet P-40, 1 mM DTT, 62.5 μg of bovine serum albumin/ml, 0.2 mg of bromophenol blue/ml), placed in 4°C for 10 min, and fractionated by 6 or 8% native polyacrylamide gel electrophoresis. The EMSA was visualized by ethidium bromide staining.
RESULTS
Modeling of YafQ toxin active site.
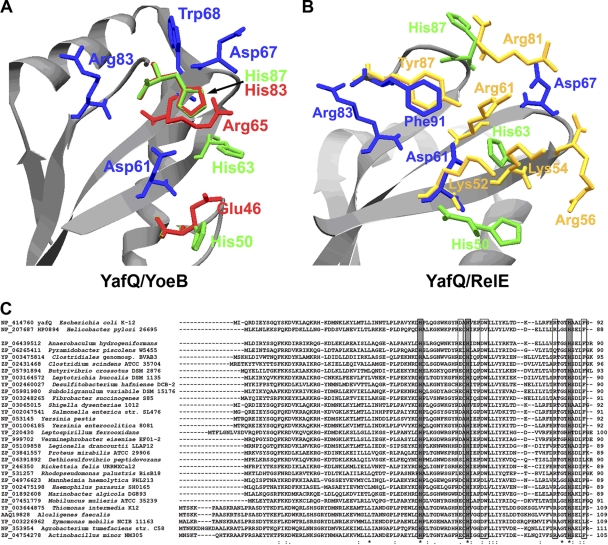
To predict the active site of YafQ, we created homology models of the protein. The models were produced using Swiss-Model protein homology modeling server (1). For homology modeling, we used the recently resolved X-ray structures of the E. coli K-12 RelE family toxins RelE and YoeB (identities of 10.75 and 20.24%, respectively) as templates (24, 35). The models produced were analyzed using ProSA-web protein structure analysis server (56). YafQ models based on the free YoeB structure (PDB code 2a6s), as well as free RelE structure (PDB code 3kha), showed scores comparable to that of experimentally determined protein structures (−4.43 and −3.03, respectively). The generated models were further used to gain information on the active site of YafQ by comparing its resemblance to the active sites of YoeB and RelE toxins.
The active sites of RelE family RNases display certain similarities as the compact, four-stranded antiparallel β sheet (2, 22, 24, 35), which is also present in YafQ models (Fig. 1A and B). Superimposition of YafQ model structures upon YoeB and RelE structures, revealed the amino acid residues of YafQ, exhibiting similar location to some catalytically important residues of YoeB and RelE RNases. His87 of YafQ matched position of His83 of YoeB, whereas Phe91 and Asp61 of YafQ corresponded to Tyr87 and Lys52 of RelE, respectively (Fig. 1A and B). Amino acids Asp61 and Asp67, able to perform as general base in the catalytic reaction, were found in the closest proximity of the presumed active site of YafQ, as well as Arg83, which is the closest arginine to YoeB Arg65 (Fig. 1A), shown to be important for phosphate binding (24). In addition, two more histidine residues, His50 and His63, were located in the predicted active site of YafQ, with His50 being in close proximity to Glu46 of YoeB. His87, His50, and His63 are absolutely conservative among YafQ homologues (Fig. 1C). Phe91 was located in similar position as Tyr87 of RelE, which is important for substrate orientation and catalysis (35).
Fig 1.
YafQ active-site prediction. (A) Active site of YafQ structural model, created using YoeB toxin structure (PDB code 2a6s). (B) Active site of YafQ structural model, created using RelE from E. coli (PDB code 3kha). The amino acids important for YoeB catalysis are drawn in red and for RelE are drawn in yellow. The absolutely conservative histidines of YafQ are drawn in green; other conservative or partially conservative amino acids of YafQ are in blue. The structure models of YafQ were created by the Swiss-Model automated protein homology modeling server (1). (C) Sequence alignment of YafQ homologues from different bacteria. Absolutely conserved histidines are indicated by gray boxes; other possibly important active-site amino acids are indicated by boxes. Identical amino acid residues are indicated by asterisks below, and similar amino acid residues are indicated with colons or dots below. Sequences were aligned using CLUSTAL W, v1.82.
Site-directed mutagenesis of YafQ.
YafQ mutants were constructed as described in Materials and Methods. Residues, conservative across the YafQ homologues (His50, His63, Trp68, Arg83, and His87), as well as partially conservative residues, located in the predicted active site of YafQ (Asp61 and Asp67) (Fig. 1C), were changed to alanines, except for Trp68, which was changed to alanine and phenylalanine, and Phe91, which was changed to alanine and tyrosine. pBAD30 plasmids, coding for yafQ gene with desired mutations, were introduced into BW25113 cells, and the toxicity of YafQ was monitored.
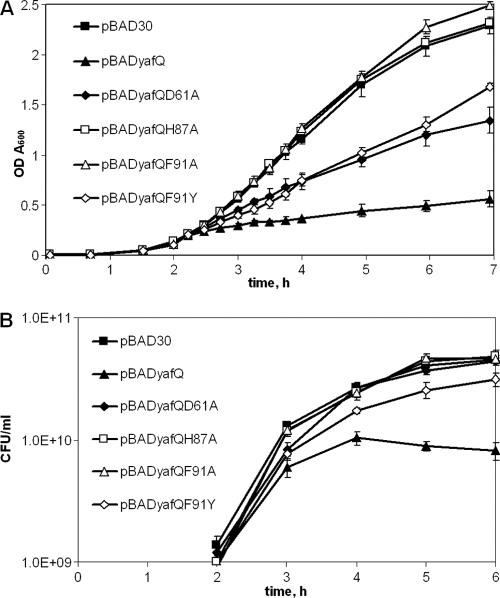
Effect of YafQ mutations on E. coli growth and viability.
