Abstract
Using a sensitive assay, we observed low levels of an unknown surfactant produced by Pseudomonas syringae pv. syringae B728a that was not detected by traditional methods yet enabled swarming motility in a strain that exhibited deficient production of syringafactin, the main characterized surfactant produced by P. syringae. Random mutagenesis of the syringafactin-deficient strain revealed an acyltransferase with homology to rhlA from Pseudomonas aeruginosa that was required for production of this unidentified surfactant, subsequently characterized by mass spectrometry as 3-(3-hydroxyalkanoyloxy) alkanoic acid (HAA). Analysis of other mutants with altered surfactant production revealed that HAA is coordinately regulated with the late-stage flagellar gene encoding flagellin; mutations in genes involved in early flagellar assembly abolish or reduce HAA production, while mutations in flagellin or flagellin glycosylation genes increase its production. When colonizing a hydrated porous surface, the bacterium increases production of both flagellin and HAA. P. syringae was defective in porous-paper colonization without functional flagella and was slightly inhibited in this movement when it lacked surfactant production. Loss of HAA production in a syringafactin-deficient strain had no effect on swimming but abolished swarming motility. In contrast, a strain that lacked HAA but retained syringafactin production exhibited broad swarming tendrils, while a syringafactin-producing strain that overproduced HAA exhibited slender swarming tendrils. On the basis of further analysis of mutants altered in HAA production, we discuss its regulation in Pseudomonas syringae.
INTRODUCTION
Biosurfactants are biologically produced amphiphilic compounds that display surface activity by lowering the tension at interfaces such as that between oil and water. A number of bacterial surfactants have been investigated extensively, but many more biosurfactants probably remain to be discovered. Even the true physiological functions of the best characterized biosurfactants have only recently been uncovered. Originally, biosurfactants were thought to be produced for the purpose of oil emulsification and degradation (34), most likely because this was a trait used to detect their production and also one of the earliest proposals for their utility. However, an increasingly sophisticated understanding of the complexities of bacterial behavior has led to additional hypothesized roles of biosurfactant production, including biofilm structure maintenance, pathogenicity, antagonistic activity against other bacteria and/or fungi, and bacterial motility (38, 40).
The biosurfactants produced by Pseudomonas aeruginosa serve as an excellent example of the complexity in determining their roles in bacterial behaviors. This bacterium produces rhamnolipids, a mixture of dirhamnolipids, monorhamnolipids, and 3-(3-hydroxyalkanoyloxy) alkanoic acid (HAA), which is the rhamnose-free lipid precursor (13). A wide range of functions have been proposed for rhamnolipids, including bacterial access to hydrophobic carbon sources, biofilm structure, biofilm departure, as well as swarming motility (34, 40, 46). Curiously, although each of these three surfactants facilitate motility on a low-agar plate, more-detailed analysis of swarming behavior revealed that HAAs actually have a repellant role, while dirhamnolipids are attractants, suggesting a more complex process by which surfactants modulate the motility of surfactant-producing cells (46). Thus, although HAAs might simplistically appear to aid bacterial motility by lowering the surface tension of surfaces traversed by cells, they probably have a more context-dependent role in in vivo motility.
Recently, while investigating the production of the lipopeptide biosurfactant syringafactin in the plant-associated bacterium Pseudomonas syringae pv. syringae B728a, it was observed that this strain produced a second surfactant detectable on agar plates (4); mutant strains in which the syringafactin biosynthetic cluster was disrupted still produced a surfactant detectable as a halo in an atomized oil assay. This second surfactant, although not produced in sufficient quantities to confer collapse of water drops, enabled swarming motility on a semisolid agar surface. Since the movement of plant pathogens as well as human pathogens on plants are of biological and practical significance, we have characterized this second surfactant in order to better understand the complex roles of surfactants on bacterial behaviors on leaf surfaces. This report addresses the identification of the remaining surfactant and a number of genes that regulate its production, in addition to demonstrating the strong environmental context-dependent production of this compound. We will show that there is an intimate link between flagellar assembly and production of this biosurfactant that suggests that it plays a specific role in motility.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. syringae pv. syringae B728a (28) was cultured on King's medium B (KB) plates with 1.5% agar (technical; Difco) (25) at 28°C. Escherichia coli strains DH5α, S17-1 (42), and SM10(λpir) (12) were cultured on Luria agar at 37°C. Antibiotics were used at the following concentrations: kanamycin, 25 μg/ml for P. syringae and 50 μg/ml for E. coli; rifampin, 100 μg/ml; gentamicin, 75 μg/ml; tetracycline, 15 μg/ml; and spectinomycin, 100 μg/ml.
Biosurfactant detection.
The atomized oil assay was performed as in previous studies (4). Briefly, bacteria were spotted onto agar plates solidified with 1.5% agar (technical; Difco) and grown overnight. An airbrush (type H; Paasche Airbrush Co., Chicago, IL) was used to apply a fine mist of mineral oil (light paraffin oil, Fisher Scientific) on the plate. The radius of a “halo” of oil droplets having altered shapes that caused them to appear brighter when visualized with an indirect source of bright light were then measured. The surface tensions of surfactant extracts were determined using the pendant drop method. The extract was analyzed with a FTA 4000 video analysis instrument (First Ten Angstroms Inc., Portsmouth, VA). Droplets were produced using a 22-gauge blunt needle, and the values reported represent an equilibrium surface tension determined 60 s after the formation of a droplet.
Random chromosomal insertional mutagenesis.
The production of transposon mutants was done as in previous studies (4). Briefly, a ΔsyfA deletion mutant of P. syringae B728a was mated with E. coli strain SM10(λpir) harboring pUT mini-Tn5 Sm/Sp (12), and putative P. syringae transposon mutants were screened for biosurfactant production by the atomized oil assay. Mutants that exhibited substantially larger (over 20%) or smaller halos than the wild-type (WT) strain were retested. Mutants with obvious growth defects were discarded. The genes into which the transposon had inserted in these mutants was determined using arbitrarily primed PCR similar to the method of O'Toole et al. (36). Mutations generated by the transposon from pUT mini-Tn5 Sm/Sp were characterized using primers complementary to the 5′ end of the transposon; primer tn5sm-ext (ext stands for exterior) (for primer sequences, see Table S1 in the supplemental material) was used in the initial PCR, and primer tn5sm-int (int stands for interior) was used in a second reaction to amplify sequences 5′ to the insertion site. The PCR product was directly purified (QIAquick PCR purification kit; Qiagen) and submitted for sequencing using primer tn5sm-int. Sequencing of the conglomerate PCR product ensured that only single transposon events were present in a given mutant strain, since multiple insertions would have resulted in mixed signals. The locations of the sequenced fragments were determined directly by a BLAST search on the Pseudomonas genome database (50) and compared to the published sequence of P. syringae B728a (16).
Motility assays.
The swarming motility of P. syringae B728a was assessed on semisolid KB plates containing 0.4% agar (technical; Difco) as in previous studies (37). The cells were grown for 1 day on KB and then harvested and washed in potassium phosphate buffer (10 mM, pH 7.5). The cells were resuspended in buffer to an optical density at 600 nm (OD600) of 0.27, and 5 μl (approximately 2.5 × 106 cells) of the appropriate bacterial strain was pipetted onto each plate and incubated for 24 h at room temperature. Swimming motility was assessed with KB plates solidified with 0.25% agar (technical; Difco) inoculated by stab inoculation. Swimming distance was measured as the distance from the point of inoculation to the bacterial front within the agar. Movement of cells through hydrated paper discs (1/4-in. diameter) (Schleicher & Schuell; grade 740-E) was also determined. Bacteria were either inoculated onto the top of the filter disc with a toothpick, or 1 μl of cell suspension was applied to the discs. Longer-distance movement through paper was assessed by placing filter paper (Whatman no. 1) cut into 1.5- by 4.5-cm strips on KB plates and inoculating them at a distance of 0.5 cm from the end by toothpick application. Strips were removed after 16 h by carefully lifting them so as to minimize manual spreading of the bacteria that had entered the strips. The plates on which the strips had rested were then incubated for at least 2 days at room temperature, and the amount of bacterial movement was measured as the most distal extent of bacterial growth.
Construction of biosurfactant deletion mutants.
A deletion mutant of syfA was constructed by cloning approximately 1-kb fragments upstream and downstream of syfA into pENTR/D-TOPO:MCS-Kan (14). The region downstream of syfA was amplified by the primers syfAe-xhoF and syfAe-xbaR, digested with XhoI and XbaI, and ligated into pENTR/D-TOPO:MCS-Kan. The region upstream of syfA was amplified by the primers syfAs-avrF and syfAs-bamR, digested with AvrI and BamHI, and ligated into pENTR/D-TOPO:MCS-Kan. The resulting region containing both flanking sequences and npt2 driving kanamycin resistance were transferred to pLVC/D (29) via a clonase LR reaction (Invitrogen). The resulting plasmid was isolated and electroporated into E. coli S17-1 for conjugal transfer. Both E. coli and P. syringae were grown individually overnight on plates and then allowed to mate overnight. Initial transformants were isolated on KB plates containing rifampin, kanamycin, and tetracycline. Deletion mutants that were kanamycin resistant but tetracycline sensitive were selected. Deletions were confirmed by PCR amplification, which verified that the kanamycin cassette had replaced syfA.
