Abstract
Porphyromonas gingivalis, the causative agent of adult periodontitis, must maintain nitric oxide (NO) homeostasis and surmount nitric oxide stress from host immune responses or other oral bacteria to survive in the periodontal pocket. To determine the involvement of a putative hydroxylamine reductase (PG0893) and a putative nitrite reductase-related protein (PG2213) in P. gingivalis W83 NO stress resistance, genes encoding those proteins were inactivated by allelic exchange mutagenesis. The isogenic mutants P. gingivalis FLL455 (PG0893∷ermF) and FLL456 (PG2213∷ermF) were black pigmented and showed growth rates and gingipain and hemolytic activities similar to those of the wild-type strain. P. gingivalis FLL455 was more sensitive to NO than the wild type. Complementation of P. gingivalis FLL455 with the wild-type gene restored the level of NO sensitivity to a level similar to that of the parent strain. P. gingivalis FLL455 and FLL456 showed sensitivity to oxidative stress similar to that of the wild-type strain. DNA microarray analysis showed that PG0893 and PG2213 were upregulated 1.4- and 2-fold, respectively, in cells exposed to NO. In addition, 178 genes were upregulated and 201 genes downregulated more than 2-fold. The majority of these modulated genes were hypothetical or of unknown function. PG1181, predicted to encode a transcriptional regulator, was upregulated 76-fold. Transcriptome in silico analysis of the microarray data showed major metabolomic variations in key pathways. Collectively, these findings indicate that PG0893 and several other genes may play an important role in P. gingivalis NO stress resistance.
INTRODUCTION
Porphyromonas gingivalis is a Gram-negative anaerobic bacterium that is a primary etiologic agent of periodontal disease. This infection-induced disease is characterized by inflammation, which can result in host-mediated destruction of tooth-supporting tissues and structures (18). Nitric oxide (NO) is a key component of the host immune response. Oral neutrophils can constitutively secrete low concentrations of NO via the involvement of multiple NO synthases. However, with activation, these neutrophils have increased secretion of NO with antimicrobial effects. As in other host-pathogen interactions (22), P. gingivalis has been shown to trigger the production of NO in immune and nonimmune host cells by activating the expression of inducible nitric oxide synthases (9, 54) and can survive in NO concentrations ranging from 4.9 μM to 19.2 μM (47). Elevated NO concentrations are reported to cause vasodilatation and a decrease in platelet aggregation, which may contribute to gingival bleeding (18), and to have cytotoxic effects on surrounding host tissue that can lead to alveolar bone loss (7). NO is present in the saliva and gingival fluid of periodontitis patients (7, 18, 57). Further, saliva NO concentrations have been shown to increase with the severity of periodontitis (47).
P. gingivalis, as an asaccharolytic microorganism, metabolizes nitrogenous compounds as a source of energy and generates a microenvironment abundant in ammonia and other important metabolic by-products (55). This nitrate-nitrite-ammonia conversion process involves the production of NO from nitrite reduction (38, 49, 60). NO is a small, lipophilic, freely diffusible gas and free radical which is highly reactive and cytotoxic. Its effects are caused by its reaction with oxygen and other molecules (e.g., thiols, metal centers, nucleotide bases, and lipids), resulting in the formation of reactive nitrogen species (32) that differ in properties and activities but have a broad spectrum of antimicrobial activity (23). Because excess NO is toxic and to prevent the bacteria from “committing suicide,” enzymes, including nitrite and NO reductases, are tightly regulated to maintain a steady-state level of free NO at nanomolar concentrations (63). This mechanism for homeostasis, which may include an ability to downregulate nitrite reduction (62), involves the expression of the several genes known to be induced by nitrogen oxides and low oxygen tension (60, 63).
An important component of NO stress resistance in P. gingivalis is its ability to maintain nontoxic NO intracellular concentrations. There is a gap in our understanding of the mechanism(s) for NO homeostasis and stress resistance in P. gingivalis. An interrogation of the P. gingivalis genome did not reveal any NO reductase; however, NO detoxification is predicted to occur via a hydroxylamine intermediate during nitrite ammonification (49). The hybrid cluster protein (HCP), a 4Fe-4S cluster binding oxidoreductase, is found in many bacteria to catalyze the reduction of hydroxylamine to form NH3 and H2O and is mostly induced under conditions of nitrite, S-nitrosoglutathione, or nitrate stress (8, 15, 28, 49). HCP was associated with a putative NADH oxidoreductase, displaying an oxygen-sensitive hydroxylamine reductase activity in facultative anaerobes like Escherichia coli, where both proteins are encoded by the hcp-hcr operon (15). The E. coli HCP was also induced by hydrogen peroxide and displayed involvement in oxidative stress protection under the regulation of the peroxide regulator OxyR (3). In P. gingivalis, HCP is predicted to be encoded by PG0893 (www.oralgen.lanl.gov); however, its role in NO stress resistance is unclear.
