Abstract
Legionella pneumophila, the causative agent of a severe pneumonia known as Legionnaires' disease, intercepts material from host cell membrane transport pathways to create a specialized vacuolar compartment that supports bacterial replication. Delivery of bacterial effector proteins into the host cell requires the Dot/Icm type IV secretion system. Several effectors, including SidM, SidD, and LepB, were shown to target the early secretory pathway by manipulating the activity of the host GTPase Rab1. While the function of these effectors has been well characterized, the role of another Rab1-interacting protein from L. pneumophila, the effector protein LidA, is poorly understood. Here, we show that LidA binding to Rab1 stabilized the Rab1-guanosine nucleotide complex, protecting it from inactivation by GTPase-activating proteins (GAPs) and from nucleotide extraction. The protective effect of LidA on the Rab1-guanine nucleotide complex was concentration dependent, consistent with a 1:1 stoichiometry of the LidA-Rab1 complex. The central coiled-coil region of LidA was sufficient for Rab1 binding and to prevent GAP-mediated inactivation or nucleotide extraction from Rab1. In addition, the central region mediated binding to phosphatidylinositol 3-phosphate and other phosphoinositides. When bound to Rab1, LidA interfered with the covalent modification of Rab1 by phosphocholination or AMPylation, and it also blocked de-AMPylation of Rab1 by SidD and dephosphocholination by Lem3. Based on these findings, we propose a role for LidA in bridging the membrane of the Legionella-containing vacuole (LCV) with that of secretory transport vesicles surrounding the LCV.
INTRODUCTION
Over the past decade, Legionnaires' disease, a severe pneumonia caused by the bacterium Legionella pneumophila, has become an increasingly prominent infectious disease (10). L. pneumophila is ubiquitously found in natural and man-made aquatic environments, where it is a parasite of freshwater amoebae (19). When inhaled by humans, L. pneumophila is ingested by alveolar macrophages and enclosed in a membrane-bound phagosome. Unlike avirulent bacteria, L. pneumophila avoids fusion of its surrounding phagosome with destructive endosomes and lysosomes (24). Instead, the pathogen hijacks proteins and membrane organelles from the host cytosol to slowly transform its Legionella-containing vacuole (LCV) into a specialized membrane compartment with morphological features reminiscent of rough endoplasmic reticulum (ER) (23, 44, 47). Within this protective niche, L. pneumophila replicates to high numbers, lyses the host cell, and infects neighboring macrophages.
Replication vacuole formation depends on the ability of the pathogen to divert host cell vesicle flow to its LCV. For that, L. pneumophila delivers over 300 effector proteins across the vacuolar membrane into the host cytosol, where they manipulate host signaling processes (17). L. pneumophila mutants with a defective Dot/Icm system are unsuccessful in transforming their vacuole into a camouflaged replication compartment and are trafficked along the endolysosomal route for degradation, underscoring the importance of effector proteins for bacterial virulence (2, 5, 7, 38, 40, 44, 47, 51). Predicting their molecular activity has proven challenging because many effector proteins are unique to the genus Legionella and lack significant homology to known proteins.
Consistent with the establishment of a membrane organelle reminiscent of the ER, marker proteins of this host compartment, such as calnexin, BIP, and Sec22, were found to localize to LCVs during infection (13, 27, 44). Another host protein hijacked by L. pneumophila during infection is Rab1, a small GTPase that regulates transport of vesicular cargo from the ER to the Golgi compartment (36, 37, 48, 52). Rab1 cycles between an active GTP-bound and an inactive GDP-bound conformation, the latter being associated with the chaperone GDP dissociation inhibitor (GDI) in the cytosol. Transition between these two conformations is controlled by upstream regulatory factors. GDP/GTP exchange factors (GEFs) activate Rab1 by replacing GDP against GTP. In its active conformation, GTP-Rab1 is membrane associated and interacts with downstream effectors involved in vesicle tethering, docking, and fusion. GTPase-activating proteins (GAPs) stimulate the intrinsic GTP hydrolysis activity of Rab1, thereby converting the GTPase back into its inactive GDP-bound form, which is recognized and extracted from the membrane by GDI.
We and others recently showed that several L. pneumophila Dot/Icm-translocated substrates are molecular mimics of host regulatory factors and specifically target Rab1 to modulate its activity (25, 30, 31, 34). Recruitment of Rab1 to the LCV membrane and activation of Rab1 require SidM (DrrA), an effector protein with multiple activities. SidM exhibits GDI displacement factor (GDF) activity as well as GEF activity, resulting in the generation of GDI-free active GTP-Rab1. Additionally, SidM possesses adenylylation (AMPylation) activity, covalently attaching AMP to tyrosine 77 of GTP-Rab1. Active AMPylated Rab1 is released by SidM and thought to mediate tethering of early secretory vesicles to the LCV, thereby promoting vacuolar transformation. The efficiency of this process is believed to be enhanced by AMPylation because Rab1 in the AMP-modified form cannot be inactivated by the L. pneumophila effector LepB and remains active for an extended period of time (33). When replication vacuole formation has progressed sufficiently, Rab1 is no longer needed for vesicle recruitment and it is de-AMPylated by the effector protein SidD (35, 46). Once AMP-free, GTP hydrolysis in Rab1 is triggered by the GAP LepB and Rab1 is removed from the LCV membrane by GDI.
During infection of host cells by L. pneumophila, Rab1 can be the substrate of yet another covalent modification, phosphocholination, which is catalyzed by AnkX (also known as LegA8) (32). This event involves the addition of a phosphocholine moiety to a serine residue in the switch II region of Rab1 adjacent to the tyrosine that is the target of AMPylation. However, it is not yet clear when and where this modification takes place and what its impact, if any, is on the activity of Rab1.
While the effects of SidM, SidD, and LepB on Rab1 have been well studied, the role of the L. pneumophila effector protein LidA during Rab1 manipulation is unclear. LidA was initially identified as a “gatekeeper” for the L. pneumophila Dot/Icm type IV secretion system (T4SS) (12). LidA is translocated into the host cell throughout the infection cycle of L. pneumophila (14). Indirect immunofluorescence studies showed that early during infection, LidA is associated with the cytosolic surface of the vacuole (12), whereas at later times, LidA is also dispersed throughout the infected cell and associated with membrane compartments of unknown identity (14). Subsequent studies revealed that, after delivery into host cells, LidA interacts with Rab1 and other Rab GTPases (31). In contrast to other Rab ligands described to date, LidA binds the GTP-bound active and the GDP-bound inactive form of Rab1 with almost equal affinity (31). However, unlike SidM and LepB, which alter the guanine nucleotide binding state of Rab1, LidA does not exhibit any detectable GEF or GAP activity toward this GTPase. In addition, despite the fact that AMPylation of Rab1 blocks its inactivation by L. pneumophila LepB and interaction with the host cell ligand MICAL-3, LidA is still able to interact with AMPylated Rab1 (33).
LidA seems to fulfill a critical yet unknown function during L. pneumophila infection. Deletion mutants of lidA show a kinetic defect in Rab1 recruitment, consistent with the hypothesis that LidA promotes Rab1 accumulation on the LCV by enhancing the process of SidM-mediated Rab1 recruitment and/or by antagonizing Rab1 removal from LCVs. In the present study, we have performed a detailed analysis of the effect of LidA on Rab1 function and propose a model that predicts a role for LidA in Rab1-mediated vesicle tethering to LCVs.
MATERIALS AND METHODS
Strains, media, and plasmids.
L. pneumophila strains were grown and maintained in medium containing thymidine as previously described (18, 20). L. pneumophila strain Lp02 (thyA hsdR rpsL) is a thymidine auxotroph derivative of Philadelphia-1. The strain Lp02ΔlidA was a kind gift from Zhao-Qing Luo (Purdue University, IN), and Lp02ΔsidM harboring an in-frame deletion in sidM was described previously (16, 29). The Lp02ΔlidAΔsidM mutant was generated in the ΔlidA mutant background by allelic exchange using plasmid pSR47S-sidM as described before (16). Plasmids were introduced into L. pneumophila by natural transformation (43).