An unmarked deletion mutant of rhlA was constructed by a modified overlap extension PCR (8). Briefly, the 5′ and 3′ regions flanking rhlA were amplified in a first round of PCRs, in addition to a kanamycin resistance cassette flanked with FLP recombination target (FRT) sites from pKD13 (10). The rhlA-F1 and rhlA-R1 primers (F stands for forward, and R stands for reverse) were used to amplify the region upstream of rhlA, while the rhlA-F2 and rhlA-R2 primers were used to amplify the region downstream of rhlA, and FRT-KM-F and FRT-KM-R were used to amplify the FRT-flanked kanamycin resistance cassette. In a subsequent PCR, all three fragments were combined and amplified for 15 cycles without added primers, followed by addition of rhlA-F1 and rhlA-R2 for 20 more PCR cycles to amplify the combined fragment. The resulting fragment was cloned into the suicide vector pTOK2T (7) and transferred into P. syringae by triparental mating (6). Initial transformants were isolated on KB plates containing rifampin, kanamycin, and tetracycline. Double-crossover mutants that were kanamycin resistant but tetracycline sensitive were selected. The kan cassette was excised by introduction of plasmid pFLP2 (21) in which the omega fragment conferring spectinomycin resistance had been added (R. Scott, unpublished data), followed by replica plating to cure the ΔrhlA strain of pFLP2-omega. Final markerless deletions were confirmed by PCR. In order to generate a ΔsyfA ΔrhlA double deletion mutant, unmarked ΔrhlA was first generated before deleting the syfA gene in order to avoid redundant kanamycin resistance genes.
Site-directed chromosomal disruptions.
Site-directed mutagenesis of fleQ, fliA, fliF, and flgD was achieved with single-crossover plasmid insertion events. Fragments of the genes of interest were amplified from P. syringae genomic DNA by PCR with primers fleQ-KO-F (KO stands for knockout) and fleQ-KO-R, fliA-KO-F and fliA-KO-R, fliF-KO-F and fliF-KO-R, and flgD-KO-F and flgD-KO-R. The resulting four inserts were subcloned into pENTR/D-TOPO and subsequently transferred into pLVC/D by clonase LR reactions. Plasmids were isolated and electroporated into E. coli S17-1 for conjugal transfer. P. syringae transconjugants were isolated on KB plates containing rifampin and tetracycline. Knockouts were confirmed by PCR amplification.
Construction of pRHLA2, a RhlA complementation vector.
Full-length rhlA was amplified by PCR from genomic DNA with primers rhlA-nco-F and rhlA-xho-R. The 5′ primer contained the ATG start codon within the NcoI site. PCR conditions were as follows: 28 cycles, with 1 cycle consisting of 1 min at 95°C, 1 min at 59°C, and 1 min at 72°C, with a final extension time of 10 min at 72°C. The resulting fragment was digested with NcoI and XhoI and cloned into pMF54-omega. The expression plasmid pRHLA2 was electroporated into wild-type and mutant strains of P. syringae B728a with selection for spectinomycin resistance.
Construction of transcriptional fusion reporters.
Green fluorescent protein (GFP)-based reporter plasmids to assess transcription of rhlA, fliC, fliE, and flgB were constructed in a manner similar to that described by Burch et al. (4). The upstream promoter region of the P. syringae B728a rhlA gene was amplified by PCR from genomic DNA with primers rhlA-pro-F (pro stands for promoter) and rhlA-pro-R to generate a 495-bp promoter region. The upstream promoter region of the fliC gene was amplified with primers fliC-pro-F and fliC-pro-R to generate a 321-bp promoter region. The upstream promoter region of the fliE gene was amplified with primers fliE-pro-F and fliE-pro-R to generate a 235-bp promoter region. The upstream promoter region of the flgB gene was amplified with primers flgB-pro-F and flgB-pro-R to generate a 278-bp promoter region. The PCR products were first cloned into pTOPO Blunt (Invitrogen) and transformed into E. coli DH5α. Each insert was sequenced to verify its identity. pTOPO-PrhlA was digested with HindIII and EcoRI, pTOPO-PfliC was digested with PstI and BamHI, and pTOPO-PfliE and pTOPO-PflgB were digested with BamHI and EcoRI. The resulting fragments were cloned into pPROBE-GT (32), which contains a promoterless gfp gene in order to generate pPrhlA-gfp, pPfliC-gfp, pPfliE-gfp, and pPflgB-gfp. Promoter reporter plasmids were electroporated into P. syringae B728a as well as mutant strains altered in biosurfactant production.
GFP fluorescence was determined in strains grown overnight on KB plates and then suspended in phosphate buffer (10 mM, pH 7.5) to an OD600 of about 0.2. Cells from hydrated paper discs were inoculated by using a toothpick onto paper discs and grown overnight, and the paper discs were then transferred into phosphate buffer and vortexed to suspend the cells. GFP fluorescence intensity was determined using a TD-700 fluorometer (Turner Designs, Sunnyvale, CA) with a 486-nm bandpass excitation filter and a 510- to 700-nm combination emission filter. A relative fluorescence unit (RFU) was defined as the fluorescence of the suspensions normalized for the suspension turbidity measured as OD600.
Extraction of HAA.
Crude biosurfactant-containing extracts were prepared using modifications of a general HAA extraction protocol (13). Agar plates with confluent lawns of the P. syringae B728a ΔsyfA mutant harboring plasmid pRHLA2 were grown for 48 h. The cells were harvested by washing four plates in 90 ml H2O, and cells were removed by centrifugation (5,000 × g, 10 min). The supernatant was filter sterilized (opaque even after filtration), brought to pH 2 with concentrated HCl, mixed with 150 ml chloroform-methanol (2:1), and separated overnight. The lower organic fraction was later dried to completion. Additionally, a white precipitate that collected between the aqueous and organic layers was saved and tested for surfactant activity, along with the dried organic fraction. The chloroform-methanol-soluble fraction of the precipitate contained large quantities of HAA, as did the original dried organic fraction but to a lesser extent.
Mass spectrometry (MS).
Extracted mixtures were analyzed with a quadruple time of flight (Q-TOF) mass spectrometer (Applied Biosystems/MDS Sciex Qstar/Elite) via nanospray (49) ionization in the negative mode using uncoated capillaries with a 1-μm orifice (New Objective, Woburn, MA) at an approximate flow rate of 40 nl/s. Samples were dissolved in water: acetonitrile (3:7). Fragmentation was attained using collision energy (collisionally activated dissociation gas setting [CAD] = 5 and collision energy [CE] = −25). Fragmentation energies were maintained at a level that maintained a large percentage of the parent ion peak in order to minimize the generation of secondary fragments. Tandem mass spectra were collected isolating the 1− charge state, as these were the most prevalent ions.
RESULTS
P. syringae B728a produces two motility-enabling surfactants.
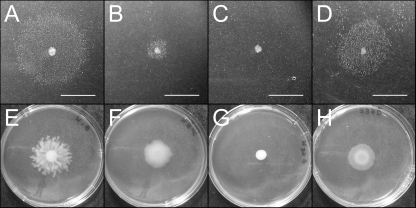
To discount the possibility that the small amount of surfactant observed in syfA and syfB insertional mutants was due to residual syringafactin production, we constructed a deletion mutant of syfA, the first of two genes in the syringafactin biosynthetic cluster. As observed in syfA and syfB insertional mutants (4), the ΔsyfA mutant retained the ability to produce a small amount of surfactant detectable with the atomized oil assay (Fig. 1B) and was still capable of limited swarming (Fig. 1F). Thus, although syringafactin is the main motility-enabling surfactant produced by P. syringae B728a on plates, a second unidentified surfactant is also produced which contributes to motility.
Fig 1.
Comparison of surfactant-induced halos around bacterial colonies grown on hard agar plates visualized in the atomized oil assay (A to D) and swarming motility for the same strains on 0.4% soft agar plates (E to H). The strains are a WT strain of P. syringae (A and E), a ΔsyfA mutant (B and F), a ΔsyfA Psyr_3129 double mutant (C and G), and a ΔsyfA Psyr_3129 double mutant harboring pRHLA2, a plasmid constitutively expressing Psyr_3129 (D and H). Bars, 1 cm (A to D).