The growth of BW25113 cells harboring plasmids with the mutations His50Ala, His63Ala, Asp67Ala, Trp68Ala, Trp68Phe, Arg83Ala, and His87Ala, in the presence of 0.2% of l-arabinose, resulted in a complete loss of growth inhibition compared to that of cells which harbored pBADyafQ with the wild-type yafQ gene (Fig. 2A [representative growth data of cells with plasmid yafQHis87Ala are shown]). The observed growth pattern indicates that introduced mutations of the predicted active site abolished YafQ toxicity and therefore its activity in vivo. Of the yafQ mutations tested, the expression of the YafQ with Asp61Ala mutation in BW25113 cells still resulted in significant growth inhibition upon addition of l-arabinose, albeit to a lesser extent compared to pBADyafQ plasmid with the wild-type yafQ. Also, substitution of YafQ Phe91 with the structurally similar Tyr residue led to only partial growth inhibition after induction with l-arabinose, whereas the Phe91 change to Ala resulted in a complete loss of arabinose-dependent YafQ toxicity in vivo (Fig. 2A). All yafQ mutant variants were transcribed at comparable levels after the addition of l-arabinose (data not shown). In order to test whether the introduction of a single point mutation does not alter YafQ protein structure, thus hindering its toxicity, a mutation of nonconservative residue Gln 20 in the N-part of the YafQ protein was introduced. The induction YafQ protein with Gln20Ala substitution resulted in growth inhibition similar to the wild-type YafQ (data not shown).
Fig 2.
Growth of E. coli BW25113 after induction of YafQ mutant proteins. Cells with pBADyafQ, pBAD30 and pBADyafQ plasmids with Gln20Ala, Asp61Ala, His87Ala, Phe91Ala, and Phe91Tyr mutations in yafQ gene were grown in liquid LB medium to an A600 of 0.1 as described in Materials and Methods. Then synthesis of the proteins was induced by adding 0.2% of l-arabinose (2-h time point). The growth was monitored at the selected time points as the OD600 (A) or the CFU/ml (B). The data are means of at least three independent experiments. Bars indicate the standard error.
Next, we determined the viability of BW25113 cells containing plasmids with all introduced yafQ mutations and with the wild-type yafQ at time intervals after induction with l-arabinose (Fig. 2B). After 1 h of induction, bacterial strains showed differences in viability, which resembled those observed for bacterial growth: cells expressing YafQ mutants were as viable as cells containing empty plasmid pBAD30, with the exception of cells expressing Asp61Ala and Phe91Tyr YafQ proteins, which showed an intermediate decrease in CFU compared to the wild-type YafQ-producing cells and the control cells with empty plasmid (Fig. 2B [growth data of cells containing plasmids pBADyafQ and pBAD30 and plasmids with the yafQ mutations Asp61Ala, His87Ala, Phe91Ala, and Phe91Tyr are shown). After longer time periods upon arabinose induction, marginal but reliable decreases in viability were observed only for cells expressing the Phe91Tyr YafQ mutant. Notably, cells expressing wild-type YafQ showed only a <6-fold loss in CFU if compared to control cells with empty vector, confirming that YafQ expression is not detrimental to cells differently to other TA system toxins (Fig. 2B) (32).
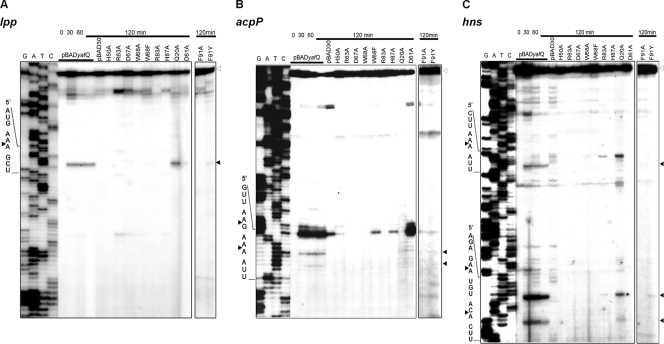
Effect of YafQ mutations on mRNA cleavage in vivo.
We have shown previously that when ectopically expressed in bacterial cells, YafQ specifically inhibits translation (33). Translation is inhibited by YafQ-mediated cleavage of cellular mRNAs in the ribosomal A site (42). Therefore, next we investigated the role of the introduced mutations on the functional activity of YafQ RNase in vivo. The wild-type YafQ and YafQ with mutations of the predicted active-site residues were expressed in BW25113 cells by the addition of l-arabinose, and at time intervals of 30, 60, and 120 min their total RNA was isolated. Primer extension analysis was performed with oligonucleotide primers, specific to the 5′ ends of three abundant cellular mRNAs: lpp, acpP, and hns. As can be seen in Fig. 3, after the induction of YafQ synthesis, truncated RNA fragments were observed when total RNA from l-arabinose-induced cells carrying plasmid pBADyafQ was used for analysis. The cleavage products were attributed to YafQ overproduction since no similar products were observed after the longest induction time (120 min) for the cells carrying an empty pBAD30 vector (Fig. 3). The amount of intact acpP mRNA (indicated by open arrowheads in Fig. 3B) was decreasing upon the induction of the YafQ toxin, indicating the shorter RNA fragments are the cleavage products. For the lpp and hns transcripts the reduction of intact transcript was less pronounced. A single cleavage site was observed for lpp, two sites were noted for acpP, and three cleavage sites were found in hns mRNAs. The Gln20Ala substitution did not alter the ability of YafQ to cleave mRNA at all sites, except for first hns cleavage site, where the RNA cleavage was not observed (Fig. 3C).