Plasmids for production of recombinant tagged proteins in Escherichia coli are listed in Table 1. Antibodies were purchased from Santa Cruz Biotechnology (Rab1a), Abcam (Giantin, GM130, and p115), BellBrook Labs (AMP), and Sigma (TEPC-15). SidM- and LidA-directed antibodies were described previously (12, 31).
Table 1.
Microbial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| L. pneumophila strains | ||
| Lp02 | Philadelphia-1, serogroup 1, salt sensitive, restriction deficient, thymidine auxotroph; Smr | 6 |
| Lp02ΔlidA | Philadelphia-1, serogroup 1, salt sensitive, restriction deficient, thymidine auxotroph, lacking lidA; Smr | Zhao-Qing Luo |
| Lp02ΔlidAΔsidM | Philadelphia-1, serogroup 1, salt sensitive, restriction deficient, thymidine auxotroph, lacking lidA and sidM; Smr | This study |
| E. coli strains | ||
| GC5 | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1λ−tonA | Genesee |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm(DE3) | Novagen |
| Plasmids | ||
| pGEX-6p-1 | GE Healthcare | |
| pGEX-6p-1-lidA | pGEX6p1 with L. pneumophila lidA (aa 1–729); Ampr | This study |
| pGEX-6p-1-N-lidA | pGEX6p1 with L. pneumophila lidA fragment (aa 1–189); Ampr | This study |
| pGEX-6p-1-M-lidA | pGEX6p1 with L. pneumophila lidA fragment (aa 190–600); Ampr | This study |
| pGEX-6p-1-R-lidA | pGEX6p1 with L. pneumophila lidA fragment (aa 190–400); Ampr | This study |
| pGEX-6p-1-C-lidA | pGEX6p1 with L. pneumophila lidA fragment (aa 601–729); Ampr | This study |
| pGEX-6p-1-sidM | pGEX6p1 with L. pneumophila sidM flanking fragments; Ampr | 31 |
| pGEX-6p-1-rab1a | pGEX6p1 with human rab1a; Ampr | 31 |
| pGEX-6p-1-rab1aQ70L | pGEX6p1 with human rab1a; Ampr | 31 |
| pGEX-6p-1-rab1aS25N | pGEX6p1 with Human rab1a; Ampr | 31 |
| pSR47S-sidM | pSR47S with L. pneumophila sidM flanking regions; Ampr | 31 |
| pQE80L-TBC1D201–364 | pQE80L with human TBC1D20 (Thr364stop); Ampr | 35 |
| pQE80L-lepB1-1232 | pQE80L with L. pneumophila lepB (Trp1232stop); Ampr | 35 |
| pDEST17-ankX | pDEST17 with L. pneumophila ankX; Ampr | This study |
| pQE80L-LidA | pQE80L with L. pneumophila lidA; Ampr | This study |
Abbreviations: Ampr, ampicillin resistance; Smr, streptomycin resistance.
The plasmids for production and purification of recombinant LidA, SidM, Rab1, Rab1 variants, LepB1232, and TBC1D20364 in E. coli were described before (12, 31, 35) and are listed and described in Table 1. Plasmids for production of LidA variants were generated by subcloning PCR fragments into pGEX-6P-1 (GE Healthcare) using oligonucleotides Fw-N (5′-GAAGAATTCATGGCAAAAGATAACAAATCACATCAAG-3′) and Rv-N (5′-CTCCTCGAGTTAGGATGAAGTGGACTGTTGGCTTG-3′) to amplify the sequence encoding the N-terminal fragment, Fw-M (5′-GAAGAATTCATGACTTCGCAAGCTGATAAAGAAATTC-3′) and Rv-M (5′-CTCCTCGAGTTATTCAGTATCTAAAGTTGGTTGGTTG-3′) to amplify the sequence encoding the middle fragment, Fw-M and Rv-R 1-2 (5′-CTCCTCGAGTTATTCCAAACTAAGCTTATGAATTAGAGC-3′) to amplify the sequence encoding the R 1-2 fragment, and Fw-C (5′-GAAGAATTCATGTCACCTGTCTTAACACCTTCTGGAG-3′) and Rv-C (5′-CTCCTCGAGTTATGATGTCTTGAATGGAGATAAAGAC-3′) to amplify the sequence encoding the C-terminal fragment. Recombinant His-tagged LidA was generated by cloning the lidA gene into the pQE80L plasmid. For production of recombinant His-tagged AnkX, the ankX gene was cloned into the Gateway pDEST17 vector (Invitrogen) from pDONOR221-AnkX (a kind gift from Ralph Isberg, Tufts University) using the LR clonase (Invitrogen). For production of recombinant Lem3, a DNA fragment containing the lpg0696 open reading frame (lem3) was amplified by PCR using the forward primer MMO105 containing an SgfI restriction site (5′-GATCGCGATCGCCATGATGAAGTTACGCTATATTATTAATG-3′) and the reverse primer MMO106 containing a PmeI restriction site (5′-GATCGTTTAAACTTTCAATTCATTTTTATTTTCTATTTCACTATCCAA-3′). The PCR fragment was digested with the specified enzymes and cloned into the respective sites in pFN22K HaloTagCMVd1 Flexi vector (Promega) to generate an N-terminal HaloTag fusion.
Production and purification of recombinant proteins.
LidA and its variants (N-LidA, M-LidA, R-LidA, and C-LidA), SidM, Rab1a, Rab1aS25N, and Rab1aQ70L were produced as glutathione S-transferase (GST) fusion proteins in E. coli BL21(DE3) (Stratagene) and purified as described before (12, 31). LepB1232 (amino acids [aa] 1 to 1232) and TBC1D20364 (aa 1 to 364) lacking the transmembrane region, as well as AnkX and LidA, were produced as 6×His fusions and were purified using TALON metal affinity resin as previously described (35).
Recombinant HaloTag-Lem3 was produced in the Single Step (KRX) competent E. coli strain (Promega), and Lem3 was purified using the HaloTag protein purification system according to the manufacturer's instructions. Briefly, E. coli cells producing HaloTag-Lem3 were harvested and resuspended in protein purification buffer (50 mM HEPES, 150 mM NaCl, pH 7.5) followed by lysis using a Microfluidics M-110P microfluidizer. The cell lysate was centrifuged at 25,000 × g for 20 min, and the supernatant was incubated with preequilibrated HaloLink resin for 2 h at 4°C. The resin was then washed a total of 3 times using the protein purification buffer, and Lem3 was cleaved off the resin using tobacco etch virus (TEV) protease (Promega) for 2 h at 4°C. HisLink resin (Promega) was used to remove the TEV protease from the supernatant, and its removal was verified by SDS-PAGE.
GTP hydrolysis assays.
Rab1a was loaded with [γ-32P]GTP by first extracting MgCl2 with 5 mM EDTA for 5 min at room temperature to release the Rab1a-bound nucleotide. Nucleotide-free Rab1a (12 μM) was then loaded with 20 nM [γ-32P]GTP in phosphate-buffered saline (PBS) buffer for 5 min at room temperature, followed by the addition of 20 mM MgCl2. Rab1a (2 μM) was incubated at room temperature with either 200 nM LepB1232 or 180 nM TBC1D20364. For samples containing LidA, the GAPs were added to the reaction mixture after preincubation of LidA with Rab1 for 15 min at room temperature. At the time intervals indicated in Fig. 1, samples were transferred to PBS buffer containing 20 mM MgCl2 (PBS-M) and immediately applied to a prewetted nitrocellulose filter (0.45 μm; Millipore) on a vacuum filtration manifold (model 1225; Millipore). Filters were washed with 2 ml of PBS-M. Nitrocellulose filters were transferred to vials containing scintillation liquid, and radioactivity was measured by using a Beckman LS 6500 liquid scintillation counter.
