Abstract
Streptococcus suis serotype 2 (S. suis 2) is an important swine and human pathogen responsible for septicemia and meningitis. A novel gene, designated atl and encoding a major autolysin of S. suis 2 virulent strain HA9801, was identified and characterized in this study. The Atl protein contains 1,025 amino acids with a predicted molecular mass of 113 kDa and has a conserved N-acetylmuramoyl-l-alanine amidase domain. Recombinant Atl was expressed in Escherichia coli, and its bacteriolytic and fibronectin-binding activities were confirmed by zymography and Western affinity blotting. Two bacteriolytic bands were shown in the sodium dodecyl sulfate extracts of HA9801, while both were absent from the atl inactivated mutant. Cell chains of the mutant strain became longer than that of the parental strain. In the autolysis assay, HA9801 decreased to 20% of the initial optical density (OD) value, while the mutant strain had almost no autolytic activity. The biofilm capacity of the atl mutant was reduced ∼30% compared to the parental strain. In the zebrafish infection model, the 50% lethal dose of the mutant strain was increased up to 5-fold. Furthermore, the adherence to HEp-2 cells of the atl mutant was 50% less than that of the parental strain. Based on the functional analysis of the recombinant Atl and observed effects of atl inactivation on HA9801, we conclude that Atl is a major autolysin of HA9801. It takes part in cell autolysis, separation of daughter cells, biofilm formation, fibronectin-binding activity, cell adhesion, and pathogenesis of HA9801.
INTRODUCTION
Bacteria produce several peptidoglycan hydrolases, some of which are autolysins able to disintegrate their own peptidoglycan saccules and lead to bacterial cell lysis in unfavorable conditions (44). Autolysins have been implicated in various biological functions, such as cell wall turnover, cell separation, cell division and antibiotic-induced autolysis (48, 55). In addition to their biological functions, bacterial autolysins are also involved or implicated in the pathogenicity of Gram-positive bacteria. Intact autolytic function is required for full virulence in Streptococcus pneumoniae (7). Autolysin-deficient mutants, including a LytA mutant of S. pneumoniae (8), an AtlE mutant of Staphylococcus epidermidis (46), and Ami (36), Auto (11), p60 (41), and MurA (28) mutants of Listeria monocytogenes are less virulent in animal models than their parental strains.
Streptococcus suis is an important zoonotic pathogen that causes a variety of serious diseases, including meningitis, arthritis, and septicemia and even sudden death in pigs and humans (35, 49). Among the 35 serotypes of S. suis that have been described, S. suis serotype 2 (S. suis 2) is the most virulent and most frequently isolated serotype. Although a set of virulence factors have been identified, the pathogenic mechanisms of S. suis 2 are still unclear. Autolysins are believed to play an important role in cell wall metabolism and in the pathogenicity of bacteria (7). However, the concentration of autolysins in S. suis 2 is rather low. Recently, a novel S. suis 2 gene (GenBank accession no. EF563971) was identified using in vivo-induced antigen technology (IVIAT) (17). When analyzed by software, the protein designated Atl was found to contain six repeated GBS_Bsp-like domains and one N-acetylmuramoyl-l-alanine amidase domain.
In this study, we cloned the autolysin gene, atl, from the S. suis 2 virulent strain HA9801 and characterized its activity and function using an atl deletion mutant and complementation strain compared with the wild type. The bacteriolytic and fibronectin-binding activity as well as the biofilm capacity of the three strains were evaluated. In addition, the in vitro adherence to eukaryotic HEp-2 cells and in vivo virulence in zebrafish were determined.
MATERIALS AND METHODS
Bacterial strains and cell culture.
Bacterial strains and plasmids used in this study are listed in Table 1.S. suis 2 strains were grown in Todd-Hewitt broth (THB) (Difco Laboratories, Detroit, MI) medium or plated on THB agar containing 5% (vol/vol) sheep blood. Escherichia coli strains were cultured in Luria broth (LB) liquid medium or plated on LB agar. S. suis 2 strains were grown in THB supplemented with 2% yeast extract (THY) for preparation of competent cells. When necessary, antibiotics were added to the plate or broth at the following concentrations: chloramphenicol (Sigma, St. Louis, MO), 5 μg/ml for S. suis, 10 μg/ml for E. coli; spectinomycin (Sigma), 100 mg/ml for S. suis, 50 mg/ml for E. coli; ampicillin (Sigma), 100 μg/ml for E. coli.
Table 1.
Summary of bacterial strains and plasmids
| Strain or plasmid | Characteristic or function | Source or reference |
|---|---|---|
| Bacterial strains | ||
| HA9801 | Virulent strain of SS2 isolated from a pig that died from acute septicemia | Collected in our laboratory |
| HA9801 Δatl mutant | Deletion mutant of atl with HA9801 background, Cmr | This study |
| Δatl complement strain | HA9801 Δatl complemented with the vector pSET2::atl, Cmr Spcr | This study |
| E. coli DH5α | Cloning host for maintaining the recombinant plasmids | Invitrogen |
| E. coli BL21 | Host for expressing the recombinant protein Atl | Invitrogen |
| Plasmids | ||
| pET32a | Expression vector, Ampr | Invitrogen |
| pSET4S | Thermo-sensitive suicide vector for gene replacement in S. suis replication of pG + host3 and pUC19, lacZ′ Spcr | 50 |
| pR326 | E. coli plasmid, Ampr Cmr | 12a |
| pSET2 | E. coli-S. suis shuttle vector; Spcr | 51 |
| pSET2::Atl | Complementation vector with the pSET2 background, containing the promoter followed by the full-length atl ORF | This study |
| pET32a::Atl | pET32a inserted in-frame with the atl gene for expressing autolysin Atl | This study |
| pSET4S::Atl | Recombinant vector with the pSET4S background, designed for knockout of Atl, Cmr Spcr | This study |
Cloning, expression, and purification of recombinant protein.
The atl sequence (residues 673 to 3081), including the six repeated GBS_Bsp-like domains and the N-acetylmuramoyl-l-alanine amidase domain, was amplified by PCR using HA9801 genomic DNA as the template and Atlex-F and Atlex-R as the primers (Table 2). The PCR product was cloned in-frame into the pET32a vector to generate the expression plasmid pET32a::atl. E. coli BL21 competent cells were transformed with the resulting plasmid and cultured in 100 ml of LB broth containing ampicillin until the optical density at 600 nm (OD600) reached 0.6. At that time, the recombinant E. coli was induced by adding 1 mM isopropyl-β-d-thio-galactoside (IPTG) and was cultured for another 5 h. The cells were then harvested by centrifugation, washed twice with phosphate-buffered saline (PBS), and resuspended in 8 ml of 50 mM sodium phosphate (pH 7.5) containing 500 mM NaCl and 20 mM imidazole. The bacterial cells were disrupted by sonication on ice and the lysate centrifuged. The supernatant was saved for subsequent purification of the 6×His-tagged fusion proteins using a HisTrap FF column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) according to the manufacturer's instructions.
Table 2.