Fig 3.
Primer extension analysis of cellular transcripts after induction of YafQ mutant proteins in vivo. BW25113 strain containing plasmids pBADyafQ, pBAD30, or pBADyafQ with Gln20Ala, His50Ala, Asp61Ala, His63Ala, Asp67Ala, Trp68Ala, Trp68Phe, Arg83Ala, His87Ala, Phe91Ala, and Phe91Tyr substitutions were grown to mid-exponential phase, and the synthesis of YafQ was induced by adding 0.2% of arabinose as described in Materials and Methods. At selected time points, the total RNA was extracted and used for primer extension reactions with lpp-, acpP-, and hns-specific 5′-32P-labeled primers. The sequence ladder was obtained by dideoxy DNA sequencing reactions using the corresponding primers for primer extension. The cleavage sites of the YafQ toxin are indicated by full arrowheads, and the full-length RNAs are indicated by empty arrowheads.
The cleavage of lpp, acpP, and hns mRNA was abolished in cells with YafQ harboring His50Ala, His63Ala, Asp67Ala, Trp68Ala, Trp68Phe, His87Ala, and Phe91Ala mutations. Notably, the YafQ with Asp61Ala mutation cleaved lpp and acpP mRNA, albeit with considerably lower efficiency compared to wild-type YafQ (the Asp61Ala YafQ-mediated second cleavage site of hns transcript could only be clearly observed after prolonged exposition [data not shown]). No Asp61Ala YafQ-mediated cleavage was observed in the first and third hns mRNA cleavage sites (Fig. 3C). Interestingly, weak cleavage of the hns transcript was observed after the induction of Arg83Ala mutant of yafQ (Fig. 3C). Phe91Tyr YafQ mutant was able to cleave all three analyzed transcripts, but with lower efficiency (Fig. 3).
Sequence specificity of YafQ endoribonuclease in vivo.
The observed total six cleavage sites in analyzed transcripts (lpp, AA/A; acpP, AA/G and AA/A; hns, AA/A, GA/A, and AC/A) showed that YafQ preferentially cleaved RNA at the 5′ side to A (in one case, 5′ to G), between the second and third nucleotides in the codon. The RNA cleavage sites were found to be distributed relatively rarely. The alignment of all observed cleavage sites revealed the preferred cleavage sequence for YafQ as 5′-AA↓A −3′. However, the hns transcript was also cleaved at 5′-GA↓A-3′ and 5′-AC↓A-3′ positions, and acpP was also cleaved at the 5′-AA↓G-3′ position, suggesting that YafQ might display a specificity other than just for AAA codons.
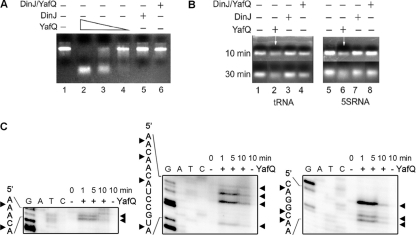
YafQ RNase activity analysis in vitro.
We performed in vitro reactions with the purified YafQ(His)6 protein and various RNA substrates. The YafQ(His)6 protein was obtained from purified DinJ-YafQ(His)6 complex as described earlier (33). We first analyzed whether YafQ cleaved E. coli asr mRNA in vitro as described in Materials and Methods. RNA was incubated with various concentrations of YafQ toxin or DinJ-YafQ(His)6 protein complex for 30 min at 37°C. As shown in Fig. 4A, lanes 2 to 4, YafQ efficiently cleaved asr mRNA in a dose-dependent manner. When present in purified DinJ-YafQ(His)6 complex, YafQ did not exhibit RNase activity (Fig. 4A, lane 6). Purified DinJ(His)6 had no activity on RNA (Fig. 4A, lane 5). We then sought to determine whether YafQ is active against small RNAs with a stable secondary structure such as tRNA and 5S rRNA. Incubation for 30 min at 37°C with increasing amounts of purified YafQ(His)6 did not result in tRNA and 5S rRNA cleavage (data not shown), indicating that YafQ toxin might be active against single-stranded RNA. Indeed, YafQ cleaved both RNA substrates only after their partial denaturation by heating for 10 min at 70°C (Fig. 4B, lanes 2 and 4). Again, the RNase activity of YafQ was blocked in the DinJ-YafQ protein complex (Fig. 4B, lanes 4 and 8). These results indicated that YafQ RNase has preference for single-stranded RNA cleavage.
Fig 4.