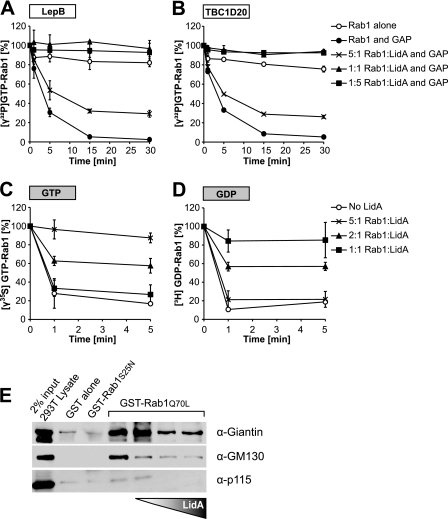
Fig 1.
LidA maintains Rab1 in an active conformation. (A, B) LidA prevents inactivation of GTP-Rab1 by LepB1232 and TBC1D20364. Rab1 (2 μM) loaded with [γ32P]GTP was preincubated with LidA at the indicated molar ratios or with buffer as negative control, followed by the addition of LepB1232 (A) or TBC1D20364 (B). Rab1-[γ32P]GTP levels were monitored over time by a filter-binding assay. Data are means ± standard deviations from three independent experiments. (C, D) LidA protects Rab1 from guanosine nucleotide extraction by EDTA. Results shown are from a guanosine nucleotide extraction assay of Rab1 loaded with [γ35S]GTP (C) or [3H]GDP (D) and preincubated for 1 min with LidA in the molar ratios indicated. The release of radiolabeled guanosine nucleotides from Rab1 was stimulated by the addition of EDTA (20 mM final concentration), and the loss of radioactive nucleotide from Rab1 was monitored by a filter-binding assay. Data are means ± standard deviations from three independent experiments. (E) LidA prevents interaction of Rab1 with downstream effectors. Magnetic beads were coated with GST, GST-Rab1aS25N, or GST-Rab1aQ70L and incubated with postnuclear supernatant (PNS) from 293T cells. Where indicated, GST-Rab1aQ70L was preincubated with increasing amounts of LidA prior to incubation with the 293T cell PNS.
Guanine nucleotide extraction assay.
For nucleotide extraction assays, 50 pmol nucleotide-free Rab1 (see above) was incubated for 2 h in loading buffer containing 1 nmol [3H]GDP or [γ-35S]GTP as indicated. The loading reaction was stopped by the addition of MgCl2 (5 mM final concentration). Rab1 loaded with [3H]GDP or [γ-35S]GTP was diluted into loading buffer containing 5 mM MgCl2 in the presence or absence of LidA (in molar ratios as indicated in the description of each experiment) or equimolar amounts of the LidA variants. Nucleotide extraction was started by the addition of EDTA (final concentration, 20 mM). Samples were removed at the time points indicated in Fig. 1 and analyzed by scintillation counting as described for GTP hydrolysis.
Pulldown of Rab1 effector proteins in the presence of LidA.
Purified GST, GST-Rab1aS25N, or GST-Rab1aQ70L proteins (30 μg) were used to coat 5 × 107 magnetic beads (Dynabeads Epoxy M270; Invitrogen) overnight at 4°C according to the manufacturer's instructions. The protein-coated beads were then incubated with 400 μg of postnuclear supernatant (PNS) for 2 h at 4°C. The PNS was generated by mechanically breaking 293T cells in PBS buffer containing 1 mM MgCl2 and 1 mM β-mercaptoethanol (PBS-MM) using a Dounce homogenizer; the cell lysate was centrifuged at 3,000 × g and the supernatant was used for pulldown studies. After incubation with PNS, the beads were washed six times with PBS-MM. Proteins bound to the beads were analyzed by Western blotting using the antibodies specified below.
In vitro AMPylation assays in the presence of LidA.
Rab1a (5 μM) was preincubated in buffer containing 20 mM HEPES, 1 mM MgCl2, 1 mM ATP, and 5 μM GTP for 15 min at room temperature with 0.5, 1, 2, and 5 μM LidA or buffer as control. AMPylation was initiated by the addition of 250 nM SidM, and the reaction was allowed to continue for a total of 90 min. The reaction was stopped after 90 min by adding SDS sample buffer and boiling the sample for 5 min. The samples were then separated on a 4 to 15% SDS–PAGE gel (Bio-Rad) and transferred to a nitrocellulose membrane (iBlot; Invitrogen) for immunoblot analysis (Fast Western; Pierce) using polyclonal AMP antibody to detect Rab1-AMP. The intensities of the protein bands detected by Western blot were quantified using ImageJ software (http://rsb.info.nih.gov/ij; National Institutes of Health, Bethesda, MD).
In vitro de-AMPylation assays in the presence of LidA.
Rab1a (6 μM) preloaded with GTP was first AMPylated in a master reaction catalyzed by 1.5 × 108 SidM-coated beads (Dynabeads M-270 Epoxy, Invitrogen) in PBS-M buffer in the presence of 50 nM [α32P]ATP. The reaction was allowed to proceed for 3 h at room temperature to achieve complete AMPylation of Rab1, after which SidM-coated beads were removed. Subsequently, 2 μM Rab1a-[32P]AMP was incubated with 0.5, 1, or 2 μM LidA or buffer as control for 15 min at room temperature to allow binding of LidA to AMPylated Rab1. De-AMPylation was initiated by the addition of 100 nM purified GST-SidD to each of the four samples. Loss of [32P]AMP from Rab1 was monitored by nitrocellulose filter-binding assays as described for GTP hydrolysis.
Protein-protein binding assays.
For pulldown experiments to identify the Rab1 binding domain, purified recombinant LidA or LidA variants (N-LidA, M-LidA, and C-LidA) were immobilized on Affigel beads (Bio-Rad) according to the manufacturer's recommendations. Beads were incubated for 2 h at 4°C in PBS containing a molar excess of either dominant active Rab1aQ70L or dominant inactive Rab1aS25N. Beads were washed 5 times with cold PBS and resuspended in SDS sample buffer. Rab1 bound to bead-immobilized proteins was detected by Western blotting.
For pulldown of covalently modified Rab1, glutathione magnetic beads (Pierce) were coated with GST or GST-LidA and incubated with purified AMPylated Rab1a (Rab1-AMP) or phosphocholinated Rab1 (Rab1-PC) generated as described below. Rab1a or SidM was used as the positive or negative control, respectively.
Protein-lipid overlay assay.
Phosphoinositide binding of full-length LidA and the four truncated variants of untagged LidA (N-LidA, R-LidA, M-LidA, and C-LidA) were tested in vitro using commercially available PIP Strips (Echelon), which are nitrocellulose membranes spotted with the phosphoinositides specified in Fig. 3. The membranes were blocked with 2% fat-free milk in TBST (50 mM Tris, 150 mM NaCl, 0.1% Tween 20 [vol/vol], pH 7.4) for 1 h at room temperature prior to incubation with 4 μg of full-length LidA or the LidA variants for 1 h at room temperature. The membranes were then washed with TBST and incubated with LidA-directed antibody for 1 h at room temperature, followed by a 1-h incubation with a peroxidase-labeled secondary antibody in TBST. After a final wash, proteins bound to the membrane were detected by chemiluminescence.
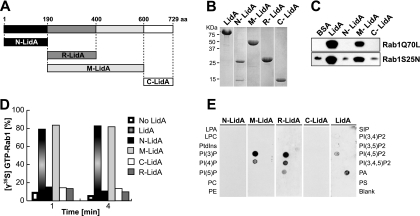
Fig 3.