Primers used for PCR amplification
| Primer | Primer sequence (5′–3′) | Restriction site | Function | |
|---|---|---|---|---|
| LA-F | CAGAAGCTTAGAAATAGTCTACAGGAGATG | HindIII | Upstream border of atl | |
| LA-R | CGCCTCGAGCTCCTTTATGTATTTCACATG | XhoI | ||
| RA-F | ATCCTGCAGAAATTGCAGGTACAGGTATTG | PstI | Downstream border of atl | |
| RA-R | CCGGAATTCAGTCAGCATTAATGGTTCAAC | EcoRI | ||
| Cm-F | GGCCTCGAGCTTGATGAAAATTTGTTTGA | XhoI | Cmr gene cassette | |
| Cm-R | GGCCTGCAGGTAAAAAGTACAGTCGGCAT | PstI | ||
| Atl5′-F | ACGTTATTATTTCTTTCTTTG | The fragment within atl near the start codon | ||
| Atl5′-R | CGTTGAGCTTGTAGTAGTCGA | |||
| Atl3′-F | GGGCGAGACTCGGGTGCTTCT | The fragment within atl near the stop codon | ||
| Atl3′-R | AACTCCGAAGGATTATCCATA | |||
| Atlex-F | ACCGAATTCTCGGTCAATGTTACACAGCCA | EcoRI | A fragment for expressing recombinant Atl | |
| Atlex-R | ACGAAGCTTCTAATTCATTACACCTTGTGCCAA | HindIII | ||
| CAtl-F | ATACTGCAGCGACAGATTGCAGAACTAACTG | PstI | A fragment for complementation of atl | |
| CAtl-R | AGTGGATCCCGCTCGAAAGGATAGAATTAGC | BamHI |
Preparation of purified cell wall and bacteriolytic enzyme.
Purified cell walls were prepared as described previously (6), with some modifications. Briefly, bacteria were grown in 500 ml of THB and harvested at an OD600 of 0.8. The cells were sedimented and washed twice with PBS. The pellets were suspended in a 4% (wt/vol) sodium dodecyl sulfate (SDS) solution in distilled water, boiled for 30 min, and the SDS heat-treated cell walls were washed six times with distilled water and lyophilized.
The SDS-stable bacteriolytic enzyme of S. suis 2 was prepared as follows: SDS cell extracts were obtained from harvested cells by centrifugation and suspending them in 4% (wt/vol) SDS (0.6 ml/g [wet weight] of cells) for 2 h at room temperature.
Zymography analysis.
Recombinant Atl and SDS-stable bacteriolytic enzymes were analyzed by SDS-polyacrylamide renaturing gel electrophoresis, as previously described (6, 15). Briefly, an SDS-polyacrylamide gel containing 0.1% (wt/vol) purified HA9801 cell walls was used for detection of lytic activity. Following electrophoresis, the gel was soaked for 30 min in 200 ml of distilled water at room temperature with gentle shaking. The gel was then transferred to 200 ml of renaturation solution (unless otherwise mentioned, 0.1% Triton X-100, 10 mM MgCl2, and 25 mM Tris-HCl [pH 7.5]), gently shaken for 30 min at room temperature, transferred to 200 ml of the same solution, and incubated for 16 h at 37°C with gentle shaking. After incubation, the gel was washed in distilled water carefully, stained with 0.1% methylene blue in 0.01% KOH for 4 h, and destained in distilled water. Lytic activity appeared as a transparent band on the blue background.
Western affinity blotting.
A Western affinity blotting assay was carried out as described previously (4). Proteins (S. suis SDS extracts and E. coli lysates) were transferred electrophoretically onto polyvinylidene difluoride (PVDF) membranes (Amersham, Bucks, England) using a semidry blotting apparatus (Amersham Pharmacia Biotech). The blotting buffer contained 2.5 mM Tris-HCl, 192 mM glycine, and 15% methanol. Membranes were incubated in Tris-buffered saline (TBS) blocking reagent (20 mM Tris-HCl, 500 mM NaCl [pH 7.5 to 8.0]) containing 2.5% bovine serum albumin (BSA). Binding proteins were detected by incubating the membranes with 30 μg fibronectin ml−1 for 1 h at room temperature and then with rabbit anti-human fibronectin polyclonal antibody. Antibodies were detected with the DAB peroxidase substrate kit (Tiangen, Beijing, China) and SPA-horseradish peroxidase conjugate.
MS analysis.
Proteins corresponding to the lytic bands in the zymograms were excised from the SDS-polyacrylamide gels and sent to Nanjing Medical University for tryptic in-gel digestion and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis. Data from MALDI-TOF MS acquisitions were used in a combined search against the NCBInr protein database using Mascot (Matrix Science). Mascot search parameters were trypsin digestion, a maximum of one missed cleavage, variable modification of oxidation (M), and peptide mass tolerance for monoisotopic data of 120 ppm. The Mascot server was used against the NCBInr for peptide mass fingerprinting (PMF).
Construction and confirmation of the HA9801 Δatl mutant.
atl was inactivated by allelic exchange with a constitutive chloromycetin resistance (Cmr) expression cassette. First, the DNA sequences flanking atl were amplified from the genomic DNA of HA9801 using specific primers carrying HindIII/XhoI and PstI/EcoRI restriction enzyme sites, and the Cmr gene cassette was amplified from pBR326 with primers carrying XhoI/PstI restriction enzyme sites. The three digested DNA fragments were joined by incubation with T4 DNA ligase for 24 h at 16°C, and PCR was carried out with primers LA-F and RA-R (Table 2), resulting in the LA-Cmr-RA DNA fragment. The DNA fragment was digested with HindIII/EcoRI and cloned into the pSET4S vector digested with the same restriction enzymes to generate the atl knockout vector pSET4S::atl. To obtain the isogenic mutant Δatl, the HA9801 competent cells were electrotransformed with pSET4S::atl as described by Takamatsu et al. (50). For all of the Cmr transformants, colony PCR was performed to detect the presence of atl in the genome. Candidate mutants in which the atl gene failed to be amplified were further verified by multiple PCR assays with a series of specific primers listed in Table 2 (29).
Complementation of mutants.
The HA9801 Δatl mutant was complemented by the atl gene in the E. coli/Streptococcus shuttle vector pSET2 (51) in which the atl gene is under the control of its native promoter. For cloning atl, the primers CAtl-F and CAtl-R (Table 2) were used. The PCR product was cut with PstI/BamHI and ligated into the vector pSET2 digested with the same enzymes to generate the recombinant plasmid pSET2::atl, which was then electrotransformed into the atl mutant to produce the complementation strain (the Δatl complement strain) screened on THB agar under double selection pressure with spectinomycin resistance (Spcr) and Cmr.
qRT-PCR.
Quantitative real-time reverse transcription-PCR (qRT-PCR) was carried out as described previously (54). S. suis cultures were grown overnight in THB at 37°C and then diluted 100-fold into fresh THB. The subcultures were collected at the logarithmic phase, and RNA was isolated with an E.Z.N.A. bacterial RNA isolation kit (Omega, Beijing, China). The RNA was treated with DNase I (Promega, Madison, WI) to exclude genomic DNA contamination. The cDNA synthesis was performed using the PrimeScript RT reagent kit (TaKaRa, Dalian, China) according to the manufacturer's instructions. mRNA levels were measured using two-step relative qRT-PCR. The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) housekeeping gene was amplified as an internal control. The specific primers used for the qRT-PCR assays are listed in Table 3. The SYBR green PCR method was used according to the SYBR Premix Ex Taq kit (TaKaRa). Reactions were carried out in triplicate. An ABI 7300 RT-PCR system (Applied Biosystems) was used for relative qRT-PCR. Dissociation analysis of amplification products was performed at the end of each PCR to confirm that only one PCR product was amplified and detected. The comparative cycle threshold method (2−ΔΔCT method) (32) was used to analyze the mRNA levels.
Table 3.
Primers used for qRT-PCR
| Name | Oligonucleotide sequence (5′–3′) | Target gene |
|---|---|---|
| Atl-up-F | ATGTTTGTGAAAAAAAGAACG | Fragment starts from start codon of atl |
| Atl-up-R | AACCTGAGATGAAGAAACTTG | |
| Atl-down-F | ATAATCCTTCGGAGTTGCAAG | Fragment ends with terminator of atl |
| Atl-down-R | CTACTTTCCCTCAACAGCACC | |
| GAPDH-F | CTTGGTAATCCCAGAATTGAACGG | gapdh |
| GAPDH-R | TCATAGCAGCGTTTACTTCTTCAGC |
Microscopy.
Microscopy was performed as described previously (42). Cells were grown to an OD600 of 0.8 in THB at 37°C, and Gram stained cells were examined with a light microscope equipped with a 100× oil immersion objective. Furthermore, wild, mutant, and complemented strains were cultured in THB at 37°C for 16 h before the bottoms of the tubes were photographed.