Analysis of RNA cleavage by YafQ toxin in vitro. (A) In vitro-synthesized substrate (asr) mRNA (7.5 μg) was incubated for 30 min in 37°C in 50 mM Tris-HCl (pH 7.0) with, respectively, 0.5, 0.25, and 0.1 μM YafQ(His)6 (lanes 2 to 4), 0.5 μM DinJ(His)6 (lane 5), and 0.5 μM DinJ-YafQ(His)6 protein complex (lane 6). (B) 5S rRNA or tRNA (0.16 μg) (lanes 1 and 5) was denatured at 70°C for 10 min and then incubated for 30 min in 37°C in 50 mM Tris-HCl (pH 7.0) with 1 μM YafQ(His)6 (lanes 2 and 6), 1 μM DinJ(His)6 (lanes 3 and 7), 1 μM DinJ-YafQ(His)6 complex (lanes 4 and 8). The sample volume was 10 μl, and samples were analyzed in a 2% agarose gel, run for 10 or 30 min as indicated on the left (in panel B). (C) In vitro-synthesized substrate (hns) mRNA (0.2 μg) was incubated for 1, 5, or 10 min in 37°C in 50 mM Tris-HCl (pH 7.0) with 2 μM YafQ(His)6 (“+” lanes) or without YafQ(His)6 (“−” lanes); the sample volume was 5 μl. After incubation, the samples were heat inactivated and used for primer extension reactions with hns-specific 5′-32P-labeled primers. The sequence ladder was obtained by dideoxy DNA sequencing reactions with corresponding primers used for primer extension. The cleavage sites of the YafQ toxin are indicated by full arrowheads.
To define the YafQ cleavage specificity in vitro, we next performed primer extension reactions after incubation of the hns mRNA with purified YafQ(His)6 protein as described in Materials and Methods. In the presence of 2 μM YafQ(His)6, hns mRNA was cleaved at multiple sites (Fig. 4C, “+” lanes), most frequently before the adenine (A) residue (Fig. 4C).
DinJ-YafQ protein complex and DinJ bind a hypothetical promoter/operator region of dinJ-yafQ operon.
We also analyzed whether DinJ and YafQ proteins bind to the promoter region of dinJ-yafQ, as has been proposed for many of TA systems (25, 29, 45). The 376-bp DNA fragment, used for assay, included the whole intergenic region between dinJ-yafQ operon and yafL gene and also the predicted LexA binding site (28) (Fig. 5A). We analyzed by EMSA whether purified DinJ-YafQ(His)6 protein complex and DinJ protein with a histidine tag located at its N or C terminus were able to bind specifically to target DNA. As can be seen in Fig. 5B, the presence of 0.5 pmol of DinJ-YafQ(His)6 complex (lanes 2 to 5) or 10 pmol of DinJ-(His)6 protein (Fig. 5B, lanes 6 to 9) caused band shift of 376-bp DNA fragment. The observed shift was more pronounced with increasing amounts of DinJ-YafQ(His)6 protein complex and DinJ-(His)6. No shift was observed for DNA, incubated with up to 80 pmol of (His)6-DinJ protein (Fig. 5B, lanes 10 to 13), suggesting that the N terminus of DinJ might by involved in the DNA binding. Secondary structure prediction of DinJ using PSIPRED server (5) revealed a ribbon-helix-helix (RHH) motif in the N-terminal part of the protein, which was shown to be involved in DNA binding of RelB and ParD (36, 39). YafQ(His)6 protein did not bind the target DNA (data not shown). In control experiment, the binding of DinJ-YafQ(His)6 to the 376-bp DNA fragment was effectively outcompeted in the presence of an excess of unlabeled dinJ-yafQ promoter DNA but not the nonspecific DNA (data not shown).
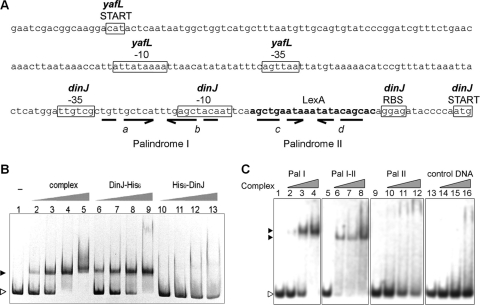
Fig 5.
Analysis of dinJ-yafQ promoter region and its interaction with DinJ-YafQ(His)6 protein complex. (A) E. coli intergenic region containing dinJ-yafQ promoter and palindromic sequences. The important predicted (BPROM) promoter sequences of dinJ and yafL genes are boxed, the putative LexA binding site is indicated in boldface, broken arrows indicate palindromic sequences, and letters in italics indicate the palindrome sequence regions. (B) EMSA analysis of interaction of DinJ-YafQ(His)6 complex with 376-bp dinJ-yafQ promoter DNA fragment. A total of 8 pmol of PCR-amplified dinJ-yafQ promoter DNA fragments was incubated in 10 μl of binding buffer (10 mM Tris-HCl, 50 mM NaCl, 1 mM DTT, 5 mM MgCl2, 2.5% glycerol [pH 7.5]) with increasing amounts of protein at 22°C for 30 min. The EMSA was visualized in a 6% polyacrylamide gel stained with ethidium bromide. Lanes 1 to 5, promoter DNA incubated with 0, 0.5, 1, 2, and 4 pmol of DinJ-YafQ(His)6 protein complex; lanes 6 to 9, promoter DNA with 10, 20, 40, and 80 pmol of DinJ(His)6, lanes 10 to 13, promoter DNA with 10, 20, 40, and 80 pmol of (His)6DinJ. (C) EMSA analysis of DinJ-YafQ(His)6 protein complex interaction with the palindromic sequences of dinJ-yafQ promoter region. EMSA was performed to detect the interactions of purified DinJ-YafQ(His)6 protein complex with three 26-nucleotide regions of the dinJ-yafQ promoter, containing palindrome I (Pal I), 3′ part of palindrome I and the 5′ part of palindrome II (Pal I-II), and palindrome II (Pal II). Random DNA sequence was used as a control (control DNA). A total of 12.5 pmol of each DNA fragment (prepared as described in Materials and Methods) was incubated in 10 μl of binding buffer (10 mM Tris-HCl, 50 mM NaCl, 1 mM DTT, 5 mM MgCl2, 2.5% glycerol [pH 7.5]) with increasing amounts of protein (0, 8, 15, and 30 nmol) at 22°C for 30 min. EMSA results were visualized by ethidium bromide staining of an 8% polyacrylamide gel. The open arrowhead indicates free DNA; full arrowheads indicate DNA bound in complex with DinJ-YafQ(His)6.