LidA binds to Rab1 and phosphoinositides through its central coiled-coil region. (A) Schematic of LidA regions used for binding studies. (B) Purified full-length LidA and LidA variants. (C) Binding to Rab1 is mediated by the central region of LidA. Pulldown of constitutively active Rab1aQ70L or inactive Rab1aS25N by purified LidA or LidA variants (N-LidA, M-LidA, and C-LidA) immobilized on agarose beads was performed. Beads were incubated for 2 h at 4°C in the presence of a molar excess of either form of Rab1, and Rab1 bound to bead-immobilized proteins was detected by Western blotting. (D) The central region of LidA protects GTP-Rab1 from EDTA-mediated guanosine extraction. Rab1 was loaded with [γ35S]GTP and preincubated with full-length LidA or a truncated version of LidA for 1 min at room temperature. The release of radiolabeled guanosine nucleotides from Rab1 was stimulated by the addition of EDTA (20 mM final concentration). Loss of radioactive nucleotide was monitored by a filter-binding assay. Data are means ± standard deviations from three independent experiments. (E) The central region of LidA mediates binding to phosphoinositides. PIP Strip assays of full-length LidA or truncated versions of LidA (LPA, lysophosphatidic acid; LPC, lysophosphocholine; PtdIns, phosphatidylinositol; PE, phosphatidylethanolamine; PC, phosphatidylcholine; SIP, sphingosine-1-phosphate; P, phosphate; P2, biphosphate; P3, triphosphate; PA, phosphatidic acid; PS, phosphatidylserine). LidA binding was detected using anti-LidA antibody.
In vitro phosphocholination assay in the presence of LidA.
Purified Rab1a (5 μM) was preincubated with buffer or increasing amounts of LidA (0.5, 1, 2, or 5 μM) for 15 min at room temperature in phosphocholination buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM CDP-choline, 1 mM MgCl2, and 1 mM ATP. The phosphocholination reaction was initiated by the addition of His-AnkX (250 nM) as specified in Fig. 4A. The reaction was stopped after 3 h by boiling in SDS sample buffer for 5 min. Proteins were separated on a 4 to 15% SDS–PAGE gel (Bio-Rad) and transferred to a nitrocellulose membrane (iBlot; Invitrogen) for immunoblot analysis (Fast Western; Pierce) using TEPC-15 antibody to detect phosphocholinated Rab1 (Rab1-PC). The intensity of protein bands detected by Western blotting was quantified using ImageJ software.
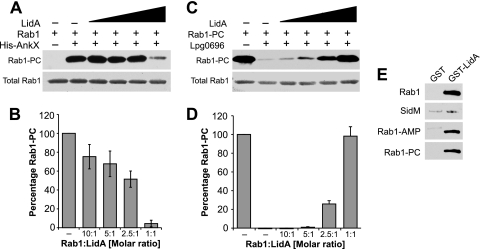
Fig 4.
LidA interferes with phosphocholination and dephosphocholination of Rab1. (A, B) LidA inhibits phosphocholination of Rab1. (A) Rab1a was preincubated with buffer or LidA at 10:1, 5:1, 2.5:1, and 1:1 molar ratios, followed by the addition of His-AnkX where indicated to initiate phosphocholination. After 3 h, Rab1-PC was detected by Western blotting using phosphocholine antibody (top); Ponceau staining shows total amount of Rab1 present on the blot (bottom). The results shown are representative of three independent experiments. (B) Quantification of Rab1-PC at 2 h after initiation of the phosphocholination reaction of Rab1 preincubated with LidA at the indicated molar ratios or with buffer as control. The intensities of the bands were determined relative to the signal obtained in the absence of LidA, which was arbitrarily set as 100%. The bar graph shows means ± standard deviations of relative band intensities obtained from three independent experiments. (C, D) LidA inhibits dephosphocholination of Rab1-PC. (C) Rab1a-PC was preincubated with buffer or His-LidA at 10:1, 5:1, 2.5:1, and 1:1 molar ratios, followed by the addition of Lem3 (Lpg0696) where indicated to initiate dephosphocholination. After 2 h, Rab1-PC was detected by Western blotting using phosphocholine antibody (top); Ponceau staining shows total amount of Rab1 present on the blot (bottom). The results shown are representative of three independent experiments. (D) Quantification of Rab1-PC at 2 h after initiation of the phosphocholination reaction of Rab1 preincubated with His-LidA at the indicated molar ratios or with buffer as control. (E) LidA binds Rab1-PC. Pulldown experiment with purified protein using beads coated with GST or GST-LidA shows binding of LidA to phosphocholinated Rab1 (Rab1-PC) and two positive controls, Rab1 and AMPylated Rab1 (Rab1-AMP) but not to SidM. These results are representative of two independent experiments.
In vitro AMPylation or phosphocholination reaction of Rab1a and purification of covalently modified Rab1a.
Rab1a was AMPylated in vitro, and Rab1-AMP was purified by gel filtration as previously described (35). Rab1a (25 μM) was phosphocholinated at room temperature for 4 h in the presence of His-AnkX (0.25 μM) in a phosphocholination buffer (described above). The reaction mixture was then incubated with 60 μl of HisLink beads (Promega) to remove His-AnkX before purification by gel filtration on a HiLoad 16/60 Superdex 75-pg column (GE Healthcare) at 4°C. Fractions containing Rab1-PC in PBS-MM were pooled, concentrated, and stored at −80°C.
In vitro dephosphocholination assay in the presence of LidA.
Rab1-PC (5 μM), purified as described above, was preincubated with buffer or increasing amounts of LidA (0.5, 1, 2, and 5 μM) for 15 min at room temperature in PBS-M buffer, followed by the addition of 250 nM Lem3. Phosphocholinated Rab1 was detected by Western blotting as described above for the phosphocholination assay.
Electron microscopy of U937 cells infected with wild type or mutants of L. pneumophila.
U937 cells were grown in RPMI 1640 supplemented with 10% fetal bovine serum in a 5% CO2 humidified atmosphere at 37°C. The cells were activated with phorbol 12-myristate 13-acetate for 48 h in a culture flask. Differentiated U937 cells were replated in 24-well tissue culture dishes containing 12-mm coverslips at a density of 5 × 105 cells per well prior to infection with L. pneumophila. The cells were challenged with post-exponential-phase L. pneumophila wild type or the ΔlidA or ΔlidA ΔsidM mutant at a multiplicity of infection of 5 in culture medium containing thymidine (100 μg/ml). At 30 min, the extracellular bacteria were washed off with warm culture medium, after which fresh medium plus thymidine was added to the wells and incubation continued until the incubation time postinfection totaled 2 h. The cells were fixed with 2% glutaraldehyde–2% formaldehyde in 0.1 M cacodylate buffer, pH 7.2, followed by postfixation in 1% osmium tetroxide–1.5% potassium ferrocyanide. Upon dehydration and embedding in EMBed-812 (EM Science, Horsham, PA), the coverslips were removed by hydrofluoric acid, cells were thin sectioned parallel to the glass, and the sections were stained with uranyl acetate and lead citrate. The samples were viewed on a Tecnai 20 transmission electron microscope (FEI, Hillsboro, OR) at 80 kV acceleration voltage.
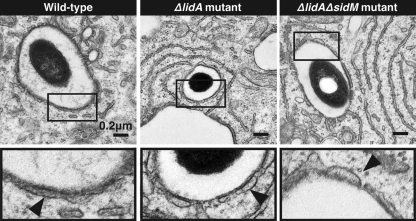
The presence of ER vesicle LCVs was quantified from electron micrographs of host cells infected with L. pneumophila for 30 min (wild type, 10 vacuoles;ΔlidA mutant, 24 vacuoles; and ΔlidA ΔsidM mutant, 9 vacuoles) and 2 h (wild type, 11 vacuoles;ΔlidA, 29 vacuoles; and ΔlidA ΔsidM mutant, 20 vacuoles). Vacuoles with a minimum of 50% surface coverage were considered positive for vesicle recruitment.
RESULTS
LidA prevents inactivation of GTP-Rab1 by Rab1 GAPs.