Autolysis assay.
The autolysis assay was carried out as previously described (19, 62). Cells were grown to an OD600 of 0.8 in THB at 37°C and pelleted by centrifugation. The cells were washed once and resuspended in 50 mM Tris-HCl (pH 7.0) containing 0.05% Triton X-100 to an OD600 of 0.6. The cell suspensions were incubated at 37°C with gentle shaking. The decrease in optical density was monitored every hour.
Biofilm assays.
The biofilm assay was performed as previously described (9, 16). An overnight culture of S. suis 2 was diluted in fresh broth to an OD600 of 0.2. Samples (100 μl) were added to the wells of a 96-well polystyrene tissue culture plate containing 100 μl of culture medium supplemented with 4 mg/ml porcine fibrinogen. After incubation for 24 h at 37°C, medium and free-floating bacteria were removed by aspiration, and the wells were washed three times with 50 mM PBS (pH 7.4). The biofilms were stained with 0.04% crystal violet (100 μl) for 10 min. The wells were washed three times with PBS to remove unbound crystal violet dye and dried for 2 h at 37°C. After adding 100 μl of 95% (vol/vol) ethanol to each well, the plate was shaken for 10 min to release the stain from the biofilms, and the absorbance at 575 nm (A575) was recorded. Wells with sterile broth medium served as controls. All biofilm assays were run in triplicate, and the mean values of three independent experiments were calculated.
LD50 determinations.
Care and feeding of zebrafish (Pearl River Fishery Research Institute, Chinese Academy of Fishery Science) were performed with some modifications, as described previously (38). Challenge with S. suis 2 strains in zebrafish was followed as described previously (60, 61). Briefly, log phase cultures of S. suis 2 strains were centrifuged at 10,000 × g for 10 min at 4°C. The pellets were washed twice in PBS (pH 7.4). Zebrafish were anesthetized with tricaine methanesulfonate (MS-222) (Hangzhou Animal Medicine Factory) at a concentration of 90 mg/liter. Several groups of 10 zebrafish were intraperitoneally injected with 10-fold serially diluted suspensions containing 103 to 107 CFU of bacteria in sterile PBS. Control fish were injected with sterile PBS. The fish were observed for 7 days until mortality was reached. The 50% lethal dose (LD50) values were calculated by the Reed and Muench method (43).
Adherence assay.
The adherence assay was performed as previously described (5, 12), with some modifications. Log phase bacteria were pelleted, washed twice with PBS (pH 7.4), and resuspended in fresh cell culture medium without antibiotics. Bacteria (107 CFU) were added to wells of a 24-well tissue culture plate containing a monolayer (105 cells) of HEp-2 cells in 0.2 ml of medium (multiplicity of infection [MOI] of 100 bacteria/cell). The plates were incubated for 3 h at 37°C in 5% CO2 to allow cell adherence by the bacteria. The monolayers were then washed five times with PBS, digested with 100 μl of trypsin for 15 min at 37°C, and then disrupted by the addition of 900 μl of sterile deionized water following repeated pipetting to release all bacteria. Aliquots (100 μl) diluted 1:100 and 1:1,000 in PBS were used for quantitative plating. All assays were performed in duplicate and repeated three times.
Statistical analysis.
The data were analyzed using SPSS version 17.0 (SPSS Inc., Chicago, IL). The difference between mean values among groups was first evaluated by one-way analysis of variance (ANOVA) and then by pairwise comparison of the mean values between the two groups, followed by Tukey's student rank test. Differences with a P value of <0.05 were considered significant, and a P value of <0.001 was considered greatly significant.
RESULTS
Cloning, expression, and purification of recombinant protein.
The atl open reading frame (ORF) was amplified by PCR from HA9801 genomic DNA. The identity of the insert and the fidelity of the amplification were confirmed by DNA sequencing. To facilitate the high-level inducible expression of the cloned fusion gene, plasmid pET32a::atl was transformed into E. coli BL21. The Atl fusion protein was extracted from a mid-log culture, which had been induced with 1 mM IPTG for 5 h. The IPTG-inducible expression plasmid pET32a contained a start codon followed by a 6×His tag sequence, allowing purification of the recombinant autolysin by using a HisTrap FF column. As shown in Fig. 1, an apparent protein band of approximately 96 kDa appeared in the induced recombinant E. coli, while it was absent in the uninduced recombinant E. coli or induced E. coli containing control plasmid.
Fig 1.
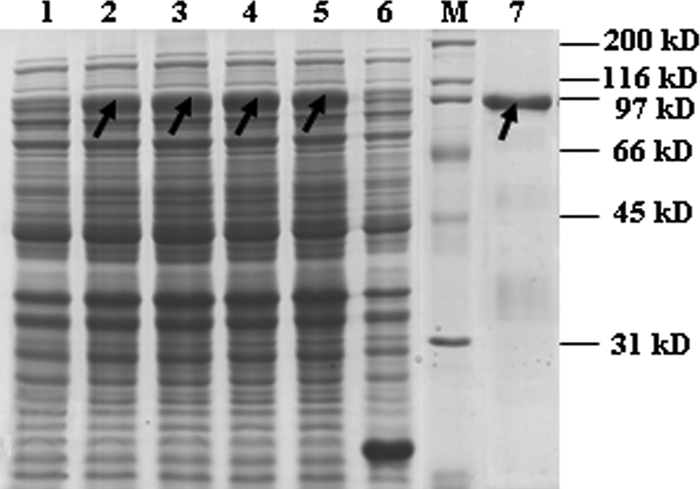
Analysis of recombinant Atl expressed from E. coli in an SDS polyacrylamide gel stained with Coomassie brilliant blue R-250. Lane M, protein molecular mass marker; lane 1, crude extract from uninduced cells carrying expression vector; lanes 2, 3, 4, and 5, crude extracts from cells induced for 2, 3, 4, and 5 h, respectively; lane 6, crude extract from cells transformed with an empty expression vector; lane 7, purified 6×His-tagged fusion proteins using a HisTrap FF column. The arrows indicate the recombinant Atl.
Zymography and Western affinity blotting.
The purified protein was assayed using zymography to confirm its lytic activity. On the zymogram of purified HA9801 cell walls, a transparent band was detected near the 96 kDa protein standard, which was consistent with the hypothesis that Atl has typical autolysin activity (Fig. 2). There were two lytic bands with the wild-type strain, which were absent with the atl mutant strain. The complementation strain could restore both lytic bands, regardless of the antibiotic pressure; however, the larger band appeared very weak compared with the smaller one (Fig. 2). The SDS extracts of S. suis 2 (including wild-type, mutant, and complemented strains) and crude cell lysates of IPTG-induced E. coli were tested to determine whether Atl could adhere to fibronectin. After the chromogenic reaction, one protein band corresponding to Atl in the recombinant E. coli lysates gave a strong fibronectin-binding signal, while there was no such signal in the negative control E. coli (Fig. 3). Two protein bands gave a strong fibronectin-binding signal in the wild-type strain, while they were absent in the atl mutant strain. The complemented strain also had the two positive bands, although the signal of the larger one was very weak (Fig. 3).
Fig 2.