In order to determine the identity of this surfactant, the atomized oil assay was used to screen a library of Tn5 mutants created in a ΔsyfA background. Over 4,500 random mutants were screened for those with significantly more or less surfactant production as evidenced by larger or smaller halos on agar plates when assayed with this high-throughput method. After excluding mutants having obvious growth defects, 4 strains were identified that exhibited a complete loss of surfactant production. Additionally, 25 other strains were found to consistently produce significantly (P < 0.01) more or less surfactant than the ΔsyfA parental strain (Table 1). The genes disrupted by transposon insertion were identified, revealing that 4 genes were required for surfactant production, while disruption of 9 different genes conferred less surfactant production and disruption of 6 genes upregulated production. Interestingly, a number of genes were identified by multiple independent insertion events (from separate matings and with insertions into different loci) (Table 1). This finding confirmed the significance of these genes for surfactant production, as well as suggested that the insertion events were not entirely random.
Table 1.
Insertional mutants of Pseudomonas syringae with altered surfactant production
| Locus of Tn5 insertion | Predicted function | No. of individual transposon hitsa | Surfactant halo radius (mm)b ± SD | Swarming diam (mm)c ± SD | Swimming diam (mm)d ± SD |
|---|---|---|---|---|---|
| Psyr_3129e (rhlA) | Acyltransferase | 1 | 0 ± 0.0** | 9.0 ± 0.0** | 8.0 ± 1.0 |
| Psyr_3698 (gacS) | Sensor kinase | 1 | 0 ± 0.0** | 9.5 ± 0.6** | 8.3 ± 0.6 |
| Psyr_0258 (ompR) | Response regulator | 1 | 0 ± 0.0** | 9.5 ± 1.0** | 6.0 ± 0.0** |
| Psyr_3461 (fleQ) | NtrC-like flagellar regulator | 1 | 0 ± 0.0** | 9.5 ± 0.7** | 0 ± 0.0** |
| Psyr_4446 (osmE) | Osmotically inducible lipoprotein | 1 | 0.7 ± 0.6** | 14.4 ± 1.5** | 5.3 ± 0.6** |
| Psyr_0936 (wbpY) | O-polysaccharide glycosyltransferase | 3 | 0.7 ± 0.6** | 11.0 ± 0.0** | 0 ± 0.0** |
| Psyr_0219 (algC) | Phosphomannomutase | 1 | 0.7 ± 0.6** | 12.3 ± 2.1** | 1.0 ± 0.0** |
| Psyr_0918 (wzt) | A-band LPS export | 2 | 1.0 ± 0.0** | 11.5 ± 0.6** | 0 ± 0.0** |
| Psyr_2083 | Unknown | 1 | 1.0 ± 0.0** | 16.8 ± 3.4 | 6.3 ± 0.5* |
| Psyr_0270 (polA) | DNA polymerase | 2 | 0.8 ± 0.4** | 13.6 ± 0.8** | 8.0 ± 0.0 |
| Psyr_1981 | PAS:GGDEF | 1 | 1.2 ± 0.4** | 15.3 ± 2.4* | 9.0 ± 1.0 |
| Psyr_3669 | Outer membrane protein | 2 | 1.3 ± 0.6** | 15.8 ± 3.0 | 8.7 ± 0.6 |
| Psyr_3480 (flgC) | Flagellar assembly | 1 | 1.7 ± 0.6** | 10.0 ± 0.0** | 0 ± 0.0** |
| No insertion (ΔsyfA) | 5.3 ± 0.5 | 19.4 ± 2.3 | 8.3 ± 0.6 | ||
| Psyr_3466 (fliC) | Flagellin | 1 | 6.7 ± 0.6** | 10.4 ± 0.5** | 0 ± 0.0** |
| Psyr_3469 (fgt1) | Flagellar glycosyltransferase | 3 | 7.7 ± 1.2** | 9.0 ± 1.0** | 2.0 ± 1.0** |
| Psyr_3468 (fgt2) | Flagellar glycosyltransferase | 1 | 7.0 ± 0.9** | 17.5 ± 2.1 | 8.3 ± 1.5 |
| Psyr_2979 (gor) | Glutathione reductase | 1 | 10.3 ± 1.2** | 27.8 ± 8.9* | 9.0 ± 1.0 |
| Psyr_0263 (algB) | NtrC-like response regulator | 4 | 11.3 ± 1.2** | 26.8 ± 2.3** | 9.7 ± 2.5 |
| Psyr_1350 (mucP) | Peptidase | 1 | 14.3 ± 0.6** | 28.8 ± 6.8** | 9.3 ± 0.6* |
Number of times that independent mutants were identified as insertions in the same gene.
Halos with significantly smaller or larger radii than the halo of the WT (for all, P < 0.01 [**] by a t test).
Bacterial motility across semisolid 0.4% agar plates. The swarming motility was significantly different from the motility of the wild type at P < 0.05 (*) or P < 0.01 (**) as determined by a t test.
Bacterial motility through semisolid 0.25% agar plates. The swimming motility was significantly different from the motility of the wild type at P < 0.05 (*) or P < 0.01 (**) as determined by a t test.
Insertion is immediately upstream of the gene, and the gene name is assigned in this article.
When these mutants were tested for swarming motility, similar to what we have observed previously (4), there was a strong positive correlation between the surfactant halo radius for a given mutant strain and its swarming diameter (R2 = 0.78, P < 0.001). None of the mutants that were deficient in surfactant halos were observed to swarm (swarm diameters were equivalent to the initial inoculum spread of 9 to 10 mm), while many mutants with significantly larger surfactant halos were observed to be hyperswarmers (swarm diameters more than 25 mm). The main outliers to this trend had insertions into flagellar components; because swarming is typically dependent on both flagella and surfactant production (24), it is of little surprise that these mutants failed to swarm despite exhibiting surfactant production. On the other hand, swimming motility, which is a form of individual motility that does not require surfactant production, was poorly correlated with the observed surfactant halos (R2 = 0.33, P = 0.16). Thus, in addition to the previously identified surfactant syringafactin, this second unidentified surfactant produced by P. syringae B728a contributes to the surface motility of this organism.
An rhlA homolog implicated in biosurfactant production.
Of the four mutants identified as being completely blocked in biosurfactant production, three of the insertions were in the global regulatory genes gacS, ompR, and fleQ. GacS is a global regulator of secondary metabolites and extracellular enzymes (19), while an OmpR homolog has recently been hypothesized to be a membrane stress sensor in P. aeruginosa (26), and FleQ is the initial regulatory element of flagellar biosynthesis (9). The remaining gene, Psyr_3129, encoding a predicted acyltransferase having 48.5% sequence identity to rhlA and 49% identity to phaG in P. aeruginosa PAO1, seemed likely to be involved directly in surfactant biosynthesis. RhlA is responsible for production of 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursor to rhamnolipids in P. aeruginosa, and this compound is independently recognized as a biosurfactant that promotes swarming motility (13). PhaG is involved in synthesis of polyhydroxyalkanoic acid (PHA), a carbon and energy storage molecule (39). Both enzymes divert hydroxydecanoic acids from fatty acid de novo synthesis and exhibit similar and sometimes overlapping polymerization functions (43).
Because the transposon insertion was in the promoter region immediately upstream of Psyr_3129, we confirmed that a knockout of this gene also blocked surfactant production by constructing a chromosomal deletion of Psyr_3129 in the ΔsyfA background of P. syringae (Fig. 1C). This double mutant was also incapable of swarming (Fig. 1G). To ensure that disruption of Psyr_3129 and not genomic changes elsewhere was responsible for abrogating biosurfactant production, we complemented this gene in trans. Psyr_3129 was inserted into pMF54 (17) to form plasmid pRHLA2 where it was driven by an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible trc promoter. When this plasmid was introduced into a ΔsyfA Psyr_3129 double mutant, abundant biosurfactant was produced (Fig. 1D) and swarming was restored (Fig. 1H) even without IPTG addition, suggesting that even low levels of expression of Psyr_3129 are sufficient for biosurfactant production. In fact, increased expression of Psyr_3129 upon addition of IPTG did not result in more surfactant production beyond that observed in uninduced cells. Thus, either the Psyr_3129 gene product synthesizes the surfactant or is essential for its expression. Significantly, rhlA from P. aeruginosa has been shown to be sufficient for HAA production in E. coli (13, 51), and a rhlA homolog in Serratia sp. strain ATCC 39006 was also linked to production of an unidentified biosurfactant (48). We thus tested whether our potential rhlA homolog was sufficient to confer biosurfactant production in E. coli. E. coli DH5α harboring plasmid pRHLA2 produced a large amount of surfactant when examined by the atomized oil assay (data not shown). We thus tentatively considered Psyr_3129 to be a rhlA homolog.