LidA plays a supportive role in Rab1 recruitment to the LCV during infection, but it is not clear how LidA contributes to enhancing the presence of Rab1 on the vacuole. Prior studies have shown that LidA interacts with high affinity with Rab1 in its GTP-bound form and that it binds GDP-Rab1 with slightly lower affinity (31). However, to date there is no evidence of LidA being either a GEF or a GAP for Rab1 (31). Given its tight binding to GTP-Rab1 in vitro and its effect on Rab1 recruitment in vivo, we wanted to test whether interaction of LidA with GTP-Rab1 prevents inactivation by the L. pneumophila-encoded Rab1-GAP LepB and the mammalian Rab1-GAP TBC1D20. Rab1 loaded with radioactive [γ32P]GTP was preincubated with increasing amounts of LidA, and GTP hydrolysis was initiated by the addition of truncated versions of LepB1232 or TBC1D20354 that lack the transmembrane domain. In the absence of LidA, the two GAPs efficiently stimulated GTP hydrolysis, whereas LidA present in an equimolar or larger amount completely blocked GTP hydrolysis by Rab1 in the presence of either type of GAP. At a subequimolar concentration (2-fold excess of Rab1), LidA prevented GTP hydrolysis in approximately half of the Rab1 molecules, consistent with a 1:1 stoichiometry of the LidA-Rab1 complex (Fig. 1A and B). Thus, the presence of LidA negatively affected GTP hydrolysis triggered by either GAP, probably by directly binding Rab1.
LidA prevents dissociation of guanosine nucleotides from Rab1.
To further investigate how LidA exerts its protective effect over Rab1, we set out to explore the possibility that LidA binding to Rab1 sterically hinders GAP access to the nucleotide binding pocket. In previous analyses, we noted that spontaneous nucleotide dissociation from Rab1 appeared to be reduced in the presence of LidA, which led us to further analyze a possible effect of LidA on the guanosine nucleotide-Rab1 complex. Magnesium (Mg2+) is an essential cofactor for high-affinity binding of guanosine nucleotides to GTPases (11), whereas EDTA causes nucleotide dissociation by chelating Mg2+ ions (9, 26). Rab1 loaded with [γ-35S]GTP, a nonhydrolyzable GTP analog, was preincubated with LidA in different molar ratios, followed by the addition of EDTA. In the absence of LidA, radiolabel was rapidly lost from Rab1, indicating that [γ-35S]GTP was released from Rab1 after Mg2+ extraction by EDTA (Fig. 1C). In contrast, LidA efficiently prevented nucleotide release from Rab1 in the presence of EDTA. No interaction between LidA and or any other radiolabeled guanosine nucleotide was detectable (data not shown). The inhibitory effect of LidA on [γ-35S]GTP dissociation from Rab1 was proportional to the concentration of LidA, with the greatest protection at an equimolar ratio of LidA to Rab1. Similar results were obtained in EDTA extraction assays using Rab1 loaded with radioactive [3H]GDP (Fig. 1D). Thus, LidA binds Rab1 in a 1:1 ratio and blocks guanine nucleotide dissociation from Rab1.
LidA prevents interaction of active Rab1 with its downstream effectors.
Since LidA not only stabilized GTP-Rab1 but also prevented Rab1 inactivation by GAPs, we hypothesized that LidA may keep Rab1 in an active conformation to allow it to bind its downstream ligands. To test this hypothesis, we performed a pulldown experiment from 293T cell lysate in which Rab1aS25N (constitutively inactive) or Rab1aQ70L (constitutively active) bound to beads was incubated with increasing amounts of LidA to assess its effect on the binding of Rab1 interacting partners (Fig. 1E). In the absence of LidA, Rab1aQ70L interacted with tethering proteins p115, giantin, and GM130 while Rab1aS25N did not, consistent with earlier studies (1, 4, 50). However, in the presence of LidA, interaction with these three Rab1 effectors was disrupted in a concentration-dependent manner (Fig. 1E). Contrary to our hypothesis, these results seem to support an inhibitory role of LidA on Rab1 interaction with the downstream effectors tested.
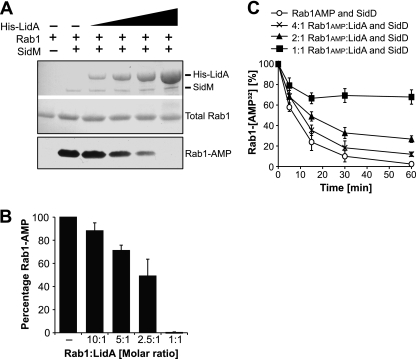
LidA binding to Rab1 inhibits AMPylation and de-AMPylation.
SidM preferentially catalyzes AMPylation of the GTP-bound form of Rab1 (33). Given that LidA binding keeps Rab1 in an active conformation even in the presence of GAPs, we wanted to determine if LidA would still allow Rab1 AMPylation by SidM. In the absence of LidA, Rab1 was efficiently AMPylated by SidM (Fig. 2A and B). When LidA was preincubated with Rab1, it inhibited Rab1 AMPylation in a concentration-dependent manner. After 90 min, the most severe attenuation of AMPylation was detectable at an equimolar amount of LidA relative to the amount of Rab1, indicating that LidA binding to Rab1 interfered with the posttranslational modification of this GTPase by SidM (Fig. 2A and B). We also tested the effect of LidA on the reverse reaction catalyzed by SidD. Rab1 AMPylated by SidM was preincubated with increasing amounts of LidA followed by the addition of SidD. When AMPylated Rab1 and LidA were preincubated at a 4:1 molar ratio, loss of [32P]AMP was reduced in about 25% of [32P]AMP-Rab1, whereas de-AMPylation was completely inhibited when the two proteins were present at equimolar amounts (Fig. 2C). A 2:1 ratio of Rab1 to LidA resulted in the protection of approximately half of the Rab1 molecules from de-AMPylation by SidD. These results show that LidA interfered with both AMPylation and de-AMPylation of Rab1, probably by binding to Rab1 and preventing access of SidM as well as SidD.
Fig 2.
LidA inhibits AMPylation and de-AMPylation of Rab1. For both assays, data are means ± standard deviations from three independent experiments. (A) LidA interfered with AMPylation of Rab1 by SidM. Rab1a was preincubated with His-LidA at a 10:1. 5:1, 2.5:1, or 1:1 molar ratio or with buffer as control, followed by the addition of SidM to initiate AMPylation. After 90 min, the accumulation of Rab1-AMP was determined by Western blotting. Ponceau staining shows total amounts of His-LidA, SidM, and Rab1 present on the blot (top); Rab1-AMP signal was detected using an AMP antibody (bottom). (B) Quantification of Rab1-AMP at 2 h after initiation of the AMPylation reaction of Rab1 preincubated with His-LidA at the indicated molar ratios or with buffer as control. The intensities of the bands were determined relative to the signal obtained in the absence of His-LidA, which was arbitrarily set as 100%. (B) LidA blocks de-AMPylation of Rab1-AMP by SidD. Rab1a-[32P]AMP was incubated with LidA at the indicated molar ratios or with buffer as control. De-AMPylation was initiated by the addition of GST-SidD to each of the four samples. Loss of [32P]AMP from Rab1 was monitored by filter-binding assays.
Identification and functional analysis of LidA domains.
Many bacterial effector proteins characterized so far are composed of multiple domains that are functionally autonomous. For example, SidM binds phosphatidylinositol 4-phosphate [PI(4)P] through its C-terminal domain to mediate its attachment to the LCV, and it catalyzes GDP/GTP exchange of Rab1 through its central part and Rab1 AMPylation through its N-terminal part (21). Because LidA also binds Rab1 and phosphoinositides, we wanted to identify the domains that are responsible for binding these molecules. We initially divided LidA into three regions: an N-terminal part (N-LidA, amino acids [aa] 1 to 189), a middle region predicted to be rich in coiled-coil structures (M-LidA, aa 190 to 600), and a C-terminal part (C-LidA, aa 601 to 729) (Fig. 3A). The borders of the LidA regions were chosen based on the proteolytic degradation pattern we observed during long-term storage of LidA at 4°C, which resulted in the accumulation of an approximately 48-kDa degradation fragment that was identified as the central region of LidA (data not shown). The purified truncated LidA variants were water soluble and resistant to proteolytic degradation during production in E. coli, suggesting that they in fact represent independent domains of the full-length protein (Fig. 3B).