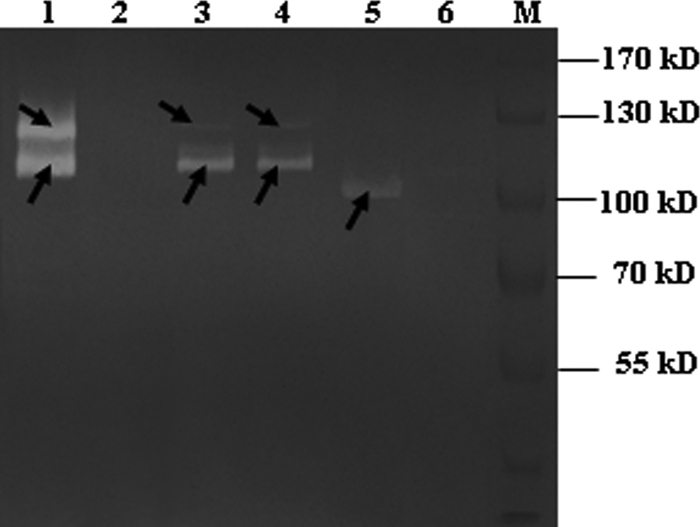
Autolysin profiles of the SDS extracts of the mutant, complementation, and wild-type strains. The recombinant Atl protein was also analyzed by renaturing gel electrophoresis. Lane 1, autolysin profile of the wild-type strain; lane 2, autolysin profile of the mutant strain; lane 3, autolysin profile of atl complementation strain under antibiotic pressure; lane 4, autolysin profile of atl complementation strain without antibiotic pressure; lane 5, purified Atl protein exhibiting lytic activity in a gel containing HA9801 PCW; lane 6, crude extract from E. coli transformed with an empty expression vector; lane M, prestained protein marker. The arrows indicate the autolysins.
Fig 3.
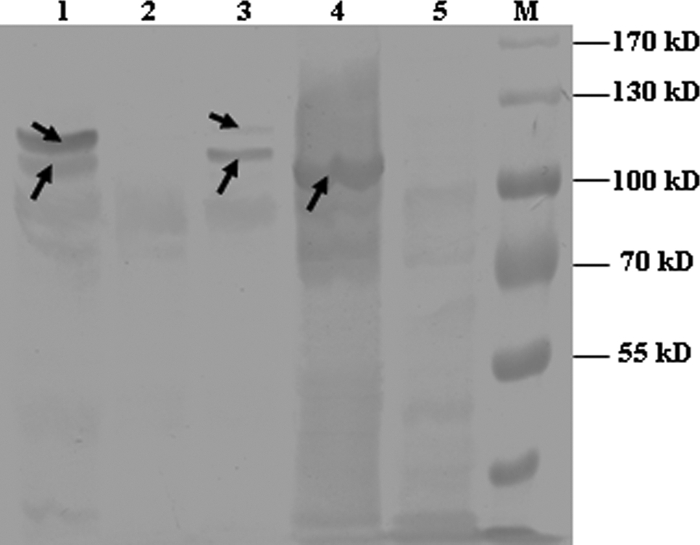
Confirmation of fibronectin-binding capacity of recombinant Atl and Atl expressed in S. suis by Western affinity blotting. Lane 1, SDS extracts of wild-type strain HA9801; lane 2, SDS extracts of the Δatl mutant strain; lane 3, SDS extracts of the Δ atl complement strain; lane 4, crude cell lysate of IPTG-induced E. coli expressing recombinant Atl; lane 5, crude extract from E. coli transformed with an empty expression vector; lane M, prestained protein marker. The arrows indicate signals representing positive fibronectin binding in the Western affinity blot.
MS analysis.
Two lytic protein bands appeared in the wild-type strain, which were not present in the atl mutant strain. The two protein bands were excised from the SDS-PAGE gel and characterized by MALDI-TOF MS. The data were compared to those in the NCBI sequence database. The probability score for the match, molecular weight (MW), number of peptide matches, and the percentage of the total translated ORF sequence covered by the peptides were used for protein band identification. The Mascot search results are summarized in Table 4. The two proteins corresponding to the lytic bands were both identified as N-acetylmuramoyl-l-alanine amidase in S. suis 05ZYH33. Furthermore, the matched peptides of the larger lytic protein started from the 7th amino acid, while the smaller one started from the 111th amino acid of the N-acetylmuramoyl-l-alanine amidase protein.
Table 4.
Proteins corresponding to the lytic band identified by MALDI-TOF MS
| Protein no.a | Identified protein |
Theoretical mol mass (Da) | Mascot scorec | No. of peptides matchedd | Coverage (%)e | |
|---|---|---|---|---|---|---|
| Mascot resultsb | Annotation/species | |||||
| 1 | gi|146318948 | N-Acetylmuramoyl-l-alanine amidase/Streptococcus suis 05ZYH33 | 113,193 | 78 | 12 | 12 |
| 2 | gi|146318948 | N-Acetylmuramoyl-l-alanine amidase/Streptococcus suis 05ZYH33 | 113,193 | 118 | 14 | 18 |
1, protein corresponding to the lytic band with the larger molecular mass; 2, protein corresponding to the lytic band with the smaller molecular mass.
gi number in NCBI.
Protein score greater than 76 was considered significant in this study (P < 0.05).
Number of peptides that matched the predicted protein sequence.
Percentage of predicted protein sequence covered by matched peptides.
Construction of S. suis atl mutant.
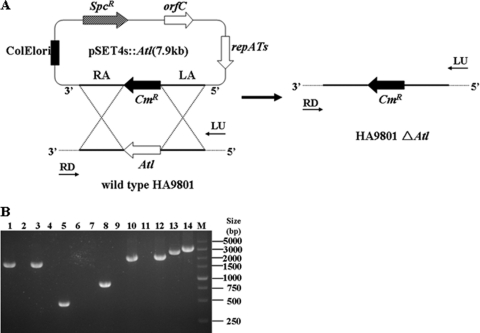
To determine the roles of atl in S. suis 2, a homologous suicide plasmid, pSET::atl, was constructed with a Cmr cassette (Fig. 4A) and electrotransformed into HA9801 competent cells. Transformants were screened on THB agar plates under chloramphenicol pressure. Colony PCR was performed to detect whether atl was still present in the genome as previously described (29). One candidate mutant in which the atl gene failed to be amplified was further verified. The allelic replacement of atl genes by the Cmr cassette in the candidate mutant was confirmed by multiple PCR analysis with specific primers (Fig. 4B). The results indicated that an isogenic knockout mutant of atl (the HA9801 Δatl mutant) was successfully constructed.
Fig 4.
Construction and confirmation of the HA9801 Δ atl knockout mutant strain. (A) Strategy for deletion mutagenesis of the Δ atl mutant in S. suis HA9801 by allelic exchange with a chloramphenicol resistance cassette and schematic representation of the chromosomal structures before (left) and after (right). The dotted lines represent the chromosomal sequences flanking the left and right arms of atl. (B) Multiple PCR analysis of the Δatl mutant. The primer combinations used in the PCR were as follows: lanes 1 and 2, LA-F/Atl5′-R; lanes 3 and 4, Atl3′-F/RA-R; lanes 5 and 6, Atl3′-F/Atl3′-R; lanes 7 and 8, Cm-F/Cm-R; lanes 9 and 10, LA-F/Cm-R; lanes 11 and 12, Cm-F/RA-R; lanes 13 and 14, LA-F/RA-R. Genomic DNA from the following strains was used as the template: wild-type strain HA9801 (lanes 1, 3, 5, 7, 9, 11, and 13) and the Δatl mutant (lanes 2, 4, 6, 8, 10, 12, and 14). The 5 kb DNA ladder marker is shown on the left (M).
Expression profiling of the atl gene.
When we first carried out the zymography analysis, the complementation strain could only restore the smaller autolysin, although the entire atl gene was complemented (data not shown). Therefore, we examined the transcriptional profile of the full-length atl by qRT-PCR. The expression levels of the Atl-up (containing start codon of atl) and Atl-down (containing terminator of atl) were quantified by qRT-PCR. Total RNA was extracted at the exponential phase with an OD600 value of 0.8. The results indicated that the mRNA levels of Atl-up and Atl-down were the same. The mRNA levels of Atl-up and Atl-down in the atl mutant, the Δatl complement strain supplemented with antibiotic, the Δatl complement strain in the absence of antibiotic pressure were decreased by 1.0, 0.26, and 0.40, respectively, compared with the parental strain. The results confirmed the entire atl gene was transcribed after complementation regardless of the antibiotic pressure.