The conversion of nitrite to NO can also affect internal NO homeostasis. Excessive reductions of nitrite would result in an accumulation of NO that would be toxic to the cell. To prevent this, bacteria closely regulate the activity of nitrite and NO reductases (63). There is a putative nitrite reductase-related protein in P. gingivalis that is encoded by the PG2213 gene. This protein shows similarities with the nitrite reductase NirB and with other nitrite reductases found in Aquifex aeolicus, Staphylococcus aureus, and Bacillus subtilis (www.oralgen.lanl.gov). In addition, a nitrite reductase which was also involved in the reduction of nitrite to ammonia had similarities in its heme group arrangement to a hydroxylamine oxidoreductase, suggesting a role for such enzymes in NO detoxification (21). While a role for nitrite reductase in NO metabolism has been demonstrated in other bacteria (17, 52), the function of PG2213 in a similar role is unknown.
In this study, we evaluated the role of PG0893 and PG2213 in NO stress resistance in P. gingivalis. Moreover, the data suggest that PG0893 may play a significant role in NO homeostasis. Whole-genome profiling by DNA microarray analysis of P. gingivalis exposed to NO identified several hypothetical genes that may play an important role in NO stress resistance. Variations in the metabolome of P. gingivalis under conditions of NO stress may reveal a strategy for survival.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains and plasmids used in this study are listed in Table 1. P. gingivalis strains were grown in brain heart infusion (BHI) broth supplemented with 0.5% yeast extract (Difco Laboratories, Detroit, MI), hemin (5 μg ml−1), vitamin K (0.5 μg ml−1), and cysteine (0.1%; Sigma-Aldrich, St. Louis, MO). P. gingivalis strains were cultured in an anaerobic chamber (Coy Manufacturing, Ann Arbor, MI) in 10% H2, 10% CO2, and 80% N2. E. coli strains were grown in Luria-Bertani broth (LB). Unless otherwise stated, all cultures were incubated at 37°C. Growth rates were determined spectrophotometrically by assessing the optical density at 600 nm (OD600). Erythromycin and tetracycline concentrations used were 10 μg ml−1 and 3 μg ml−1, respectively.
Table 1.
Plasmids and strains used in this study
| Plasmid or strain | Phenotype, genotype, or description | Source or reference |
|---|---|---|
| Plasmids | ||
| pVA2198 | SprermF-ermAM | 20 |
| pT-COW | AprtetQ | 26 |
| pFLL455a | AprtetQ PG0893∷Tc | This study |
| Strains | ||
| P. gingivalis W83 | Wild type | 29 |
| P. gingivalis FLL455 | PG0893-defective mutant | This study |
| Complemented P. gingivalis FLL455 | Complemented PG0893-defective mutant | This study |
| P. gingivalis FLL456 | PG2213-defective mutant | This study |
| P. gingivalis FLL32 | recA-defective mutant | 29 |
| E. coli DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ−thi1 gyrA96 relA1 | Invitrogen |
Nitric oxide dosage to mimic physiological range.
NO was produced using the NO donor diethylamine (DEA) NONOate (Cayman Chemical, Ann Arbor, MI). Samples (13 ml each) of hypoxic BHI broth were taken from an anaerobic chamber (Coy Manufacturing, Ann Arbor, MI) and introduced through a catheter system into an airtight reaction chamber saturated with N2 and kept warm (37°C) through a water bath system (E100; Lauda, Lauda-Königshofen, Germany). The hypoxic warm BHI was continuously stirred with a helix mixer to ensure solution homogeneity in the chamber. DEA NONOate was added to the chamber through the catheter system using a 10-μl Hamilton syringe (Hamilton Co., Reno, NV). The NO levels in the BHI were detected and recorded with a computerized NO-sensing electrode (Apollo 4000/Apollo 4.1.4 software; World Precision Instruments, Sarasota, FL). To generate a standard curve for NO, a NaNO2 solution (6.5 μl, 500 μM; Sigma-Aldrich, St. Louis, MO) was added repeatedly at similar intervals to 13 ml of a H2SO4-NaI solution (50 ml double-distilled water [ddH2O], 750 mg NaI [Fischer Scientific, NJ]; 27 μl of 18.4 M H2SO4 [Mallinckrodt, Paris, KY]) in the reaction chamber.
Growth analysis under heat stress.
Based on previous heat stress studies (50), cultures of P. gingivalis strains were grown anaerobically at 37°C overnight to an OD600 of 1.5 and were appropriately diluted with BHI to an OD600 of ∼0.2. The latter were incubated at 37°C (for controls) and 42°C for 24 h under anaerobic conditions. The OD600 was measured at various intervals (1 to 6 h) for 24 h.