Although production of this surfactant in a ΔsyfA strain of P. syringae is readily detected with the atomized oil assay, it was not detectable with other assays such as the drop collapse assay or by direct chemical detection. This suggested that either the molecule had properties such as low water solubility that prevented its detection with the water drop collapse assay, or that it was made in relatively small amounts that are not easily detected by assays with low sensitivity (5). Indeed, a ΔsyfA strain harboring pRHLA2 in which RhlA was constitutively expressed caused a drop collapse (data not shown); therefore, we presume that low rates of production in native strains explain its lack of detection in a ΔsyfA strain in a drop collapse assay. Using a modified protocol for HAA extraction (13), we extracted the unidentified surfactant from plate-grown cultures of the ΔsyfA mutant harboring plasmid pRHLA2. The resulting extract yielded an opaque solution in water, indicative of a surfactant with low water solubility which aggregates in water (33). This concentrated surfactant lowered the surface tension of water to 29 mN m−1 when measured in a pendant drop assay, a value indicative of a potent surfactant and the same value observed for HAAs (13).
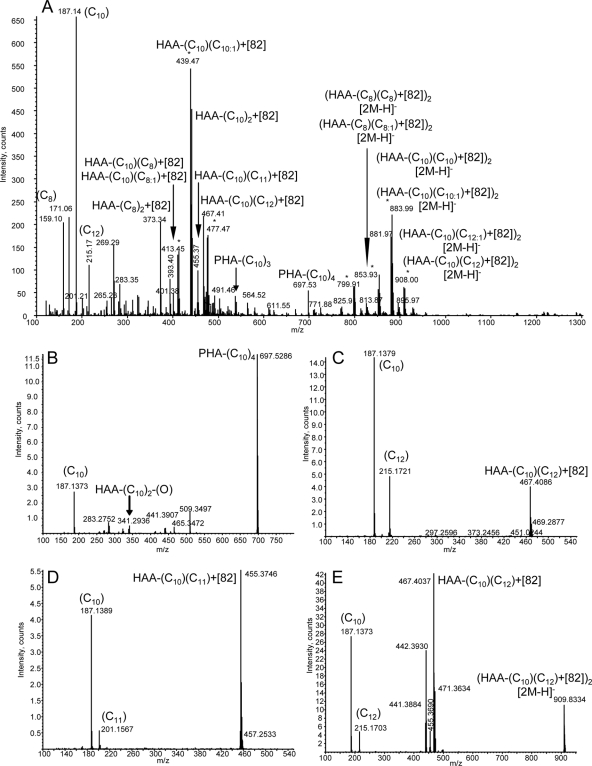
The concentrated biosurfactant mixture was analyzed by mass spectrometry (MS). Samples were subjected to negative mode static nanospray ionization. The mass spectrum (Fig. 2A) of the mixture revealed a large number of components, several of which could be assigned as HAA monomers [m/z 159 (C8), 187 (C10), and 215 (C12)] and PHA oligomers [m/z 527 (C10)3, 697 (C10)4]; however, many additional peaks present at m/z 400 to 480 and 800 to 910 did not correspond to HAAs or PHAs by mass alone but had the expected pattern of HAA ions with repeating units differing by 28 Da, indicating increasing chain lengths. These peaks existed as pairs of ions separated by a mass of 2 Da, denoted by asterisks in Fig. 2A, perhaps indicating the presence of unsaturated bonds in the structures. The unassigned masses corresponded to HAA masses with a consistent additional 82-Da modification, where the larger mass ions corresponded to [2M+82-H]1− ion clusters. For simplicity, these ions have been labeled as the most simple compositions, i.e., HAA-(C10)2 also contains a portion of HAA-(C8)(C12); this is confirmed by the presence of both ions in many tandem MS experiments. To confirm the presence of HAAs in the observed ions, a series of tandem mass spectrometry experiments were run to ascertain whether the characteristic tandem mass spectrometric fragmentation patterns of HAAs were present (27, 41). Fragmentation of the ion corresponding to PHA(C10)4 revealed a pattern of ions consistent with the repeating structure of C10 units, including the strong peak at m/z 187 (Fig. 2B). Of greater importance were the fragmentation characteristics of ions containing the consistent 82-Da mass shift due to their prevalence in the spectrum. Fragmentation of m/z 467 (Fig. 2C), corresponding to HAA-(C10)(C12)+82, yielded fragment ions of HAA-(C10) and HAA-(C12), where the 82 adduct appeared to be lost as a neutral ion; this was potentially an adduct of two acetonitrile molecules generated during ionization. All ions within the mass range of 400 to 480 assignable to HAA variations contained the expected fragment ions corresponding to HAA-C8, HAA-C10, and HAA-C12 and unsaturations for ions where the mass was 2 Da lower than the calculated saturated HAA ion. Interestingly, the ion present at m/z 455, corresponding to HAA-(C10)(C11)+82, yielded a spectrum that contained the fragment ions of an HAA chain containing an odd number of carbons (Fig. 2D). Fragmentation of larger ion masses corresponding to [2M+82-H]1− showed the presence of HAA-C8, HAA-C10, and HAA-C12 ions (Fig. 2E). The use of tandem mass spectrometry showed that the ions observed in the mass spectrum were predominately derived from HAAs, PHAs, and/or adducts of these components. Thus, although similar extracts from wild-type and ΔsyfA strains of P. syringae yielded insufficient quantities of surfactant for detection by mass spectrometry, a strain with upregulated transcription of the P. syringae rhlA homolog demonstrates that this gene indeed directs the synthesis of HAA in P. syringae B728a, as in Pseudomonas aeruginosa (13, 43). Additionally, the detection of small quantities of PHAs in this strain with upregulated rhlA transcription demonstrates some enzymatic overlap between HAA and PHA production (43).
Fig 2.
(A) Negative mode static nanospray mass spectrum of extracted biosurfactant. (B to E) Tandem mass spectra of representative ions corresponding to HAA-containing ions.
Flagellar assembly classes and biosurfactant production.
An unexpected finding in the analysis of mutants with altered biosurfactant production was that many of these mutants (with both increased and decreased surfactant production) harbored disruptions in genes encoding members of the flagellar biosynthetic pathway (Table 1). Flagella are assembled in a multitiered temporal manner, where the expression of successive components of the flagellum are dependent on completion of the previous steps. In P. aeruginosa, which is representative of most gammaproteobacteria, this is a four-step process (30). Initially, the master regulator FleQ (class I) controls the expression of class II genes, which include genes necessary for site selection and initialization of the flagellar basal body as well as the two-component system FleSR. FleR in turn controls the class III genes which encode proteins that complete the basal body structure. Once the basal body is completed, an anti-sigma factor FlgM is exported through it, which frees its cognate sigma factor FliA to activate the class IV genes, including flagellin (FliC) and chemotaxis genes (9). When the different flagellar mutants from our screen were ordered by their flagellar assembly classes, a consistent pattern emerged: the lower the class of the flagellar gene that was disrupted, the smaller the observed surfactant halo was (Table 2). An insertion into the master regulator fleQ resulted in a total loss of surfactant production, disruption of the class III flagellar assembly gene flgC resulted in a large (3-fold) reduction in the surfactant halo, while an insertion in fliC resulted in significantly larger surfactant halos. Furthermore, insertions in fgt1 and fgt2, two genes involved in flagellar glycosylation that have been shown in P. syringae pv. tabaci 6605 to be important for flagellar function and stability (45), both also resulted in enlarged surfactant halos. These results led us to hypothesize that assembly of the flagellar basal body is an important trigger for the production of HAA and that once the flagellar basal body is assembled and flagellin synthesis is initiated, mutations that hinder flagellar assembly or functionality serve to upregulate the production of HAA.
Table 2.
Flagellar assembly classes and their effects on surfactant production
| Source | Assemblya | Gene | Relative surfactant production ± SDb |
|---|---|---|---|
| From screen | Class I | fleQ | 0 ± 0.00** |
| Targeted mutant | Class I | fleQ | 0 ± 0.00** |
| Targeted mutant | Class I and IV | fleQ or fliC | 0 ± 0.00** |
| Targeted mutant | Class I | fliA | 0.36 ± 0.13** |
| Targeted mutant | Class II | fliF | 0.27 ± 0.16** |
| From screen | Class III | flgC | 0.36 ± 0.13** |
| Targeted mutant | Class III | flgD | 0.33 ± 0.15** |
| From screen | Class IV | fliC | 1.22 ± 0.29* |
| From screen | fgt1 | 1.19 ± 0.27* | |
| From screen | fgt2 | 1.25 ± 0.26** |
Assembly class designations are given according to those given by Dasgupta et al. (9) for Pseudomonas aeruginosa.
Relative surfactant production is expressed as the average surfactant halo radius measured for the mutant divided by the average halo size measured for the ΔsyfA mutant; all averages were calculated from six replicate cultures. Halos were significantly different from the wild type at P < 0.05 (*) or P < 0.01 (**) as determined by a t test.