Microscopy.
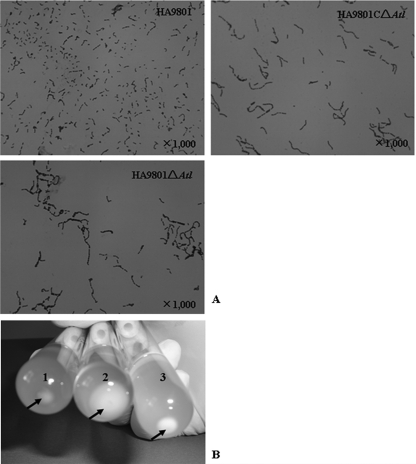
The growth curve of S. suis 2 was not affected by the atl mutation (data not shown). However, the mutant cells formed longer cell chains, and the cell chain lengths were nearly restored to that of the wild-type strain by atl complementation (Fig. 5A). This result suggests that the longer chains formed in the mutant arose from inactivation of the atl gene encoding autolysin of HA9801.
Fig 5.
(A) Microscopy of atl mutant, complementation, and wild-type S. suis 2. Log phase bacteria were treated with Gram stain, and the three strains were imaged under the same magnification scale. (B) Sedimentation of bacteria cultured in THB for 16 h. The arrows indicate the sedimented cells at the bottom of tubes. Tube 1, wild-type strain HA9801; tube 2, the Δatl mutant strain; tube 3, the Δatl complement strain.
Autolysis assay.
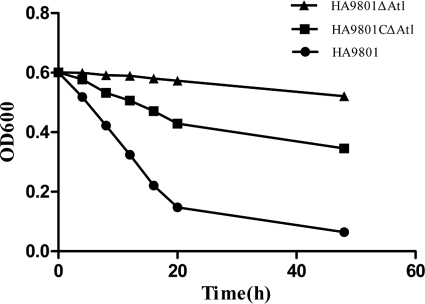
The autolytic activities of HA9801, the Δatl mutant, and the Δatl complement strain were measured. The autolysis assay results showed a decrease in optical density over time for whole-cell suspensions in buffer. There was an obvious difference among the three autolysis curves. HA9801 decreased to 20% of the initial OD value, the Δatl complement strain came to 58% of the initial OD value, while the mutant strain had almost no autolysis ability after 48 h of incubation (Fig. 6).
Fig 6.
Autolytic activity of atl mutant, complementation, and wild-type S. suis 2. Cells were suspended in autolysis buffers and incubated at 37°C. Lytic activity of the cell suspensions was monitored at OD600. Data are representative of mean values of three independent assays performed in triplicate.
Biofilm assays.
The capacities of HA9801, the Δatl mutant, and the Δatl complement strain to form biofilms in 96-well microtiter plates were evaluated. Biofilm formation by the atl mutant in THB supplemented with fibrinogen was significantly decreased compared to that of the parental strain HA9801, which was 0.73-fold of HA9801 (P < 0.05). Furthermore, the biofilm formation by the Δatl complement strain was restored, but it did not reach the level of the parental strain (Fig. 7). The results revealed that the inactivation of atl reduced the biofilm formation of S. suis 2 HA9801.
Fig 7.
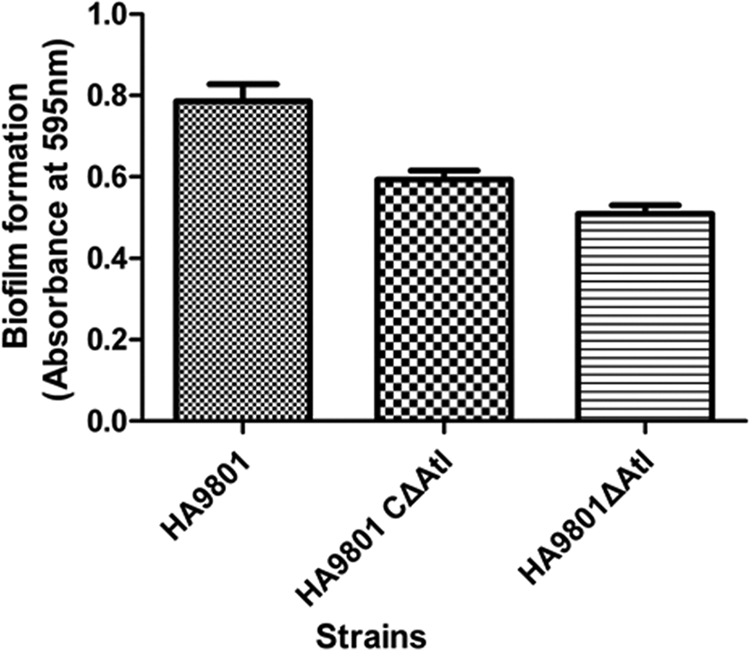
Biofilm assays of cells grown in THB supplemented with fibrinogen on the surfaces of 96-well microtiter plates. Biofilms were grown and quantified as described in the text. The results shown are mean values of three independent experiments carried out in triplicate. The error bars indicate standard deviations. The expression of atl led to significantly increased biofilm formation (P < 0.05) by atl positive strain HA9801 and the Δatl complement strain compared to the Δatl mutant strain.
LD50 determinations.
To study the effect of the atl mutation on S. suis virulence, we injected zebrafish via the intraperitoneal route with either HA9801, the Δatl mutant, or the Δatl complement strain. The mortality of zebrafish was observed within 7 days after the challenge. As shown in Table 5, the LD50 was 3.37 × 105 CFU/fish for the atl mutant, 5.28 × 104 CFU/fish for HA9801, and 1.15 × 105 CFU/fish for the Δatl complement strain. These results indicated a reduction in the virulence of S. suis 2 in the mutant strain (P < 0.001), while that of the complementation strain was restored to 2-fold higher than the mutant strain. The results suggested that the inactivation of atl could attenuate the virulence of S. suis 2 in zebrafish to some extent.
Table 5.
LD50 of wild-type, complementation, and atl mutant S. suis 2 strains
| Dose | Average mortality |
||
|---|---|---|---|
| HA9801 | HA9801 Δatl complement strain | HA9801 Δatl mutant | |
| 2.0 × 107 CFU/fish | 15/15 | 15/15 | 15/15 |
| 2.0 × 106 CFU/fish | 13/15 | 12/15 | 10/15 |
| 2.0 × 105 CFU/fish | 9/15 | 8/15 | 6/15 |
| 2.0 × 104 CFU/fish | 7/15 | 6/15 | 4/15 |
| 2.0 × 103 CFU/fish | 2/15 | 0/15 | 0/15 |
| LD50 CFU/fish | 5.28 × 104 | 1.15 × 105 | 3.37 × 105 |
Adherence assay.
The capacity of adhesion to HEp-2 cells was compared between the parental, mutant, and complementation strains under the same conditions. Consistent with the hypothesis that autolysin of S. suis 2 may be involved in cell adherence, mutation of atl was shown to affect adherence to HEp-2 cells. As shown in Fig. 8, there was a reduction of 50% in the adherence of the Δatl mutant strain compared with the parental strain HA9801 (P < 0.001). The adherence of the Δatl complement strain was approximately 1.5 times that of the Δatl mutant, which was about 75% of the parental strain.
Fig 8.