We have identified several palindromic sequences in the promoter region of dinJ-yafQ by sequence analysis (BPROM; Softberry, Inc., Mount Kisco, NY) (Fig. 5A). The palindrome II overlaps with the DNA sequence, which shows similarity to a consensus LexA binding site. Also, palindromes I and II share sequence similarities enabling them to form a palindrome with 3′ part of palindrome I and 5′ part of palindrome II (Fig. 5A, indicated by “b” and “c”, respectively). We next examined the role of palindromes in the specific binding of DinJ-YafQ(His)6 protein complex. DNA fragments containing inverted repeats, were prepared as described in Materials and Methods. Incubation of DNA fragments harboring palindromes I and II with 8 to 30 nmol of purified DinJ-YafQ(His)6 protein complex resulted in the band shift of palindrome I DNA (Fig. 5C, lanes 2 to 4). No interaction of toxin-antitoxin complex was observed for palindrome II and control DNA (Fig. 5C, lanes 10 to 12 and lanes 14 to 16, respectively). The DNA fragment containing 3′ part of palindrome I and 5′ part of palindrome II also resulted in DNA shift when incubated with DinJ-YafQ(His)6 protein complex. Lower concentrations of protein complex were required to bind all palindrome I-II DNA compare to palindrome I DNA (Fig. 5C, lanes 4 and 6). Also, a difference in DNA-protein complex migration was observed (Fig. 5C, indicated by black arrowheads).
DISCUSSION
In this study we have determined amino acids important for E. coli YafQ toxin functional activity by YafQ active-site structure modeling and site-directed mutagenesis. We also present new data on mRNA cleavage specificity by YafQ and new features of a regulatory region of dinJ-yafQ locus.
YafQ exhibits high structural similarity to bacterial and archaeal TA toxins, which belong to RelE superfamily of RNases (E. coli YoeB, Pyrococcus horikoshii and E. coli RelE, bacteriophage T4 RegB) and also possesses a structural fold, similar to some genuine bacterial RNases such as barnase, T1, colicins D and E5, and Sa2 RNase (24, 33, 37, 46). Since the structure and the organization of the active site of YafQ is still unknown, we compared several models of YafQ with RelE family RNases with resolved X-ray structures. The YafQ active-site region showed similarities with several RNases, none of the known active-site composition matching perfectly to YafQ. The active-site residues involved in catalysis are generally glutamate for the general base and histidine for the general acid, as has been described for RelE family bacterial RNases (2, 22, 24). The active-site residues of E. coli YoeB are similar to RNase Sa2 (2) and T1 (22), involving Glu and His as the general base and acid, respectively (24). Similar catalytic amino acid residues were proposed for Helicobacter pylori (strain 26695) protein HP0894 (19, 20), the recently described RelE family toxin, which exhibits the highest homology to YafQ (29.54%) among RelE family toxins (Fig. 1C) (18). The amino acid residues responsible for the catalysis can differ, as has been shown recently for RelE toxin (35). The predicted active site of RelE, although similarly positioned, is based on Tyr, which acts as a general base, due to a high local concentration of positive charge, and also participates in the substrate base orientation (35).
When analyzing the active-site area of YafQ, we observed a higher similarity to YoeB than to RelE toxin (Fig. 1A and B). Thus, His87, the possible candidate for general acid of YafQ, is at the similar location as His83 of YoeB (Fig. 1A) and is also homologous to catalytically important His84 from H. pylori HP0894 protein (18). His87 has been previously proposed to be important for YafQ catalytic activity (33). Indeed, Prysak et al. (42) have shown recently that mutation of His87 of YafQ abolished its RNase activity while retaining the ability to bind the ribosome. In our study, replacement of all three conservative histidine residues (His87, His63, and His50) of YafQ with alanines abolished its RNase activity, indicating their functional importance. His63 does not colocalize with any of the catalytically important residues of RelE and YoeB toxins, except for RelE Lys54 (Fig. 1B), which might participate in the stabilization of the transition state. Surprisingly, mutation of YafQ Asp61 was not crucial for the catalysis, although position of Asp61 overlaps with Lys52 of RelE, which is proposed to act as general base (35) and also is homologous to Glu58 of HP0894 (Fig. 1C), which probably has the same role (18). Moreover, homology modeling showed that His50 from YafQ is in close position to Glu46 from YoeB, which is general base here (Fig. 1A). Therefore, it is possible that due to local organization of the active site, histidines might have capacity to act as a general base and a general acid in the YafQ-mediated catalysis. The role of general base for Asp67 should not be excluded also, since it was crucial for the toxicity of YafQ. Interestingly, Asp67 is not absolutely conserved across YafQ homologues, although in most cases is replaced by Glu residue, suggesting their similar functional role (Fig. 1C).