To further support our hypothesis that expression of HAA is dependent on flagellar assembly itself and not the expression of a few specific flagellar genes, we constructed targeted knockouts in additional flagellar genes involved at different stages of flagellar assembly (Table 2). A directed knockout mutant of fleQ was deficient in surfactant production, confirming our earlier observations of an insertional mutation of this gene. Additionally, a directed knockout mutant of fleQ in a strain harboring a transposon insertion in fliC was also deficient in surfactant production. Although the initial screen did not identify any insertions in class II genes, important for the initial establishment of the flagellar apparatus, a directed knockout of fliF exhibited a dramatic decrease in surfactant production. Furthermore, a knockout of flgD, a class III flagellar gene in an operon downstream of flgC, resulted in a similar 3-fold reduction in the size of the HAA halo (Table 2). Disruption of fliA encoding the sigma factor responsible for initiating transcription of class IV genes also conferred a 3-fold reduction in the surfactant halo. Thus, since FliA is necessary for expression of late-stage flagellar genes, the residual HAA production in a ΔsyfA fliA mutant strain signifies that rhlA is not directly under FliA control and should not be considered a class IV flagellar gene.
Because the establishment of the flagellar basal body appears important for production of this surfactant, we postulated that perhaps the flagellum is in some way necessary for the export of HAA. In order to test this model, we introduced plasmid pRHLA2, conferring constitutive RhlA expression, into a ΔsyfA fleQ double mutant of P. syringae. This strain, despite lacking flagella, exhibited unaltered surfactant production (data not shown), indicating that flagella are not necessary for surfactant export. Thus, it appears that the flagellar assembly process most likely influences HAA at the production level.
Coordinate regulation of flagellin and HAA.
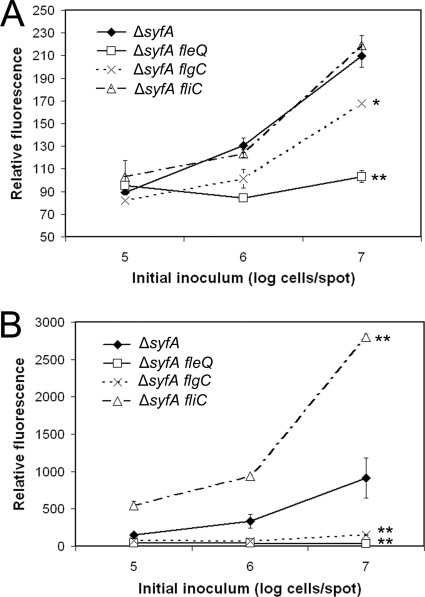
To investigate the contribution of flagellar assembly to transcriptional regulation of rhlA, we linked a gfp reporter gene to the promoter-containing region 5′ to rhlA in the stable plasmid vector pPROBE-GT (32) to produce reporter plasmid pPrhlA-gfp. We introduced pPrhlA-gfp into the mutants blocked at different stages of flagellar assembly and observed that, as was indicated by the atomized oil assay, the expression of rhlA was significantly lower in ΔsyfA fleQ and ΔsyfA flgC mutant strains than in a ΔsyfA strain (Fig. 3A). This difference was most notable in cells grown to high cell densities, whereas all strains at lower cell densities and earlier growth stages shared similarly low rhlA transcription levels. Unexpectedly, rhlA expression in a ΔsyfA fliC mutant was indistinguishable from a ΔsyfA strain at all growth stages. This lack of transcriptional difference was also true of the glycosylation mutants (data not shown). Although the radius of the surfactant halo of a ΔsyfA fliC mutant is only 1.22 times the radius of a ΔsyfA strain, the three-dimensional diffusion of surfactants on agar plates contributes to a nonlinear relationship between the spread radius and surfactant abundance, and thus, a ΔsyfA fliC mutant produces considerably more HAA than its parent strain. However, actual estimation of that difference is not possible, since the surfactant concentration is not evenly distributed across the observed halo, production and diffusion of surfactants occur simultaneously, and time and concentration are both positively correlated with surfactant halo sizes (4). Furthermore, this difference was large enough to identify the flagellin mutant, as well as four independent flagellin glycosylation mutants with halos of similar sizes, in a blind screen of insertional mutants. Therefore, we believe that the increase in HAA production in these late-stage flagellar genes is not accurately reflected by rhlA transcription levels and is likely being regulated posttranscriptionally in these mutants. Further research is warranted to determine the nature of this regulation.
Fig 3.
Relative GFP fluorescence of flagellar mutant strains of P. syringae B728a harboring either a plasmid in which GFP expression is dependent on the promoter of rhlA (A) or on the promoter of fliC (B). Promoter strains were inoculated at a range of 105 to 107 cells in 5-μl suspensions onto agar plates and incubated for 20 h to yield a range of cell densities at the time of sampling. The error bars represent the standard deviations of the mean cell-normalized GFP fluorescence. Promoter expression was significantly different in the mutant strain compared to the ΔsyfA strain at the highest cell density at a confidence level of P < 0.05 (∗) or P < 0.01 (∗∗) as determined by a t test.
We also constructed reporter plasmid pPfliC-gfp in which a gfp reporter gene was fused to the promoter-containing region of fliC to provide estimates of the expression of the gene encoding flagellin in the mutants in which we had determined rhlA expression (Fig. 3B). Similar to the expression of rhlA, the expression of fliC was significantly reduced in both a ΔsyfA fleQ and a ΔsyfA flgC mutant background. In contrast to rhlA expression levels but similar to observed HAA production, fliC was overexpressed relative to the level in a ΔsyfA background in a ΔsyfA fliC mutant. Furthermore, fliC expression levels were significantly higher in both glycosylation mutants than in the ΔsyfA parent strain, although the levels were about half that observed in the ΔsyfA fliC mutant (data not shown). Thus, the transcription of fliC closely mirrors the observed production levels of HAA in P. syringae, although evidence from rhlA transcription levels suggests that this is not coordinated strictly at the transcriptional level.
Feedback of glycosylation defects on flagellin transcription.
As far as we are aware, flagellar glycosylation has not been documented to have a feedback role in flagellin biosynthesis. Although it is intuitive that a loss of flagellin production might result in constitutive activation of the late-stage flagellar genes through FliA (via continual FlgM export), it is less obvious how flagellar glycosylation mutations upregulate flagellin production, especially in the case of the fgt2 mutant, which does not exhibit any impairment of flagellar function. In order to investigate the feedback process, we constructed transcriptional reporters of both flgB, a class II flagellar gene, and fliE, a class III flagellar gene, in addition to the fliC class IV reporter. Reporter plasmids pPflgB-gfp and pPfliE-gfp were separately introduced into the original ΔsyfA strain and a ΔsyfA fgt2 double mutant, respectively, so that the effect of flagellar glycosylation on the expression of the three classes of flagellar genes could be observed. The loss of flagellar glycosylation resulted in upregulation only of the late-stage flagellin gene fliC and not of fliE or flgB (see Fig. S1 in the supplemental material). Loss of glycosylation most likely affects the flagella in such a way as to encourage the export of the anti-sigma factor FlgM, either through increased flagellar breakage or increased export within the flagella, thus releasing FliA from FlgM control.
Mutants that influence HAA production through flagella.
Many mutations could potentially disrupt flagellar assembly, and any mutation disrupting flagellar assembly will likely also have an effect on HAA production. Therefore, we postulated that some of the mutants recovered in our screen might indirectly influence production of our surfactant by modulating the function of flagella. Motility assays were therefore analyzed to determine whether any mutants displayed potentially altered flagellar function (Table 1). Mutants in which Psyr_0936 (wbpY), Psyr_0219 (algC), and Psyr_0918 (wzt) were disrupted exhibited nearly abolished swimming and swarming motility. Interestingly, these three mutations are all also expected to change the surface morphology of P. syringae, as their gene homologs are all involved in lipopolysaccharide (LPS) synthesis or export. These mutants were examined for their ability to express flagellin biosynthesis genes; any mutations that affect rhlA expression via altered flagellar function should exhibit low levels of fliC expression. However, only a Psyr_0936 (wbpY) mutant, encoding a glycosyltransferase, exhibited lower fliC expression than the WT strain did (Table 3). Thus, we conclude that Psyr_0936 (wbpY), but not Psyr_0219 (algC) or Psyr_0918 (wzt), is affecting rhlA expression indirectly via an inhibitory effect on flagella.
Table 3.