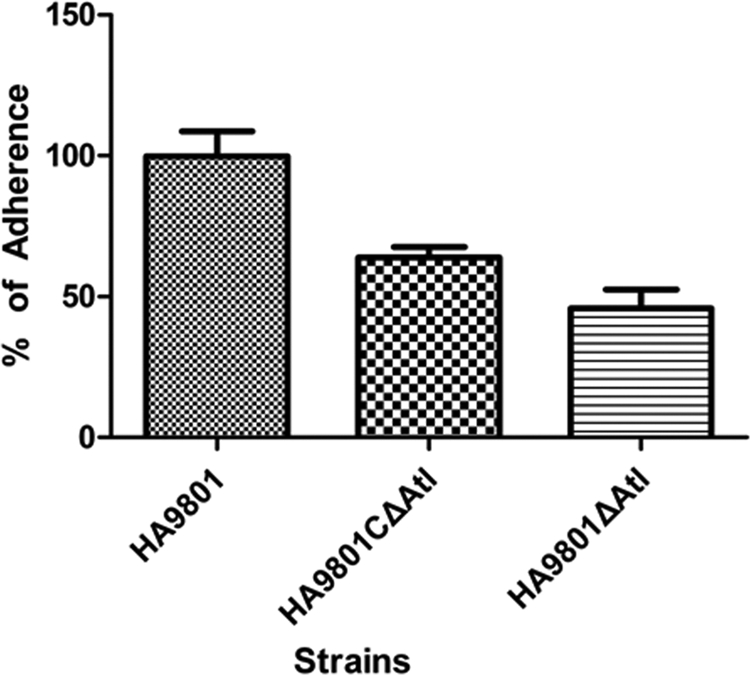
Effects of atl on S. suis 2 adherence to HEp-2 cells (MOI, 100). Results are presented as percent adherence recovered from the inoculums. The data are representative of three independent experiments carried out in triplicate. The error bars indicate standard deviations. The adherence of the parental strain HA9801 and the Δatl complement strain was significantly different compared with that of the Δatl mutant strain (P < 0.001).
DISCUSSION
Autolysin has been reported to play important roles in various biological functions, such as cell wall turnover, cell separation, cell division, and antibiotic-induced autolysis in a large range of bacterial species (48, 55). In addition to their biological functions, autolysins are also involved or implicated in the pathogenicity of Gram-positive bacteria. However, the function of the atl gene in S. suis 2 is still unclear. Thus, this study was conducted to explore the multiple roles of Atl in S. suis 2.
When we executed the CD-search tool in NCBI to search the Conserved Domain Database with the Atl protein sequence, we found that it contains six repeated GBS_Bsp-like domains, which may act as substrate binding domains, and one N-acetylmuramoyl-l-alanine amidase domain, which serves as the catalytic domain. In the zymography assay, Atl containing the six repeated GBS_Bsp-like domains and the N-acetylmuramoyl-l-alanine amidase domain (residues 673 to 3081) could lyse the HA9801 cell walls. As shown in Fig. 2, a transparent band was detected near the 96 kDa protein standard where the recombinant Atl should be. This result is consistent with the function of autolysin AtlA in Streptococcus mutans. By expressing a series of truncated derivatives of AtlA in S. mutans, Ahn and Burne (1) demonstrated that full activity of AtlA required the C terminus and repeat regions. Since parts of Atl showed lytic activity, there was no doubt that the whole Atl protein would also exert lytic activity. These results confirmed the peptidoglycan hydrolysis activity of Atl.
In our study, to detect autolytic activity in a denaturing polyacrylamide gel, two lytic bands were found in SDS extracts of HA9801 cells by using HA9801 cell wall as the substrate. Comparison of the wild-type strain with an atl deletion mutant revealed that the two autolytic protein bands originated from atl, as neither band was present in the atl mutant strain. The two autolytic protein bands were analyzed by MS analysis and were identified as the same protein, Atl. Furthermore, peptides were matched in the larger protein starting from the 7th amino acid and from the 111th amino acid in the smaller protein. From the Mascot search results, we presumed that the smaller protein band was a proteolytic product of the larger protein Atl, created by degradation of the N-terminal portion of the Atl, as found in S. mutans (47). By MS analysis, we found that the larger protein had a molecular mass of 113 kDa, which was lower than the estimate of 130 kDa previously attributed to this protein on the basis of mobility. Overestimation of molecular mass based on mobility also occurred with Staphylococcus caprae autolysin AtlC (2). The complementation strain could restore both autolytic protein bands, although the larger one was very weak compared with the smaller one. When extracts treated with SDS for 30 min were used for zymography analysis, only the smaller autolysin appeared in the complementation strain (data not shown). That was an unexpected result, as the full-length atl sequence with its promoter was cloned in the plasmid pSET2 and introduced into the mutant strain. We confirmed by qRT-PCR analysis that the entire atl gene was successfully transcribed. We also extended the SDS incubation time from 30 min to 24 h. Interestingly, the larger autolysin appeared when the SDS incubation time was increased up to 2 h, but the amount of the larger protein remained the same with incubation time exceeding 2 h (data not shown). When analyzed by RONN to predict protein disorder, the region between residues 24 and 49 of Atl showed high probabilities of disorder. Kim et al. (27) found that disordered or unfolded proteins are prone to be degraded by the proteolytic enzyme HtrA in E. coli. Generally, chaperones and proteases are thought to bind to exposed hydrophobic sequences, present in unfolded or misfolded proteins in E. coli, and protein quality control is carried out by a protein network, comprised of chaperones and proteases (14). A critical question that immediately arises is how the unfolded larger autolysin can survive in the host cell. The most likely explanation is that the activity of the proteolytic enzyme is tightly regulated in the host cell, being carried out by the proteasomes in response to some specific signal (59). Therefore, the levels of the larger autolysin and the smaller autolysin can be rigidly controlled in the wild-type strain. After functional complementation with the recombinant plasmid pSET2:atl, it is interesting to note that the ratio of the larger autolysin to the smaller autolysin changed greatly compared with HA9801 (Fig. 2). It is believed that the activity or expression of autolysin AtlA must be strictly regulated in S. mutans (1). We inferred that the regulation of Atl expression in plasmid was not as strict as that in the genome, resulting in the sharp reduction of the larger autolysin. Ahn and Burne (1) have shown that when the atlA defect was complemented with an intact atlA gene on a plasmid, only a limited quantity of AtlA was shown to be converted to the mature form, implying that the coordinated synthesis of autolysin and other factors produced in the genome is critical to proper production of autolysin. From these observations, the most reasonable conclusion is that Atl activity is regulated through an association with factors in S. suis 2 under particular conditions.
While the atl mutant strain demonstrated almost no cell autolysis when suspended in lysis buffer, the OD600 of HA9801 was reduced to 20% of the initial value, and the complementation strain had partially restored autolysis capacity (Fig. 6). The results revealed that Atl plays a major role in HA9801 autolysis when bacteria are in adverse conditions. Having observed that recombinant Atl demonstrated peptidoglycan hydrolysis activity and that the inactivation of atl caused the disappearance of two lytic bands in the SDS extracts, we can conclude that the atl gene cloned in this study encodes the major autolysin of S. suis 2 and plays a major role in HA9801 autolysis.
Light microscopic observations revealed that the atl mutant cells connected to each other to form very long cell chains compared with those formed by the parental strain (Fig. 5A). The inactivation of the lytB gene also leads to the formation of long chains of cells in S. pneumoniae (13), and the inactivation of atlA causes the formation of long chains in S. mutans and Staphylococcus aureus (47). In this study, most atl mutant cells sedimented when grown overnight, while the broth culture of wild-type cells was turbid, with fewer sedimented cells (Fig. 5B). As the growth rate of the cells was not affected in the atl mutant (data not shown), the difference in sedimented cells could be attributed to its longer cell chain length.