The central coiled-coil region of LidA mediates Rab1 binding.
To identify the region of LidA that mediates interaction with Rab1, an in vitro binding assay was performed using purified proteins. LidA variants immobilized to agarose beads were incubated with Rab1 mutants locked either in the active (Rab1aQ70L) or in the inactive form (Rab1aS25N). The amount of Rab1 mutant protein retained by the beads was determined by Western blot analysis using Rab1-specific antibody. Both Rab1 variants efficiently interacted with beads coated with M-LidA or full-length LidA, respectively, whereas N-LidA and C-LidA showed Rab1 binding similar to the level obtained with BSA-coated control beads (Fig. 3C). Thus, the central coiled-coil region of LidA is sufficient for Rab1 binding, and any further truncation of this region abrogated interaction with Rab1 (data not shown).
Though the N- and C-terminal regions are dispensable for Rab1 binding, we tested their involvement in the other activities described above for full-length LidA. As full-length LidA protects Rab1 from nucleotide extraction by EDTA, we tested the different LidA fragments for their effect on Rab1-guanine nucleotide complexes in an in vitro nucleotide extraction assay. Purified Rab1 loaded with radiolabeled [γ35S]GTP was incubated with an equimolar amount of either full-length LidA or the truncated LidA variants prior to the addition of EDTA. The amount of γ35S-GTP–Rab1 present in the experiment was monitored over time (Fig. 3D). In the absence of LidA, [γ35S]GTP was rapidly extracted from Rab1 by EDTA, leading to a quick drop in radioactivity. In contrast, full-length LidA and M-LidA efficiently prevented [γ35S]GTP extraction from Rab1, whereas N-LidA and C-LidA showed no effect on nucleotide extraction from Rab1 (Fig. 3D). Therefore, the central coiled-coil region of LidA is sufficient to prevent EDTA-mediated nucleotide extraction, without a requirement for the N- or C-terminal domain.
The central region of LidA mediates phosphoinositide binding.
LidA was previously shown to bind PI(3)P and, less strongly, PI(4)P and PI(5)P. To identify the LidA region that is responsible for binding these phosphoinositides, we performed a protein-lipid overlay assay. In addition to the three regions tested as described above for Rab1 binding (N-LidA, M-LidA, and C-LidA), we included a subregion spanning the amino-terminal 200 amino acids of the M-LidA fragment, called R1-2-LidA (aa 190 to 400) (14). This region contained two repeats with weak homology to spectrin repeats, which are known to bind phosphoinositides (39). Of the four LidA fragments tested, M-LidA and R1-2-LidA showed strong binding to PI(3)P and reduced binding to PI(4)P (Fig. 3E). N-LidA and C-LidA did not bind any phosphoinositides. Thus, the central coiled-coil region of LidA is responsible not only for Rab1 binding but also for binding to phosphoinositides. Full-length LidA showed variable binding to phosphatidic acid depending on the antibody used to detect the binding. Overall, LidA bound to phosphatidic acid in three out of six independent experiments, whereas LidA bound to PI(3)P in each of our phosphoinositide binding experiments.
LidA interferes with phosphocholination and dephosphocholination of Rab1.
A recent study found that Rab1 can be phosphocholinated by AnkX during infection of host cells by L. pneumophila (32). Thus, we evaluated LidA's effect on this posttranslational modification. Rab1 was preincubated with LidA at different molar ratios, and AnkX was added to initiate phosphocholination in the presence of CDP-choline as substrate. We found that phosphocholination of Rab1 by AnkX was increasingly impaired when LidA was present at a 10:1, 5:1, or 2.5:1 molar ratio of Rab1 to LidA and that the reaction was almost completely inhibited by an equimolar amount of LidA (Fig. 4A and B).
We subsequently analyzed the effect of LidA on phosphocholine removal from Rab1 by L. pneumophila Lem3, a previously uncharacterized L. pneumophila effector that catalyzes the removal of phosphocholine from Rab1a (45; also M. Machner, unpublished data). Similarly to de-AMPylation, dephosphocholination of Rab1 by Lem3 was efficiently prevented by increasing amounts of LidA (Fig. 4C and D). Thus, like Rab1 AMPylation by SidM and de-AMPylation by SidD, phosphocholination by AnkX and dephosphocholination by Lem3 were attenuated by LidA binding to Rab1. We also found that phosphocholination of Rab1 did not affect its binding to LidA (Fig. 4E). Consequently, the most likely explanation for the inhibitory effect of LidA is that by binding to Rab1, it occludes the access of SidM, SidD, AnkX, or Lem3 to the residue of Rab1 that needs to be covalently modified or demodified.
LidA and Rab1 are not essential for the recruitment of host cell vesicles.
The findings that LidA is dispensable for the recruitment and activation of Rab1 suggested that this effector is involved in events downstream of Rab1 recruitment by SidM. Given that Rab1 binding occurs through LidA's central region, predicted to be enriched in coiled-coils, we hypothesized that this bacterial protein may be a molecular mimic of cellular tethering factors. Tethering factors are elongated proteins that bind Rab GTPases via coiled-coil regions, thereby facilitating the initial docking of transport vesicles to target membranes. To determine if LidA plays a similar role during replication vacuole transformation, we challenged human macrophage-like U937 cells for 30 min or 2 h with either wild-type L. pneumophila or a ΔlidA mutant strain and analyzed the colocalization of LCVs with host vesicles by electron microscopy (Fig. 5). We found no obvious difference in the ability of ΔlidA mutants to recruit host vesicles to their LCV compared to the ability of the parental strain at either 30 min postinfection (10 of 10 vacuoles for wild-type L. pneumophila versus 22 of 24 for ΔlidA mutants) or 2 h postinfection (8 of 11 for wild-type L. pneumophila versus 29 of 29 for the ΔlidA mutant). Notably, an L. pneumophila ΔlidA ΔsidM double mutant, which is unable to recruit Rab1, was equally as proficient (19 of 20) as wild-type bacteria in recruiting host vesicles to its LCV at 2 h postinfection (Fig. 5). This strengthens the hypothesis that L. pneumophila exploits vesicle transport routes other than the Rab1-regulated secretory pathway, a feature that could ensure vacuolar transformation even in the absence of SidM and LidA.
Fig 5.
Host vesicles are recruited to the LCV in the absence of LidA and SidM. (Top) Electron micrographs show host vesicles attached to the cytosolic surface of the LCVs at 2 h postinfection of U937 cells with L. pneumophila Lp02 wild-type, Lp02ΔlidA, or Lp02ΔlidAΔsidM (for quantification, see text). (Bottom) Magnification of selected regions (boxed) from the upper panel. The arrows indicate vesicles surrounding the LCV.
DISCUSSION
L. pneumophila is a pathogen that targets and modifies host Rab GTPases to ultimately manipulate vesicular trafficking for its own benefit. The organism employs multiple effector proteins that act sequentially to achieve Rab1 recruitment and manipulation of the GTPase at the LCV. Of the four effectors shown to influence Rab1 dynamics on the vacuole (SidM, LidA, SidD, and LepB), LidA's role in Rab1 manipulation is the least defined.