Relationship between HAA production of transposon mutants and the transcription of rhlA and fliC
| Locus of Tn5 insertion | Relative surfactant production ± SDa,d | GFP fluorescence (mean ± SD) of mutants containing the following reporter gene fusion: |
|
|---|---|---|---|
| pPrhlA-gfpb,d | pPfliC-gfpc,d | ||
| Psyr_3698 (gacS) | 0 ± 0.00** | 0.60 ± 0.18 | +e |
| Psyr_0258 (ompR) | 0 ± 0.00** | 0.15 ± 0.08* | + |
| Psyr_4446 (osmE) | 0.13 ± 0.11** | 1.09 ± 0.15 | + |
| Psyr_0936 (wbpY) | 0.13 ± 0.11** | 0.25 ± 0.04* | 0.01 ± 0.00** |
| Psyr_0219 (algC) | 0.13 ± 0.11** | 1.04 ± 0.23 | 0.99 ± 0.09 |
| Psyr_0918 (wzt) | 0.19 ± 0.02** | 0.60 ± 0.18 | 0.90 ± 0.22 |
| Psyr_2083 | 0.19 ± 0.02** | 0.60 ± 0.14 | + |
| Psyr_0270 (polA) | 0.16 ± 0.08** | 0.74 ± 0.11 | + |
| Psyr_1981 | 0.22 ± 0.08** | 0.87 ± 0.20 | + |
| Psyr_3669 | 0.25 ± 0.11** | 0.87 ± 0.15 | + |
| Psyr_2979 (gor) | 1.93 ± 0.29** | 1.09 ± 0.15 | + |
| Psyr_0263 (algB) | 2.13 ± 0.30** | 1.66 ± 0.40 | + |
| Psyr_1350 (mucP) | 2.69 ± 0.28** | 2.56 ± 0.66* | + |
Surfactant production of insertional mutants in a ΔsyfA mutant background is quantified as the radius of surfactant-induced halos in an atomized oil assay divided by the average halo size measured for the ΔsyfA mutant (both values from Table 1).
GFP fluorescence of mutants containing the rhlA reporter gene fusion is normalized against that obtained in the ΔsyfA mutant itself.
GFP fluorescence of mutants containing the fliC reporter gene fusion is normalized against that obtained in the ΔsyfA mutant itself.
Values that were significantly different from those of the wild type as determined by a t test are indicated as follows: *, P < 0.05; **, P < 0.01.
Expression levels of flagellin were not investigated for strains with a plus sign, because they exhibited swimming motility.
To further test the importance of flagellar function in the mutants obtained from our initial screen, we determined whether mutants with enhanced HAA production required functional flagella. In order to test this, we constructed fleQ disruptions in Psyr_2979 (gor), Psyr_0263 (algB), and Psyr_1350 (mucP) mutant backgrounds. A fleQ disruption in a strain in which glutathione reductase (Psyr_2979 [gor]) was also blocked produced no detectable HAA, while a fleQ disruption in algB and mucP mutant backgrounds (members of the AlgT extracellular stress pathway) greatly reduced but did not abolish HAA production (data not shown). This demonstrates that the AlgT stress pathway controls HAA production independently of flagellar function and furthermore that FleQ is not necessary for HAA production.
Assessment of the remaining mutants.
The levels of rhlA transcription in mutants with reduced surfactant production but having functional flagella were assessed to determine whether regulation of surfactant production was mediated at the transcriptional or posttranscriptional level (Table 3). Of all such mutants examined, only the strain with a mutation in the OmpR homolog had a pronounced effect on rhlA transcription. The levels of rhlA expression in a GacS mutant, although lower than a WT strain, could not account for the absence of any production of HAA. Thus, in agreement with the role of GacS as a posttranscriptional regulator, it appears that it is mainly affecting HAA production posttranscriptionally. OsmE and AlgC mutants also did not reduce the transcription of rhlA, and thus might act posttranscriptionally to reduce surfactant synthesis and/or export of HAA. The remaining mutants all had moderately lower levels of expression of rhlA that could have accounted for their reduced surfactant production, but the manner in which they are linked to HAA production remains unclear.
Porous surfaces upregulate flagellin and HAA.
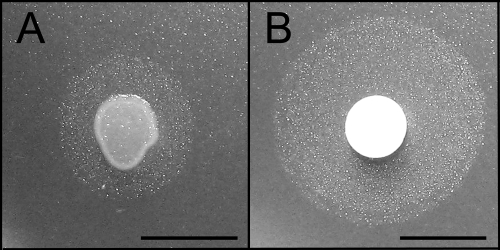
Although HAA clearly has a role in P. syringae swarming motility over a planar agar surface, it is unclear how relevant its swarming motility is in vivo. Bacterial movement over more “natural” surfaces has recently been explored, such as in a porous-surface model, where an uneven ceramic surface harbors a heterogeneous range of water film thicknesses, only some of which are thick enough to enable bacteria to swim (11). Our approach to a natural surface was to place a porous filter paper on an agar plate, which the bacteria rapidly colonize (see Movie S1 in the supplemental material). This movement occurs regardless of whether or not nutrients are present in the agar plate (data not shown). Filter paper allows rapid swimming motility through the hydrated regions of the cellulose surface, perhaps enabling even more rapid swimming than in a purely aqueous environment by the formation of narrow swimming channels (3). While comparing different methods of culturing bacteria on agar plates, it was observed that production of HAA increased dramatically when the strain was grown on the porous surface of hydrated filter paper discs, with a doubling in the HAA radius (Fig. 4). Similarly large surfactant halos were observed when cells were grown on discs of other wettable filters and materials, such as cotton and polyester fabrics, demonstrating that this is not a cellulose-specific phenomenon (data not shown). The porous-material induction of surfactant production led us to the hypothesis that the unidentified surfactant might contribute to the colonization of natural surfaces.
Fig 4.
Biosurfactant halo produced by a ΔsyfA mutant of Pseudomonas syringae grown for 16 h from a spot of approximately 2.5 × 106 cells applied directly onto an agar plate (A) or onto a filter paper disc placed on an agar plate (B). Bars, 1 cm.
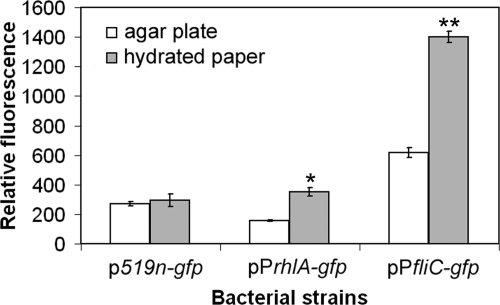
To address the process by which the hydrated porous surfaces upregulate production of HAA, we measured the expression of rhlA under various growth conditions. The GFP fluorescence of a WT strain harboring pPrhlA-gfp was compared in cells grown on filter paper discs placed on agar plates with that of cells grown directly on the agar plates. While GFP fluorescence exhibited by P. syringae harboring plasmid p519n-gfp conferring constitutive GFP expression was similar in these two growth conditions, much higher GFP fluorescence of the strain carrying pPrhlA-gfp was observed after growth on the porous paper (Fig. 5). This was true for cells at both 4 and 20 h after inoculation, although cells inoculated at high densities (107 cells on a single filter disc) no longer exhibited this upregulation by 20 h of growth (data not shown). Such apparent paper surface-induced upregulation of rhlA was observed in both the WT strain as well as a ΔsyfA strain (data not shown). No such induction of syfA was observed when strains harboring pPsyfA-gfp were grown on paper discs (data not shown), indicating that syringafactin is not similarly regulated.
Fig 5.
Upregulation of rhlA and fliC in cells of P. syringae grown on paper discs. GFP fluorescence of WT P. syringae carrying either a constitutively expressed gfp reporter gene (p519n-gfp), a fusion of a gfp reporter gene with the promoter region of rhlA (pPrhlA-gfp), or a fusion of a gfp reporter gene with the promoter region of fliC (pPfliC-gfp). Strains were assayed after overnight growth either on agar plates (white bars) or on filter paper discs placed on agar plates (gray bars). The error bars represent the standard deviations of the mean cell-normalized GFP fluorescence. Promoter expression was significantly different in the strain grown on hydrated paper compared to a plate-grown strain at a confidence level of P < 0.05 (∗) or P < 0.01 (∗∗) as determined by a t test.
Because we observed both enhanced production of HAA and elevated expression of rhlA in cells grown on hydrated paper discs, as well as a dependence of HAA production on flagellar assembly, we hypothesized that genes coding for flagellar motility would also be upregulated on the paper discs. To test this, we compared the GFP fluorescence of cells harboring the fliC reporter plasmid pPfliC-gfp when grown on agar plates and paper discs. As hypothesized, the genes encoding flagellin were upregulated in cells exploring the porous paper surface (Fig. 5). Similar to rhlA, this was not true for cells inoculated at high densities after 20 h (data not shown). This implies that flagellar motility is important for colonization of this porous surface. In order to examine the necessity of flagella for movement through hydrated paper, we compared the spread of our mutant strains on paper discs. While flagellated strains quickly spread throughout the paper discs, the nonflagellated strains remained at the site of inoculation and formed colonies only on top of the paper (see Fig. S2 in the supplemental material). This observation was true both for the ΔsyfA fleQ mutant which does not produce HAA and for the ΔsyfA fliC mutant that produces higher levels of HAA. This requirement of motility for colonization of paper discs appears very similar to that observed for exploration of a porous ceramic surface (11).
Contribution of HAA to P. syringae B728a motility.