In addition to the bacteriolytic activity, some staphylococcal autolysins have adhesive functions. The S. epidermidis autolysin AtlE mediates the primary attachment to polystyrene and binds vitronectin, whereas its binding to fibronectin is very weak and not reproducible (19). The Staphylococcus saprophyticus autolysin Aas binds fibronectin and sheep erythrocytes, and the binding domain has been mapped to the central region containing the repeats (20). Whether S. suis 2 autolysin Atl also has such an adhesive function needs to be explored. Here, we used the SDS extracts of S. suis (wild-type, mutant, and complemented strains) and crude cell lysates of IPTG-induced E. coli to test whether Atl could adhere to fibronectin. Both the natural and recombinant autolysin gave strong positive fibronectin-binding signals (Fig. 3), revealing the fibronectin-binding activity of Atl. The atl mutant strain showed greatly reduced adherence to the HEp-2 eukaryotic cells in vitro. The primary attachment of the atl mutant to HEp-2 was reduced by approximately 2-fold (P < 0.05) in comparison to that of the wild-type strain, indicating that Atl can facilitate bacterial adherence to the host cell. The other autolysins, including Aas of S. saprophyticus (22), AtlC of S. caprae (3), AtlE of S. epidermidis (21), and Ami of L. monocytogenes (36), have also been shown to be involved in bacterial adhesion. Binding to fibronectin has been suggested to play an important role in adherence of the group A Streptococcus pyogenes to host epithelial cells (20). The finding that Atl could bind to fibronectin may explain how autolysin Atl affects the S. suis 2 adherence to HEp-2 cells.
Compared with the parental strain, biofilm formation of the atl mutant was significantly decreased in a microtiter plate assay. The complementation strain restored biofilm formation at a specific concentration. In Enterococcus faecalis, the autolysin (atn) mutant also causes reduction in biofilm formation compared with the parental strain (37). Previous studies have indicated that cells in mutant biofilms (e.g., the comC mutant, the brpA mutant, and the hr11 and rr11 mutants) with abnormal structures tend to form long chains, suggesting that the variation in biofilm structure may result from the formation of extremely long chains of cells (30, 31, 56). Biofilm formation is thought to be a two-step process that requires the adhesion of bacteria to a substrate surface followed by cell-cell adhesion, forming the multiple layers of the biofilm. S. suis biofilm formation on a polystyrene surface is stimulated in a dose-dependent manner when fibrinogen is added to the culture medium (9). Perhaps fibrinogen promotes the biofilm formation by strengthening cell-cell adhesion. In this study, we found that autolysin Atl could bind to fibrinogen and contribute to the fibrinogen-mediated cell-cell adhesion, which is the main step of biofilm formation. The hypothesis was supported by the fact that the atl isogenic mutant strain had greatly reduced biofilm formation compared with the parent strain.
In vivo studies with animal models (mice, rats, and guinea pigs) of infection have shown that mutant strains defective in autolysin synthesis, including AtlE of S. epidermidis (45), p60, Ami and Auto of L. monocytogenes (11, 36, 41), and LytA of S. pneumoniae (8), are less virulent than the wild-type strains. Autolysin also plays crucial roles in the pathogenesis of pneumococcal meningitis (24). In the last few years, zebrafish has been used as a model for human disease, cancer and immunological studies, and some experimental infections have been conducted with several streptococcal species such as Streptococcus iniae, S. pyogenes (34, 38), and Streptococcus agalactiae (40). The fact that zebrafish have adaptive and innate cellular immune defenses that are not unlike those of the mammalian species most often used to model infectious diseases of humans (52) makes them highly useful as a model host organism. Fish have immunoglobulins, antigen-processing cells, T cells, and B cells (18, 57, 58) as well as complement and phagocytic cells and leukocytes capable of producing reactive species of oxygen and nitrogen (23). Thus, it is likely that the underlying principles of host-pathogen relationships in fish will be very similar to those in mammalian species. Upon infection with S. suis, the transcriptional responses in zebrafish have shown clear conservation with host responses detected in porcine cells, human cells, or mammalian models, including induction of genes involved in the immune response, inflammatory response, complement activation, and defense response, and provided further validation of this model for S. suis infection (61). That Atl may affect the virulence of S. suis was supported by observations of the LD50 in zebrafish. A 5-fold reduction of virulence was observed in vivo for the atl mutant compared with the wild type. Given that the atl mutant strain had attenuated adherence to HEp-2 cells and lower virulence in zebrafish and that the atl complementation strain could restore these phenotypes to some extent, we concluded that autolysin Atl contributes to the virulence of S. suis 2.
The mechanism by which autolysin Atl affects the virulence of S. suis 2 may be explained by the following observations of our work and others. LytA of S. pneumoniae and MurA and p60 of L. monocytogenes are thought to mediate the release of cytoplasmic toxins or proinflammatory degraded cell wall components resulting in subsequent tissue injuries (25, 28, 33, 53). When it comes to the atl mutant strain, the release process may be hindered for the loss of autolysis capacity.
In Gram-positive bacteria, the presentation or release of virulent factors is crucially dependent on the continuous remodeling of the cell wall (10), and a defect in this process in the atl mutant could have contributed somewhat to the attenuated virulence. Adherence of pathogenic microorganisms to the cell surface is also a key event during infection (36). Previous findings have suggested that certain autolysins may contribute directly to the pathogenicity of Gram-positive bacteria by mediating bacterial adherence (21). It stands to reason that the atl mutation reduced the bacterial adherence and therefore weakened the virulence of S. suis 2. AtlA, an autolysin of Streptococcus mutans, is a fibronectin-binding protein that contributes to bacterial survival in the bloodstream and virulence, causing infective endocarditis (26). The S. suis 2 autolysin Atl could also bind fibronectin and thereby escape the host cell defense.
The results presented herein provide insights into novel roles of Atl, a suspected autolysin of S. suis that takes part in the cell autolysis, separation of daughter cells, biofilm formation, fibronectin-binding activity, cell adhesion, and pathogenesis. The expression, distribution, and activation of the autolysin are likely to be tightly controlled in S. suis. Understanding this mechanism could provide unique insights into the pathogen and facilitate the development of new strategies for the treatment of S. suis infection.
ACKNOWLEDGMENTS
This work was financially supported by the Fund of the National Natural Science Foundation of China (U0931002) and the Special Fund for Public Welfare Industry of Chinese Ministry of Agriculture (200803016).
We thank Tsutomu Sekizaki and Daisuke Takamatsu for kindly providing plasmid pSET4s for this study.