LidA's early presence on the LCV seems to coincide with its role in boosting SidM-driven recruitment of Rab1 to the LCV (31). However, it is not clear how LidA is involved in this process. One possibility is that LidA does not directly recruit Rab1 but, rather, that it interferes with its inactivation and removal in order to prolong the presence of active Rab1 on the LCV. We found that LidA inhibited LepB- or TBC1D20-stimulated GTP hydrolysis by forming a complex with Rab1 (Fig. 1A and B). LidA's inhibitory effect on Rab1 inactivation is most likely due to its close interaction with active Rab1, which probably sterically hinders GAP access to the nucleotide binding pocket of active Rab1. LidA binds to Rab1 with unparalleled affinity among known Rab effectors, including GAP proteins (41). This affinity may increase the stability of the LidA-Rab1 complex to such an extent that GAP proteins could not compete for access to Rab1 (41). Thus, LidA binding locks GTP-loaded Rab1 in the active conformation, thereby extending its presence on the LCV surface by preventing its GDI-mediated extraction from the membrane. This conclusion is further supported by the fact that EDTA, a small molecule with a molecular weight of only 292 Da, was unable to cause extraction of GTP or GDP from Rab1 in the presence of LidA (Fig. 1C and D), suggesting that LidA binds Rab1 close to its guanine nucleotide binding pocket. Another bacterial effector that acts similarly is EspG, a type III effector protein of the enterohemorrhagic Escherichia coli O157:H7. EspG forms a complex with ARF6 and blocks its inactivation by GAPs (42). Though examples of cellular ligands exist that were found to decelerate GTP hydrolysis by binding to their target GTPase (22), pathogenic bacteria seem to use this mechanism in a more forceful manner to precisely control signaling events regulated by small GTPases without interference from the host.
We reasoned that LidA might keep Rab1 in an active conformation in order to present it to downstream effectors involved in vesicle tethering and fusion. However, our results so far do not support this idea, since LidA had an inhibitory rather than a stimulating effect on Rab1 interaction with GM130, giantin, and p115 (Fig. 1E). The possibility that other cellular downstream effectors might be able to interact with Rab1 even in the presence of LidA cannot be excluded. Alternatively, we speculate that while LidA interferes with binding of host downstream effectors of Rab1, it may facilitate the interaction of this GTPase with L. pneumophila effectors that mimic the function of host Rab1 ligands. The inhibition of Rab1 binding to its downstream ligands by LidA is reminiscent of the inhibitory effect of SidM-catalyzed AMPylation of Rab1 which also blocks the interaction of the GTPase with at least one of its cellular binding partners, MICAL-3 (33). L. pneumophila may use this strategy to restrict Rab1 interaction to a selected set of ligands, thereby allowing the organism to efficiently compete with host proteins on the Golgi compartment for recruitment of secretory vesicles.
Our studies revealed that LidA interfered with Rab1 AMPylation and de-AMPylation in vitro, raising the question about its effect on these posttranslational modifications during infection. Given that Rab1 recruitment to LCVs is dependent on SidM, it is most likely that LidA primarily interacts with Rab1 after it has been AMPylated by SidM. Consistent with this, LidA is the only known protein that interacts with Rab1 in its AMPylated (35) or phosphocholinated form (Fig. 4E). The inhibitory effect of LidA on the de-AMPylation (and dephosphocholination) of Rab1 is corroborated by the recently released structure of LidA in complex with another host GTPase, Rab8 (41). The serine and tyrosine residues of Rab8 that are equivalent to the ones phosphocholinated or AMPylated in Rab1 are buried within the complex with LidA and may not be easily accessible to SidD (or Lem3), respectively. It should also be noted that activation of Rab1 by SidM precedes AMPylation and, in vitro, it seems to take place at a higher rate than AMPylation. Due to SidM's low binding affinity for GTP-bound Rab1, we could envision an alternative role for LidA in capturing those Rab1 molecules on the LCV surface that have been activated by SidM but escaped AMPylation, thereby protecting them from GAP-stimulated inactivation. Overall L. pneumophila appears to delay the inactivation of Rab1 in several ways that, at a first glance, may seem redundant but that are necessary to efficiently antagonize host cell processes that may otherwise interfere with Rab1-mediated binding of ER-derived vesicles to the LCV.
The concept of functional redundancy explains why interference with individual factors or pathways within living cell systems does not always result in a detectable phenotype. The emerging consensus is that L. pneumophila infection displays a high degree of functional redundancy which makes it challenging to detect growth phenotypes when individual or even several effector proteins or host cell pathways are disrupted (15). For instance, an L. pneumophila strain lacking SidM is unable to recruit host cell Rab1, yet this mutant is as proficient as wild-type bacteria in intracellular survival and replication vacuole formation (31, 34). In fact, none of the known L. pneumophila effector proteins that modulate the activity of Rab1 are required for virulence. This suggests that L. pneumophila targets multiple vesicle-trafficking routes simultaneously to reroute membrane material to the LCV. Consistent with this, we found that LCVs containing L. pneumophila ΔlidA or ΔsidM ΔlidA mutants showed extensive colocalization with host cell vesicles (Fig. 5), presumably because deletion of sidM and lidA, alone or together, affected only the recruitment of Rab1-dependent vesicles to the LCV, leaving intact other pathways that provided material for the vacuolar transformation process. Thus, it will be interesting to identify the source(s) of the Rab1-independent membrane material and to determine which bacterial and host factors are involved in diverting those vesicles to the LCV.
LidA has characteristics of tethering factors, proteins that mediate docking of transport vesicles with the membrane of the destination compartments. Since LidA is exclusively membrane-associated within infected cells (14) but lacks an obvious transmembrane domain, it is most likely that LidA is a “peripheral” membrane protein that is recruited to the cytosolic surface of the LCV or to surrounding vesicles by binding either Rab1 or the phosphoinositide PI(3)P (8). These features may allow LidA to also bind membranes elsewhere in the cell on which this GTPase-lipid combination is found. Several effectors, including SidM, have been shown to bind PI(4)P and use this interaction for attachment to the vacuolar membrane. We found that, unlike SidM, LidA does not have separate regions for binding Rab1 and PI(3)P; instead, the same central coiled-coil region binds both Rab1 and the phosphoinositide, while the N- and C-terminal regions do not bind either (Fig. 3C to E). PI(3)P is commonly found on early endosomal membranes, and intracellular pathogens can interfere with its metabolism to avoid phagosome maturation. SapM from Mycobacterium tuberculosis is a PI(3)P phosphatase (49), and it avoids phagosome maturation by preventing the PI(3)P-dependent recruitment of lysosomal enzymes and vacuolar ATPase (28). No enzymatic activities have been described for LidA, but given its affinity for PI(3)P, it could either interfere with its metabolism or simply mask its presence on the vacuole to occlude potential binding sites for the early endosomal marker EEA1. The presence of PI(3)P on ER subdomains involved in autophagosome formation (3) suggests that additional host cell membrane compartments enriched in this phosphoinositide may be targeted by LidA.
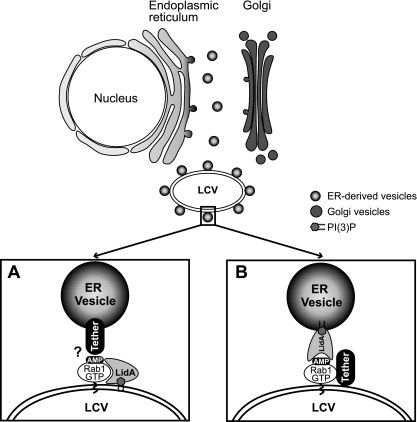
We propose a model in which LidA promotes docking of host vesicles with the LCV early during infection (Fig. 6). In one scenario, LidA could localize to the LCV membrane, where it keeps Rab1 recruited by SidM in an active conformation by blocking inactivation through GAP-stimulated GTP hydrolysis. The LidA-Rab1 complex could act as an adaptor for vesicular tethering and fusion proteins, thereby bridging the membrane of the LCV with that of secretory transport vesicles surrounding the LCV. Alternatively, upon translocation, LidA might localize to the surface of surrounding vesicles through interaction with phosphoinositides and mediate docking of the vesicles with the LCV via interaction with AMPylated Rab1 and/or other L. pneumophila effectors present on the LCV. The former model would explain how LidA enhances Rab1 accumulation on LCVs (31), whereas the latter model would be consistent with the fact that LidA is the only known protein capable of binding Rab1 in its AMPylated form (33).