To better determine the relative rate of movement of different strains through paper, we increased the distance over which the bacteria were allowed to move. After inoculating the strains with a toothpick onto large filter paper strips on an agar plate, we could observe the distance the bacteria were able to travel by removing the paper at chosen times and allowing the subsequent growth of cells that had penetrated through the paper. While the WT strain moved rapidly (0.29 cm/h), a ΔsyfA mutant strain moved at a rate of only 0.18 cm/h, while a fleQ mutant and a ΔsyfA ΔrhlA double mutant moved at rates of only 0.06 cm/h and 0.14 cm/h, respectively. Since all of the surfactant mutants moved much slower than the WT strain, both syringafactin and HAA apparently contribute to the form of motility that enables movement through porous materials. Although the surfactants are not necessary for motility through porous paper, they strongly facilitate the process.
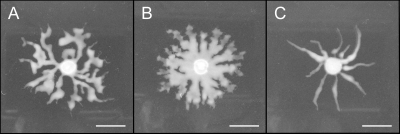
While HAA apparently aids motility both on low-agar swarming plates and on hydrated porous papers, these behaviors were always observed in a ΔsyfA mutant incapable of producing syringafactin. We therefore constructed a ΔrhlA mutant in a WT background that produces syringafactin. This strain exhibited equivalent production of syringafactin compared to a WT strain, as evidenced both by an equivalent-sized halo with the atomized oil assay and by equivalent expression of the syringafactin promoter in reporter assays (data not shown). We compared the movement of this ΔrhlA mutant with that of the WT strain to determine the relative contribution of HAA and syringafactin to cellular locomotion. The ΔrhlA strain did not differ from the WT strain in its speed of movement through porous paper (data not shown); however, it did differ from the WT strain in the manner in which it moved on swarming plates. The ΔrhlA mutant produced tendrils of cells that moved away from the point of inoculation that were much broader than those of the WT strain (Fig. 6). Such apparent movement was initially as fast as that of the WT strain, but unlike the WT strain, this mutant failed to fully explore the swarming plate; even after 4 days, a colony of the ΔrhlA mutant had not covered the agar surface, whereas the WT had fully covered the swarming plate by day 2 (picture not shown). As a further test of the role of HAA in the movement of P. syringae, we overexpressed RhlA constitutively in the WT strain capable of syringafactin production and observed its swarming motility. In contrast to the broad but short tendrils of cells produced by the ΔrhlA mutant, overexpression of RhlA led to the formation of very long and narrow tendrils that moved at the same speed as the WT strain and eventually covered the plate (Fig. 6). This behavior is similar to that observed in P. aeruginosa, where the branching and avoidance of other tendrils have been proposed to be due to the repellent effect of HAAs which serves to move the swarm front forwards (46), although P. syringae swarming colonies are not repelled by each other or by concentrated sources of HAA (data not shown).
Fig 6.
Swarming phenotypes of Pseudomonas syringae strains differing in expression of rhlA. Swarming of a WT strain of P. syringae B728a (A), a ΔrhlA mutant (B), and a WT strain harboring plasmid pRHLA2 in which rhlA is expressed constitutively at a high level (C) after 16 h of incubation on 0.4% swarm agar plates. Images are representative of at least five repetitions. Bars, 1 cm.
DISCUSSION
Although prior research has shown a linkage of biosurfactant production with flagellar motility, as far as we are aware, this is the first report to show this as a linear process, where particular and ordered stages of flagellar assembly are required for proper regulation of surfactant production. While we find genetic evidence that biosurfactants are tied to flagellar motility, this is not the first report of flagellar control of expression of nonflagellar genes. Salmonella enterica ties the expression of some virulence factors to mid-stage flagellar assembly (22). Additionally, Frye et al. identified a number of genes that are under the control of flagellar promoters but that have no apparent effect on flagellar function in S. enterica (18). Furthermore, expression of virulence factors in Proteus mirabilis was found to be tightly coregulated with FliC expression (1). Despite these reports of coregulation, to our knowledge, there has been no previous recognition that a flagellin knockout or impairment of flagellar function would serve to further upregulate production of a nonflagellar gene product.
Why does P. syringae coregulate expression of a biosurfactant with flagellar synthesis? One possibility might be that HAA is used for flagellar lubrication. In this scenario, under conditions where there is increased breakage of flagella, there will also be increased production of both flagellin and HAA. Production of HAA might help lubricate sticky surfaces and/or flagella to minimize breakage. Microscopic and immunostaining approaches might be utilized in future studies to determine whether such a model holds for P. syringae. Alternatively, there is experimental support that flagella can function as surface sensors, conveying to the bacterium positional information through the inhibition of flagellar rotation (2, 31) or moisture conditions through the export of FlgM (47). If flagellin synthesis itself is indicative of an external condition for which biosurfactant production would be beneficial, then coordinate production of biosurfactant would be an efficient response to this condition.
Given the coregulation of HAA production with class IV flagellar genes, it was initially tempting to speculate that FliA, the sigma factor that activates transcription of class IV genes, might also be directly responsible for regulating rhlA expression. However, a disruption of fliA did not abolish surfactant production, and rhlA transcription is not upregulated in a ΔsyfA fliC mutant which should exhibit increased FliA activity. It is possible that rhlA is directly regulated by FleQ, since disruptions in the flagellar biogenesis pathway downregulate fleQ transcription and thus genes under FleQ control (9), although this remains to be proven. It also remains to be determined exactly how mutations in late-stage flagellar genes affect surfactant production. Although these mutants do not exhibit a transcriptional upregulation of rhlA, their significantly larger HAA halos suggest that these late-stage mutations increase surfactant production. It is striking that flagellar glycosylation appears connected to this regulatory process: why do the glycosylation mutants, especially a mutant strain with a block in fgt2, which has sufficient flagellar function to enable unaltered swimming motility, have an equivalent effect on HAA halo size as a disruption of flagellin production itself? Glycosylation has been proposed to function in flagellar stabilization and lubrication in P. syringae pv. tabaci, where nonglycosylated flagella formed stiff flagellar bundles (44). Glycosylation might mask sticky amino acids present in flagella, preventing its binding to substrates such as cellulose, thereby preventing flagellar breakage. If lack of glycosylation makes the flagella more sticky and prone to breakage, then nonglycosylated mutants might still have functional flagella, but these flagella might break more easily, requiring an enhanced supply of fresh flagellin and/or a lubricating surfactant.
Some clues about the properties of HAA can lead us to hypothesize possible functions. The milky appearance of HAA in water is indicative of its low water solubility, suggesting that it most likely associates with surfaces such as the bacterial cell surface, instead of the bulk aqueous medium. Therefore, when HAA is produced, it likely coats the cells and modulates the surface properties of either the bacterium producing HAA or the surfaces over which the bacterium must move. Knowledge of the detailed behavior of this surfactant in aqueous environments and its effects on cell surfaces and the adhesiveness of cells should provide insight into the processes by which bacteria move across natural surfaces. In P. aeruginosa, HAAs serve to repel neighboring tendrils and maintain an outward motility during swarming (46). Although HAA does not chemically repel neighboring tendrils in P. syringae, we have observed that its production contributes to the outward motility of swarm tendrils. Such a behavior would tend to maximize the ability of a bacterial colony to explore a given habitat by suppressing inward movement, and thus enhancing only outward movement away from colonized areas; surfactin in B. subtilis has been similarly indicated to have this role (23).
Another important clue for the function of HAA in P. syringae was the finding that multiple stress pathways apparently impact its production. The OmpR homolog in P. aeruginosa, termed AmgR, has been described to function similar to CpxR in E. coli, which confers membrane stress in response to stimuli such as surface adherence (26). A knockout of this potential stress pathway in P. syringae abrogated surfactant production. In contrast, our mutant screen also revealed the role of two members of the AlgT extracellular stress pathway, both of which when knocked out resulted in an upregulation of HAA production. MucP is a peptidase that degrades the AlgT anti-sigma factor, while AlgB is a response regulator downstream of AlgT. It remains unclear why these potentially overlapping stress responses have apparently opposite effects on production of HAA. Further examination of their roles in surfactant production should help elucidate the complex interaction between these two pathways. It might be the case that the combination of these two pathways allows the cell to differentiate between subtly different stressful situations, only some of which would benefit from surfactant production.
In order to elucidate the natural roles of biosurfactants, it is important to work with a system that balances the complexity of a natural environment with the consistency and reproducibility of a laboratory experiment. Although our use of hydrated porous paper is not a refined system, we believe its resemblance to natural soil and/or moist surfaces make it a good starting point to explore these environments. There have been some reports of less effective colonization of natural surfaces by biosurfactant-deficient strains (20, 35), but the complexity of these systems made it unclear exactly how the surfactant might be functioning in nature to improve motility. Our finding that surfactant production is tied to flagellar assembly suggests that this could be an active process, whereby the surfactants lubricate the flagellum and/or sticky surfaces, enabling active motility. Alternatively, surfactant production was recently observed to reduce water retention in soils via a reduction in the soil-water surface tension (15); such a reduction might enable the passive wicking of surfactant producers through porous material. Further work with systems such as ours or the porous-surface model (11) will hopefully enable a more detailed dissection of the role of biosurfactant production in bacterial surface colonization.