Among key components of the active site of bacterial RNases, including RelE family toxins, are arginine residues, which participate in the phosphate binding (2, 24, 35). None of the YafQ Arg residues located in the proximity of the predicted active-site match Arg65 of YoeB and Arg61 of RelE (Fig. 1A and B), proposed to perform this role (24, 35). Instead, the Arg83 of YafQ is homologous to Arg80 of HP0894 toxin (Fig. 1C), which has been shown critical for catalysis and RNA specificity and suggested to play a role corresponding to Arg65 of YoeB (18). Inactivation of YafQ Arg83 led to loss of toxicity, still the toxin was able to cleave some RNA in vivo (Fig. 3C). Arg83 of YafQ could, similarly to Arg65 of YoeB, bind phosphate and stabilize the transition state.
The proposed recognition of the second position nucleotide in RelE-mediated mRNA cleavage is by nucleotide base stacking with Tyr87, which also acts as general base in catalysis (35). YafQ structural model contains Phe91 in a similar location to Tyr87 of RelE (Fig. 1B). This residue is conserved in H. pylori HP0894 toxin as Phe88 (Fig. 1C) and has been proposed to be involved in RNA substrate binding according the analysis of nuclear magnetic resonance titration with RNA substrate homologues (18). Therefore, the recognition of the second position nucleotide in the A site of the ribosome could be similar to RelE. Indeed, replacement of YafQ Phe91 by structurally similar Tyr residue resulted in partial loss of YafQ functional activity arguing its role in substrate orientation and also in catalysis.
Despite the high structural similarity, the cleavage specificity of RelE family RNases, known to date, varies between strong for bacteriophage RegB (GG↓AG) (52) to relatively weak as for E. coli toxins YoeB (A↓ or G↓) (24) and RelE (Py Pu↓G) (35, 41). When analyzing RNase activity of YafQ mutants in vivo, we observed that although YafQ shows preference for the AAA codons, cleaving between second and third positions (as has been earlier shown by Prysak et al. [42]), the cleavage site is not stringent. Importantly, AAA codons of some of the mRNA investigated (gatY), situated close to the translation start, were not cleaved (data not shown). Our observed cleavage sites were in most cases Pu Pu↓Pu, the first and the third sites mostly being A's. Since we were unable to detect any significant sequence specificity, it is possible that specificity of YafQ-mediated RNA cleavage is directed more to the position of RNA in the ribosome and not the RNA sequence itself, as suggested by Christensen-Dalsgaard et al. (9) for YafO and HigB toxins. In contrast to YafO and HigB, relatively few YafQ cleavage sites were observed, indicating that other factors such as availability of cleavage site in a single stranded structure of RNA or positioning in the ribosome are important for selection. In the model of RelE toxin, presented by Neubauer et al. (35), the specificity of the cleavage is mostly determined by positioning of the toxin in the ribosome A site, with the possibility of specific or favored recognition of the second and third positions (35). The specific cleavage of YoeB toxin is still under the question, since this toxin was earlier shown to have a weak intrinsic RNase activity for purines (24), although later it was suggested that YoeB might cleave while bound to the ribosome, in the proximity of the start codon also yielding rare cleavage sites (59). Intrinsic RNase activity was also observed for HP0894 protein by Han et al. (18). HP0894 protein cleaved target mRNA in vitro at multiple sites before adenine (A) or guanine (G) residues, A was preferred to G. The immediate upstream base of the A and G was U and C (18). Whether HP0894 toxin exhibits the same cleavage specificity in vivo remains to be determined. The purified YafQ(His)6 cleaved target hns mRNA in vitro at multiple sites, with the preferred base A after the cleavage site (Fig. 4C). In the study by Prysak et al. (42), another target, bacteriophage MS2 RNA, was cleaved by YafQ(His)6 before G (most frequently in the middle of GG) and A. Therefore, both YafQ and HP0894 RNases display clear preference for purines downstream the cleavage site.
YafQ cleavage in vitro showed a preference for the single-stranded RNA. Cleavage of stable, largely double-stranded 5S rRNA and tRNA was achieved after denaturing the RNA and the subsequent exposure of single-stranded regions. Similar effects were also noticed for MazF family toxins, where the cleavage depended on the formation of certain RNA structures (61, 62). Although RelE toxin requires ribosome 16S rRNA residue C1054 for substrate orientation (35) and is unable to cleave RNA in its free form (40), YoeB (24), HP0894 toxin (18) and YafQ might be able to cleave the substrate without ribosome assistance.
The expression of chromosomal TA systems is autoregulated in the cell without causing any effect on the cellular processes. Whenever the antitoxin is degraded for some reason (e.g., nutrient deprivation causing a halt in translation or activation of proteases due to stress), a toxin becomes active (6, 8). The amount of the TA in the cell must be regulated, which is achieved by autorepression of the promoter of TA operon (25, 29, 45). Some antitoxins alone are able to interact with the promoter sequence, although often antitoxin interaction with the toxin facilitates the binding (25, 29, 38, 39). In the case of the RelBE system, the binding activity of the protein to DNA is increased when the RelBE complex is used (29, 38, 39), similarly to YefM-YoeB and other TA proteins (25, 45). The specific interaction of DinJ-YafQ(His)6 complex with DNA was also stronger than with antitoxin alone. Similarly as described for YefM, RelB, Phd, ParD, and other antitoxins (25, 29, 36, 63), the N-terminal part of DinJ was important for DNA binding, preventing (His)6DinJ protein from protein-DNA interactions. By protein secondary structure prediction, we found an RHH motif in the N-terminal part of DinJ antitoxin; therefore, the interaction with DNA could be similar to that of ParD or RelB. The latter antitoxins possess the RHH motifs at their N-terminal ends and interact with operator sites by insertion of the antiparallel β-strand into the major groove of DNA (36, 38).