Fig 6.
Model(s) of LidA-mediated attachment of ER-derived vesicles to the LCV. (A) LidA translocated to the cytosol of infected host cells localizes to the LCV membrane by binding AMPylated Rab1 and/or phosphoinosides present on the vacuolar membrane. LidA keeps modified Rab1 in an active conformation and acts as an adaptor protein to mediate the interaction of Rab1 with tethering protein(s) located on the surface of ER-derived vesicles. (B) Upon translocation into the cytosol of infected host cells, LidA localizes to ER vesicles through phosphoinositide binding and mediates docking of these vesicles to the LCV via interaction with AMPylated Rab1 and/or other L. pneumophila effectors present on the LCV that act as tethers.
In order to fully understand the function of LidA during L. pneumophila infection, several questions remain to be answered. It is currently unclear what protein or signal causes the high-affinity LidA-Rab1 complex to dissociate in order to allow SidD-mediated de-AMPylation and subsequent inactivation of Rab1. Also, a function for the N- and C-terminal domain of LidA has yet to be determined, and the identity of other bacterial or host proteins or pathways likely to be targeted by LidA is currently unclear. Though mammalian Rab6 and Rab8 were found to bind LidA (31), the role of these interactions for L. pneumophila virulence has not been elucidated yet. Addressing these questions is essential to obtain a clear perspective on LidA's role during infection of a host cell.
ACKNOWLEDGMENTS
We thank Ralph Isberg for insightful discussion and for kindly providing reagents.
This work was supported by the Intramural Research Program of the NIH.
Footnotes
Published ahead of print 6 January 2012
REFERENCES
- 1. Allan BB, Moyer BD, Balch WE. 2000. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science 289:444–448 [DOI] [PubMed] [Google Scholar]
- 2. Andrews HL, Vogel JP, Isberg RR. 1998. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66:950–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Axe EL, et al. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beard M, Satoh A, Shorter J, Warren G. 2005. A cryptic Rab1-binding site in the p115 tethering protein. J. Biol. Chem. 280:25840–25848 [DOI] [PubMed] [Google Scholar]
- 5. Berger KH, Isberg RR. 1994. Intracellular survival by Legionella. Methods Cell Biol. 45:247–259 [DOI] [PubMed] [Google Scholar]
- 6. Berger KH, Isberg RR. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7–19 [DOI] [PubMed] [Google Scholar]
- 7. Berger KH, Merriam JJ, Isberg RR. 1994. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol. Microbiol. 14:809–822 [DOI] [PubMed] [Google Scholar]
- 8. Brombacher E, et al. 2009. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J. Biol. Chem. 284:4846–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burstein ES, Macara IG. 1992. Interactions of the ras-like protein p25rab3A with Mg2+ and guanine nucleotides. Biochem. J. 282(Pt 2):387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention 2011. Legionellosis—United States, 2000-2009. MMWR Morb. Mortal. Wkly. Rep. 60:1083–1086 [PubMed] [Google Scholar]
- 11. Coleman DE, Sprang SR. 1998. Crystal structures of the G protein Gi alpha 1 complexed with GDP and Mg2+: a crystallographic titration experiment. Biochemistry 37:14376–14385 [DOI] [PubMed] [Google Scholar]
- 12. Conover GM, Derre I, Vogel JP, Isberg RR. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305–321 [DOI] [PubMed] [Google Scholar]
- 13. Derre I, Isberg RR. 2004. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 72:3048–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Derre I, Isberg RR. 2005. LidA, a translocated substrate of the Legionella pneumophila type IV secretion system, interferes with the early secretory pathway. Infect. Immun. 73:4370–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dorer MS, Kirton D, Bader JS, Isberg RR. 2006. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dumenil G, Isberg RR. 2001. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol. Microbiol. 40:1113–1127 [DOI] [PubMed] [Google Scholar]
- 17. Ensminger AW, Isberg RR. 2009. Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 12:67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feeley JC, et al. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fields BS. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286–290 [DOI] [PubMed] [Google Scholar]
- 20. Gabay JE, Horwitz MA. 1985. Isolation and characterization of the cytoplasmic and outer membranes of the Legionnaires' disease bacterium (Legionella pneumophila). J. Exp. Med. 161:409–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goody RS, et al. 2011. The versatile Legionella effector protein DrrA. Commun. Integr. Biol. 4:72–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herrmann C, Horn G, Spaargaren M, Wittinghofer A. 1996. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J. Biol. Chem. 271:6794–6800 [DOI] [PubMed] [Google Scholar]
- 23. Horwitz MA. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horwitz MA. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ingmundson A, Delprato A, Lambright DG, Roy CR. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450:365–369 [DOI] [PubMed] [Google Scholar]
- 26. Kabcenell AK, Goud B, Northup JK, Novick PJ. 1990. Binding and hydrolysis of guanine nucleotides by Sec4p, a yeast protein involved in the regulation of vesicular traffic. J. Biol. Chem. 265:9366–9372 [PubMed] [Google Scholar]
- 27. Kagan JC, Stein MP, Pypaert M, Roy CR. 2004. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 199:1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kusner DJ. 2005. Mechanisms of mycobacterial persistence in tuberculosis. Clin. Immunol. 114:239–247 [DOI] [PubMed] [Google Scholar]
- 29. Luo ZQ, Isberg RR. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. U. S. A. 101:841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Machner MP, Isberg RR. 2007. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318:974–977 [DOI] [PubMed] [Google Scholar]
- 31. Machner MP, Isberg RR. 2006. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 11:47–56 [DOI] [PubMed] [Google Scholar]
- 32. Mukherjee S, et al. 2011. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477:103–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Muller MP, et al. 2010. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329:946–949 [DOI] [PubMed] [Google Scholar]
- 34. Murata T, et al. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8:971–977 [DOI] [PubMed] [Google Scholar]
- 35. Neunuebel MR, et al. 2011. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333:453–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nuoffer C, Davidson HW, Matteson J, Meinkoth J, Balch WE. 1994. A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J. Cell Biol. 125:225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Plutner H, et al. 1991. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J. Cell Biol. 115:31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roy CR, Berger KH, Isberg RR. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663–674 [DOI] [PubMed] [Google Scholar]
- 39. Saarikangas J, Zhao H, Lappalainen P. 2010. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 90:259–289 [DOI] [PubMed] [Google Scholar]
- 40. Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schoebel S, Cichy AL, Goody RS, Itzen A. 2011. Protein LidA from Legionella is a Rab GTPase supereffector. Proc. Natl. Acad. Sci. U. S. A. 108:17945–17950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Selyunin AS, et al. 2011. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature 469:107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sexton JA, Vogel JP. 2004. Regulation of hypercompetence in Legionella pneumophila. J. Bacteriol. 186:3814–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Swanson MS, Isberg RR. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63:3609–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan Y, Arnold RJ, Luo ZQ. 2011. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc. Natl. Acad. Sci. U. S. A. 108:21212–21217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tan Y, Luo ZQ. 2011. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475:506–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR. 2001. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 114:4637–4650 [DOI] [PubMed] [Google Scholar]
- 48. Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, Balch WE. 1992. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119:749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vergne I, et al. 2005. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 102:4033–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weide T, et al. 2001. The Golgi matrix protein GM130: a specific interacting partner of the small GTPase rab1b. EMBO Rep. 2:336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wiater LA, Dunn K, Maxfield FR, Shuman HA. 1998. Early events in phagosome establishment are required for intracellular survival of Legionella pneumophila. Infect. Immun. 66:4450–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilson BS, et al. 1994. A Rab1 mutant affecting guanine nucleotide exchange promotes disassembly of the Golgi apparatus. J. Cell Biol. 125:557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]