In this study, we have utilized an atomized oil assay to identify the biosynthetic and regulatory pathways leading to production of a biosurfactant expressed in a strongly context-dependent way in P. syringae B728a. Its coordinated expression with flagella suggests an intimate role between surfactant production and flagellar motility, but the identification of many other regulatory elements reveals a complicated mechanism of regulation. Examinations of the interaction of this surfactant with the bacterial cell, its flagella, and with the surfaces that this bacterium colonizes should illuminate its role in the epiphytic lifestyle of P. syringae.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Russell Scott and Juliana Cho for providing biological materials, Arnaud Dechesne for helpful discussions, and helpful comments from the anonymous reviewers.
This work was supported in part by the Energy Biosciences Institute, UC Berkeley.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Footnotes
Published ahead of print 22 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Allison C, Lai H, Hughes C. 1992. Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol. Microbiol. 6:1583–1591 [DOI] [PubMed] [Google Scholar]
- 2. Belas R, Suvanasuthi R. 2005. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J. Bacteriol. 187:6789–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berg HC, Turner L. 1990. Chemotaxis of bacteria in glass capillary arrays. Escherichia coli, motility, microchannel plate, and light scattering. Biophys. J. 58:919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burch AY, Shimada BK, Browne PJ, Lindow SE. 2010. Novel high-throughput detection method to assess bacterial surfactant production. Appl. Environ. Microbiol. 76:5363–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burch AY, Browne PJ, Dunlap CA, Price NP, Lindow SE. 2011. Comparison of biosurfactant detection methods reveals hydrophobic surfactants and contact-regulated production. Environ. Microbiol. 13:2681–2691 [DOI] [PubMed] [Google Scholar]
- 6. Chen C, Beattie G. 2007. Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-β-synthase domains are required for its osmoregulatory function. J. Bacteriol. 189:6901–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C, Malek AA, Wargo MJ, Hogan DA, Beattie GA. 2010. The ATP binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 75:29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dasgupta N, et al. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809–824 [DOI] [PubMed] [Google Scholar]
- 10. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dechesne A, Wang G, Gülez G, Or D, Smets B. 2010. Hydration-controlled bacterial motility and dispersal on surfaces. Proc. Natl. Acad. Sci. U. S. A. 107:14369–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Lorenzo V, Herrero M, Jakubzik U, Timmis KN. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deziel E, Lepine F, Milot S, Villemur R. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy) alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005–2013 [DOI] [PubMed] [Google Scholar]
- 14. Dulla GFJ. 2008. Bacterial babel: breaking down quorum sensing cross-talk in the phyllosphere; analysis of the contributions of abiotic and biotic factors on AHL-mediated quorum sensing to epiphytic growth and virulence in Pseudomonas syringae. Ph.D. thesis. University of California, Berkeley, CA [Google Scholar]
- 15. Fechtner J, Koza A, Sterpaio PD, Hapca SM, Spiers AJ. 2011. Surfactants expressed by soil pseudomonads alter local soil–water distribution, suggesting a hydrological role for these compounds. FEMS Microbiol. Ecol. 78:50–58 [DOI] [PubMed] [Google Scholar]
- 16. Feil H, et al. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 102:11064–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Franklin MJ, et al. 1994. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J. Bacteriol. 176:1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frye J, et al. 2006. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:2233–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heeb S, Haas D. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant Microbe Interact. 14:1351–1363 [DOI] [PubMed] [Google Scholar]
- 20. Hildebrand P, Braun P, McRae K, Lu X. 1998. Role of the biosurfactant viscosin in broccoli head rot caused by a pectolytic strain of Pseudomonas fluorescens. Can. J. Plant Pathol. 20:296–303 [Google Scholar]
- 21. Hoang TT, Karkhoff-Schweizer RAR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 22. Iyoda S, Kamidoi T, Hirose K, Kutsukake K, Watanabe H. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81–90 [DOI] [PubMed] [Google Scholar]
- 23. James BL, Kret J, Patrick JE, Kearns DB, Fall R. 2009. Growing Bacillus subtilis tendrils sense and avoid each other. FEMS Microbiol. Lett. 298:12–19 [DOI] [PubMed] [Google Scholar]
- 24. Kearns D. 2010. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. King E, Ward M, Raney D. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301–307 [PubMed] [Google Scholar]
- 26. Lee S, et al. 2009. Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. U. S. A. 106:14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lépine F, Déziel E, Milot S, Villemur R. 2002. Liquid chromatographic/mass spectrometric detection of the 3-(3-hydroxyalkanoyloxy) alkanoic acid precursors of rhamnolipids in Pseudomonas aeruginosa cultures. J. Mass. Spectrom. 37:41–46 [DOI] [PubMed] [Google Scholar]
- 28. Loper JE, Lindow SE. 1987. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77:1449–1454 [Google Scholar]
- 29. Marco ML, Legac J, Lindow SE. 2005. Pseudomonas syringae genes induced during colonization of leaf surfaces. Environ. Microbiol. 7:1379–1391 [DOI] [PubMed] [Google Scholar]
- 30. McCarter L. 2006. Regulation of flagella. Curr. Opin. Microbiol. 9:180–186 [DOI] [PubMed] [Google Scholar]
- 31. McCarter L, Hilmen M, Silverman M. 1988. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345–351 [DOI] [PubMed] [Google Scholar]
- 32. Miller WG, Leveau JHJ, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13:1243–1250 [DOI] [PubMed] [Google Scholar]
- 33. Myers D. 2006. Surfactant science and technology, 3rd ed John Wiley & Sons, New York, NY [Google Scholar]
- 34. Neu TR. 1996. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 60:151–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nielsen TH, Nybroe O, Koch B, Hansen M, Sorensen J. 2005. Genes involved in cyclic lipopeptide production are important for seed and straw colonization by Pseudomonas sp. strain DSS73. Appl. Environ. Microbiol. 71:4112–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Toole GA, et al. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91–109 [DOI] [PubMed] [Google Scholar]
- 37. Quinones B, Dulla G, Lindow SE. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant Microbe Interact. 18:682–693 [DOI] [PubMed] [Google Scholar]
- 38. Raaijmakers J, de Bruijn I, Nybroe O, Ongena M. 2010. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34:1037–1062 [DOI] [PubMed] [Google Scholar]
- 39. Rehm B, Krüger N, Steinbüchel A. 1998. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. The PhaG gene from Pseudomonas putida KT2440 encodes a 3-hydroxyacyl-acyl carrier protein-coenzyme A transferase. J. Biol. Chem. 273:24044–24051 [DOI] [PubMed] [Google Scholar]
- 40. Ron EZ, Rosenberg E. 2001. Natural roles of biosurfactants. Environ. Microbiol. 3:229–236 [DOI] [PubMed] [Google Scholar]
- 41. Shen W, Yang S, Li X. 2005. Electrospray ionization mass spectrometric detection of rhamnolipids and their acid precursors in Pseudomonas sp. BS-03 cultures. J. Chin. Biotechnol. 25:83–87 [Google Scholar]
- 42. Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 43. Soberon-Chavez G, Aguirre-Ramírez M, Sanchez R. 2005. The Pseudomonas aeruginosa RhlA enzyme is involved in rhamnolipid and polyhydroxyalkanoate production. J. Ind. Microbiol. Biotechnol. 32:675–677 [DOI] [PubMed] [Google Scholar]
- 44. Taguchi F, et al. 2008. Effects of glycosylation on swimming ability and flagellar polymorphic transformation in Pseudomonas syringae pv. tabaci 6605. J. Bacteriol. 190:764–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taguchi F, et al. 2006. Identification of glycosylation genes and glycosylated amino acids of flagellin in Pseudomonas syringae pv. tabaci. Cell. Microbiol. 8:923–938 [DOI] [PubMed] [Google Scholar]
- 46. Tremblay J, Richardson AP, Lepine F, Deziel E. 2007. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 9:2622–2630 [DOI] [PubMed] [Google Scholar]
- 47. Wang Q, Suzuki A, Mariconda S, Porwollik S, Harshey R. 2005. Sensing wetness: a new role for the bacterial flagellum. EMBO J. 24:2034–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Williamson N, Fineran P, Ogawa W, Woodley L, Salmond G. 2008. Integrated regulation involving quorum sensing, a two-component system, a GGDEF/EAL domain protein and a post-transcriptional regulator controls swarming and RhlA-dependent surfactant biosynthesis in Serratia. Environ. Microbiol. 10:1202–1217 [DOI] [PubMed] [Google Scholar]
- 49. Wilm MS, Mann M. 1994. Electrospray and Taylor-Cone theory, Dole's beam of macromolecules at last? Int. J. Mass Spectrom. 136:167–180 [Google Scholar]
- 50. Winsor G, et al. 2009. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 37:D483–D488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu K, Rock CO. 2008. RhlA converts β-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the β-hydroxydecanoyl-β-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J. Bacteriol. 190:3147–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.