A universal feature of the regulation of TA systems is by the presence of palindromic sequences in the promoter region (4, 25, 29, 39, 45). Direct and inverted repeats are also present in RHH antitoxin operator sites (36, 38). We detected two imperfect inverted repeats in the promoter region of dinJ-yafQ operon, one previously known to harbor a putative LexA box (28) and another, earlier unrecognized, palindrome situated upstream from it. The palindromes were longer and more distant between each other than the hexad repeats of relBE operator or the L and S palindromes of yefM-yoeB (25, 39). We show that palindrome I and hybrid palindrome (containing the 3′ part of palindrome I and the 5′ part of palindrome II) strongly and specifically bound the DinJ-YafQ protein complex, whereas palindrome II was unable to interact with it (only a smear was seen on EMSA). Less protein complex was required to shift DNA-containing hybrid palindrome compare to palindrome II. It is possible that the 3′ part of palindrome I is recognized first and then oligomeric protein complex is formed, recruiting DinJ antitoxins with their cognate toxins to palindrome I or palindrome II. There are, however, controversial reports regarding the involvement of the dinJ-yafQ locus in the SOS response and the regulatory role of the putative LexA box (11, 13, 42, 43). Although Prysak et al. (42) have demonstrated the binding of LexA and DinJ-YafQ complex to palindrome containing the putative LexA box, an earlier study showed no LexA-dependent regulation of this locus in a response to DNA damage (13). We noticed a similar effect when we analyzed the dinJ-yafQ-specific mRNA level in E. coli cells upon UV irradiation (data not shown). Therefore, the role of the dinJ-yafQ locus in the DNA damage-induced response, if any, remains to be clarified.
ACKNOWLEDGMENTS
This study was supported by Lithuanian State Science and Studies Foundation grant T-71/09. M.J. and G.B. were supported by a Lithuanian Science Council Student Research Fellowship Award.
We thank Česlovas Venclovas and Kęstutis Sužiedėlis for helpful comments and advice.
Footnotes
Published ahead of print 13 January 2012
REFERENCES
- 1. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The Swiss-Model Workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 2. Bauerová-Hlinková V, Dvorský R, Perecko D, Povazanec F, Sevcík J. 2009. Structure of RNase Sa2 complexes with mononucleotides: new aspects of catalytic reaction and substrate recognition. FEBS J. 276:4156–4168 [DOI] [PubMed] [Google Scholar]
- 3. Black DS, Kelly AJ, Mardis MJ, Moyed HS. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173:5732–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown BL, Page R. 2010. Preliminary crystallographic analysis of the Escherichia coli antitoxin MqsA (YgiT/b3021) in complex with mqsRA promoter DNA. Acta Crystallogr. Sect. F Struct. Biol. Crystallogr. Commun. 66:1060–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bryson K, et al. 2005. Protein structure prediction servers at University College London. Nucleic Acids Res. 33:W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. 2005. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30:672–679 [DOI] [PubMed] [Google Scholar]
- 7. Christensen SK, Pedersen K, Hansen FG, Gerdes K. 2003. Toxin-antitoxin loci as stress response elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809–819 [DOI] [PubMed] [Google Scholar]
- 8. Christensen SK, et al. 2004. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 51:1705–1717 [DOI] [PubMed] [Google Scholar]
- 9. Christensen-Dalsgaard M, Jørgensen MG, Gerdes K. 2010. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75:333–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Condon C. 2006. Shutdown decay of mRNA. Mol. Microbiol. 61:573–583 [DOI] [PubMed] [Google Scholar]
- 11. Courcelle J, Khodursky A, Peter A, Brown PO, Hanawalt PC. 2001. Comparative gene expression profiles following UV exposure in wild type and SOS-deficient Escherichia coli. Genetics 158:41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fernandez de Henestrosa AR, et al. 2002. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560–1572 [DOI] [PubMed] [Google Scholar]
- 14. Gerdes K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182:561–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gerdes K, Christensen SK, Lobner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371–382 [DOI] [PubMed] [Google Scholar]
- 16. Gotfredsen M, Gerdes K. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065–1076 [DOI] [PubMed] [Google Scholar]
- 17. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han KD, et al. 2011. Functional identification of toxin-antitoxin molecules from Helicobacter pylori 26695 and structural elucidation of the molecular interactions. J. Biol. Chem. 286:4842–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han KD, Park SJ, Jang SB, Son WS, Lee BJ. 2005. Solution structure of conserved hypothetical protein HP0894 from Helicobacter pylori. Proteins 61:1114–1116. [DOI] [PubMed] [Google Scholar]
- 20. Harrison JJ, et al. 2009. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 53:2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hayes F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496–1499 [DOI] [PubMed] [Google Scholar]
- 22. Heinemann U, Saenger W. 1983. Crystallographic study of mechanism of ribonuclease T1-catalyzed specific RNA hydrolysis. J. Biomol. Struct. Dyn. 2:523–538 [DOI] [PubMed] [Google Scholar]
- 23. Jörgensen MG, Pandey DP, Jaskolska M, Gerdes K. 2009. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 191:1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamada K, Hanaoka F. 2005. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 19:497–509 [DOI] [PubMed] [Google Scholar]