Footnotes
Published ahead of print 6 January 2012
REFERENCES
- 1. Ahn SJ, Burne RA. 2006. The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J. Bacteriol. 188:6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allignet J, et al. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun. 69:712–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allignet J, et al. 2002. Several regions of the repeat domain of the Staphylococcus caprae autolysin, AtlC, are involved in fibronectin binding. FEMS Microbiol. Lett. 213:193–197 [DOI] [PubMed] [Google Scholar]
- 4. Allignet J, et al. 1999. Tracking adhesion factors in Staphylococcus caprae strains responsible for human bone infections following implantation of orthopaedic material. Microbiology 145:2033–2042 [DOI] [PubMed] [Google Scholar]
- 5. Benga L, Friedl P, Valentin-Weigand P. 2005. Adherence of Streptococcus suis to porcine endothelial cells. J. Vet. Med. B Infect. Dis. Vet. Public Health 52:392–395 [DOI] [PubMed] [Google Scholar]
- 6. Bernadsky G, Beveridge TJ, Clarke AJ. 1994. Analysis of the sodium dodecyl sulfate-stable peptidoglycan autolysins of select gram-negative pathogens by using renaturing polyacrylamide gel electrophoresis. J. Bacteriol. 176:5225–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berry AM, et al. 1989. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect. Immun. 57:2324–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berry AM, Paton JC. 2000. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect. Immun. 68:133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonifait L, Grignon L, Grenier D. 2008. Fibrinogen induces biofilm formation by Streptococcus suis and enhances its antibiotic resistance. Appl. Environ. Microbiol. 74:4969–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bublitz M, et al. 2009. Structural basis for autoinhibition and activation of Auto, a virulence-associated peptidoglycan hydrolase of Listeria monocytogenes. Mol. Microbiol. 71:1509–1522 [DOI] [PubMed] [Google Scholar]
- 11. Cabanes D, et al. 2004. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 51:1601–1614 [DOI] [PubMed] [Google Scholar]
- 12. Charland N, et al. 2000. Streptococcus suis serotype 2 interactions with human brain microvascular endothelial cells. Infect. Immun. 68:637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a. Claverys JP, et al. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123–128 [DOI] [PubMed] [Google Scholar]
- 13. De Las Rivas B, et al. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-beta-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 184:4988–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dougan DA, Mogk A, Bukau B. 2002. Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell. Mol. Life Sci. 59:1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Foster SJ. 1992. Analysis of the autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J. Bacteriol. 174:464–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grenier D, Grignon L, Gottschalk M. 2009. Characterisation of biofilm formation by a Streptococcus suis meningitis isolate. Vet. J. 179:292–295 [DOI] [PubMed] [Google Scholar]
- 17. Gu H, Lu C. 2008. Identification and characterization of a novel infection-related factor (cell wall hydrolase/autolysin) of Streptococcus suis serotype 2. Wei Sheng Wu Xue Bao 48:68–72 (In Chinese.) [PubMed] [Google Scholar]
- 18. Haire RN, et al. 2000. Characterization of three isotypes of immunoglobulin light chains and T-cell antigen receptor alpha in zebrafish. Immunogenetics 51:915–923 [DOI] [PubMed] [Google Scholar]
- 19. Hanaki H, et al. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199–209 [DOI] [PubMed] [Google Scholar]
- 20. Hanski E, Caparon M. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 89:6172–6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heilmann C, et al. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013–1024 [DOI] [PubMed] [Google Scholar]
- 22. Hell W, Meyer HG, Gatermann SG. 1998. Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol. Microbiol. 29:871–881 [DOI] [PubMed] [Google Scholar]
- 23. Herbomel P, Thisse B, Thisse C. 1999. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126:3735–3745 [DOI] [PubMed] [Google Scholar]
- 24. Hirst RA, et al. 2008. Streptococcus pneumoniae deficient in pneumolysin or autolysin has reduced virulence in meningitis. J. Infect. Dis. 197:744–751 [DOI] [PubMed] [Google Scholar]
- 25. Jedrzejas MJ. 2001. Pneumococcal virulence factors: structure and function. Microbiol. Mol. Biol. Rev. 65:187–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jung CJ, et al. 2009. Streptococcus mutans autolysin AtlA is a fibronectin-binding protein and contributes to bacterial survival in the bloodstream and virulence for infective endocarditis. Mol. Microbiol. 74:888–902 [DOI] [PubMed] [Google Scholar]
- 27. Kim KI, et al. 1999. Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein in Escherichia coli. J. Mol. Biol. 294:1363–1374 [DOI] [PubMed] [Google Scholar]
- 28. Lenz LL, et al. 2003. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 100:12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li M, et al. 2008. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS One 3:e2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li YH, et al. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184:6333–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li YH, et al. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 33. Lock RA, Hansman D, Paton JC. 1992. Comparative efficacy of autolysin and pneumolysin as immunogens protecting mice against infection by Streptococcus pneumoniae. Microb. Pathog. 12:137–143 [DOI] [PubMed] [Google Scholar]
- 34. Lowe BA, Miller JD, Neely MN. 2007. Analysis of the polysaccharide capsule of the systemic pathogen Streptococcus iniae and its implications in virulence. Infect. Immun. 75:1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lun ZR, et al. 2007. Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect. Dis. 7:201–209 [DOI] [PubMed] [Google Scholar]
- 36. Milohanic E, et al. 2001. The autolysin Ami contributes to the adhesion of Listeria monocytogenes to eukaryotic cells via its cell wall anchor. Mol. Microbiol. 39:1212–1224 [DOI] [PubMed] [Google Scholar]
- 37. Mohamed JA, et al. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 72:3658–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neely MN, Pfeifer JD, Caparon M. 2002. Streptococcus-zebrafish model of bacterial pathogenesis. Infect. Immun. 70:3904–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reference deleted.
- 40. Phelps HA, Neely MN. 2005. Evolution of the zebrafish model: from development to immunity and infectious disease. Zebrafish 2:87–103 [DOI] [PubMed] [Google Scholar]
- 41. Pilgrim S, et al. 2003. Deletion of the gene encoding p60 in Listeria monocytogenes leads to abnormal cell division and loss of actin-based motility. Infect. Immun. 71:3473–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Priyadarshini R, Popham DL, Young KD. 2006. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J. Bacteriol. 188:5345–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reed L, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 27:493 [Google Scholar]
- 44. Rigden DJ, Jedrzejas MJ, Galperin MY. 2003. Amidase domains from bacterial and phage autolysins define a family of gamma-D,L-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28:230–234 [DOI] [PubMed] [Google Scholar]
- 45. Rupp ME, Fey PD. 2001. In vivo models to evaluate adhesion and biofilm formation by Staphylococcus epidermidis. Methods Enzymol. 336:206–215 [DOI] [PubMed] [Google Scholar]
- 46. Rupp ME, et al. 1999. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 67:2627–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shibata Y, et al. 2005. Identification and characterization of an autolysin-encoding gene of Streptococcus mutans. Infect. Immun. 73:3512–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shockman G, Holtje J. 1994. Microbial peptidoglycan (murein) hydrolases. Bacterial Cell Wall 27:131–166 [Google Scholar]
- 49. Staats JJ, et al. 1997. Streptococcus suis: past and present. Vet. Res. Commun. 21:381–407 [DOI] [PubMed] [Google Scholar]
- 50. Takamatsu D, Osaki M, Sekizaki T. 2001. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46:140–148 [DOI] [PubMed] [Google Scholar]
- 51. Takamatsu D, Osaki M, Sekizaki T. 2001. Construction and characterization of Streptococcus suis-Escherichia coli shuttle cloning vectors. Plasmid. 45:101–113 [DOI] [PubMed] [Google Scholar]
- 52. Trede NS, Zapata A, Zon LI. 2001. Fishing for lymphoid genes. Trends Immunol. 22:302–307 [DOI] [PubMed] [Google Scholar]
- 53. Tuomanen EI. 2000. Pathogenesis of pneumococcal inflammation: otitis media. Vaccine 19:S38–S40 [DOI] [PubMed] [Google Scholar]
- 54. Wang Y, et al. 2011. Functional analysis of luxS in Streptococcus suis reveals a key role in biofilm formation and virulence. Vet. Microbiol. 152:151–160 [DOI] [PubMed] [Google Scholar]
- 55. Ward JB, Williamson R. 1985. Bacterial autolysins: specificity and function, p 159–166 In Nombela C. (ed), Microbial cell wall synthesis and autolysis. Elsevier Science B.V., Amsterdam, The Netherlands [Google Scholar]
- 56. Wen ZT, Burne RA. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Willett CE, et al. 1999. Early hematopoiesis and developing lymphoid organs in the zebrafish. Dev. Dyn. 214:323–336 [DOI] [PubMed] [Google Scholar]
- 58. Willett CE, et al. 1997. Expression of zebrafish rag genes during early development identifies the thymus. Dev. Biol. 182:331–341 [DOI] [PubMed] [Google Scholar]
- 59. Wright PE, Dyson HJ. 1999. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 293:321–331 [DOI] [PubMed] [Google Scholar]
- 60. Wu Z, Zhang W, Lu C. 2008. Comparative proteome analysis of secreted proteins of Streptococcus suis serotype 9 isolates from diseased and healthy pigs. Microb. Pathog. 45:159–166 [DOI] [PubMed] [Google Scholar]
- 61. Wu Z, et al. 2010. Transcriptome profiling of zebrafish infected with Streptococcus suis. Microb. Pathog. 48:178–187 [DOI] [PubMed] [Google Scholar]
- 62. Yamada A, Tamura H, Kato H. 2009. Identification and characterization of an autolysin gene, atlg, from Streptococcus sobrinus. FEMS Microbiol. Lett. 291:17–23 [DOI] [PubMed] [Google Scholar]