- 25. Kedzierska B, Lian LY, Hayes F. 2007. Toxin-antitoxin regulation: bimodal interaction of YefM-YoeB with paired DNA palindromes exerts transcriptional autorepression. Nucleic Acids Res. 35:325–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kolodkin-Gal I, Engelberg-Kulka H. 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J. Bacteriol. 188:3420–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kolodkin-Gal I, Verdiger R, Shlosberg-Fedida A, Engelberg-Kulka H. 2009. A differential effect of Escherichia coli toxin-antitoxin systems on cell death in liquid media and biofilm formation. PLoS One 4:e6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lewis LK, Harlow GR, Gregg-Jolly LA, Mount DW. 1994. Identification of high-affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J. Mol. Biol. 241:507–523 [DOI] [PubMed] [Google Scholar]
- 29. Li GY, Zhang Y, Inouye M, Ikura M. 2008. Structural mechanism of transcriptional autorepression of the Escherichia coli RelB/RelE antitoxin/toxin module. J. Mol. Biol. 380:107–119 [DOI] [PubMed] [Google Scholar]
- 30. Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 108:13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175:6850–6856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Motiejūnaitė R, Armalytė J, Šeputienė V, Sužiedėlienė E. 2005. Escherichia coli dinJ-yafQ operon shows characteristic features of bacterial toxin-antitoxin modules. Biologija 4:9–15 [Google Scholar]
- 33. Motiejūnaitė R, Armalytė J, Markuckas A, Sužiedėlienė E. 2007. Escherichia coli dinJ-yafQ genes act as a toxin-antitoxin module. FEMS Microbiol. Lett. 268:112–119 [DOI] [PubMed] [Google Scholar]
- 34. Mutschler H, Gebhardt M, Shoeman RL, Meinhart A. 2011. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 9:e1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Neubauer C, et al. 2009. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139:1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oberer M, Zangger K, Gruber K, Keller W. 2007. The solution structure of ParD, the antidote of the ParDE toxin-antitoxin module, provides the structural basis for DNA and toxin binding. Protein Sci. 16:1676–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Odaert B, et al. 2007. Structural and functional studies of RegB, a new member of a family of sequence-specific ribonucleases involved in mRNA inactivation on the ribosome. J. Biol. Chem. 282:2019–2028 [DOI] [PubMed] [Google Scholar]
- 38. Overgaard M, Borch J, Gerdes K. 2009. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J. Mol. Biol. 394:183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Overgaard M, Borch J, Jørgensen MG, Gerdes K. 2008. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol. 69:841–857 [DOI] [PubMed] [Google Scholar]
- 40. Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pedersen K, et al. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131–140 [DOI] [PubMed] [Google Scholar]
- 42. Prysak M, et al. 2009. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame dependent mRNA cleavage. Mol. Microbiol. 71:1071–1087 [DOI] [PubMed] [Google Scholar]
- 43. Quillardet P, Rouffaud M-A, Bouige P. 2003. DNA array analysis of gene expression in response to UV irradiation in Escherichia coli. Res. Microbiol. 154:559–572 [DOI] [PubMed] [Google Scholar]
- 44. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Schmidt O, et al. 2007. prlF and yhaV encode a new toxin-antitoxin system in Escherichia coli. J. Mol. Biol. 372:894–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sevcik J, Dauter Z, Lamzin V, Wilson K. 1996. Ribonuclease from Streptomyces aureofaciens at atomic resolution. Acta Crystallogr. D 52:327–344 [DOI] [PubMed] [Google Scholar]
- 47. Shah D, et al. 2006. Persisters: a distinct physiological state of Escherichia coli. BMC Microbiol. 6:536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schumacher MA, et al. Molecular mechanisms of HipA mediated multidrug tolerance and its neutralization by HipB. Science 323:396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 50. Sužiedėlienė E, Sužiedėlis K, Garbenčiūtė V, Normark S. 1999. The acid-inducible asr gene in Escherichia coli: transcriptional control by the phoBR operon. J. Bacteriol. 181:2084–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tan O, Awano N, Inouye M. 2011. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FstZ and MreB. Mol. Microbiol. 79:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Uzan M, Favre R, Brody E. 1988. A nuclease that cuts specifically in the ribosome binding site of some T4 mRNAs. Proc. Natl. Acad. Sci. U. S. A. 85:8895–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Van Melderen L. 2010. Toxin-antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 13:781–785 [DOI] [PubMed] [Google Scholar]
- 54. Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing, and gene expression in Escherichia coli. Gene 100:195–199 [PubMed] [Google Scholar]
- 55. Wang X, et al. 2011. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7:359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wiederstein M, Sippl MJ. 2007. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35:W407–W410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yamagushi Y, Inouye M. 2011. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 9:779–790 [DOI] [PubMed] [Google Scholar]
- 58. Yamaguchi Y, Park JH, Inouye M. 2009. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 284:28746–28753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Y, Inouye M. 2009. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J. Biol. Chem. 284:6627–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Y, Inouye M. 2011. RatA (YfjG), an Escherichia coli toxin, inhibits 70S ribosome association to block translation initiation. Mol. Microbiol. 79:1418–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang Y, Zhang J, Hara H, Kato I, Inouye M. 2005. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 280:3143–5310 [DOI] [PubMed] [Google Scholar]
- 62. Zhang Y, Zhu L, Zhang J, Inoue M. 2005. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J. Biol. Chem. 280:26080–26088 [DOI] [PubMed] [Google Scholar]
- 63. Zhao X, Magnuson RD. 2005. Percolation of the Phd repressor operator interface. J. Bacteriol. 187:1901–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]