Gingipain activity.
The activities of Arg-X and Lys-X-specific cysteine proteases (Rgp and Kgp) were determined using BAPNA (Nα-benzoyl-dl-arginine-p-nitroanilide; 4 mM) and ALNA (AC-Lys-p-nitroanilide HCl; 4 mM) as substrates in an activated protease buffer (0.1 M Tris [pH 7.6], 0.2 M NaCl, 5 mM CaCl2, 10 mM NaOH, 9 mM l-cysteine). Substrates were individually added to early (OD600 ∼ 0.4)- and late (OD600 ∼ 1.7)-exponential-phase P. gingivalis culture samples, and the endpoint OD was determined at 405 nm using a microplate reader (Bio-Rad, Hercules, CA).
Hemolysis type.
P. gingivalis strains were plated on brucella blood agar and incubated anaerobically at 37°C for 7 days. The aspect and size of the colonies and surrounding hemolytic zones were determined.
Sensitivity to UV stress.
Fresh cultures of the P. gingivalis strains were grown to exponential phase (OD600 ∼ 0.6) in BHI broth from overnight cultures. A UV sensitivity test was done with 500 J and 1,000 J UV light as previously reported by Abaibou et al. (1).
Sensitivity to nitric oxide and hydrogen peroxide (H2O2).
Fresh cultures of P. gingivalis strains were grown from overnight cell cultures. P. gingivalis strains were grown to early exponential phase (OD600 ∼ 0.3) in BHI broth. For single-stress studies, a stress agent (DEA NONOate [10 μl, 36 mM], H2O2 [10 μl, 0.25 mM], or NaOH [10 μl, 0.1 M]) was added to the cultures, which were further incubated at 37°C anaerobically. For multiple-stress studies, 36 mM DEA NONOate was added (10 μl initially; after 15 min, 8 μl every 15 min) over 1 h. The OD600 was measured at various intervals (1 to 6 h) for 24 h. Untreated and NaOH-treated cell cultures grown at 37°C were used as controls. NaOH controls were used for preliminary NO sensitivity studies only to determine the effect of NaOH on cell cultures.
Microarray study.
Culture samples were taken 15 min after treatment with DEA NONOate or NaOH from growing P. gingivalis W83 cultures (as in the NO sensitivity experiment), and total RNA was extracted from them using a RiboPure kit (Ambion, Austin, TX). DNase treatment was carried out by using a DNase kit (Ambion, Austin, TX). Samples from untreated cultures of P. gingivalis W83 were processed similarly. NaOH-treated and untreated cultures were used as controls. DNA microarray gene expression was carried out using Roche NimbleGen custom arrays (100910_CW_P_ging_W83_expr_HX12; Roche, Indianapolis, IN) according to the standard NimbleGen procedure (NimbleGen arrays user's guide: gene expression analysis, v5.1). Briefly, cDNA was synthesized from the RNA samples using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). Both RNA and cDNA quality was checked using an Agilent Bioanalyzer and Agilent RNA 6000 Nano and DNA1000 chips. cDNA (0.5 to 1 μg) was used to start the amplification and labeling reaction using a NimbleGen one-color labeling kit, in which Cy3 was randomly incorporated into the newly synthesized DNA by the Klenow fragment. Labeled DNA (2 μg) derived from each RNA sample was hybridized with each array for over 16 to 18 h. The slides and arrays were washed, spun dry, and then scanned with a Roche MS200 microarray scanner with a resolution of 2 μm. The normalization was done with the NimbleScan 2.6.0.0 built-in normalization function. Microarray data analysis was performed with a Partek Genomics suite (v6.5). Differentially expressed genes were identified by determining change (≥2-fold) plus P (≤0.05), with a false-discovery rate (FDR) of 0.05. The microarray data were submitted to the Gene expression Omnibus database (www.ncbi.nlm.nih.gov/geo/).
Real-time PCR analysis.
Total RNA samples were obtained from 15-min untreated and DEA NONOate-treated (as in NO sensitivity experiments) cultures using a RiboPure kit (Ambion, Austin, TX). DNase treatment was carried out using DNase kit (Ambion, Austin, TX). cDNA was synthesized using a Transcriptor high-fidelity cDNA synthesis kit (Roche, IN). cDNA samples were processed using a QuantiTect SYBR green PCR kit (Qiagen, Valencia, CA) and Cepheid Smart Cycler II instrument. The real-time primers used are listed in Table 2. The absolute quantification method was used for data analysis, and data were normalized to data for the 16S gene.
Table 2.
Primers used in this study
| Name | Description | Sequence |
|---|---|---|
| PG0893I F | PG0893 (hcp) forward | 5′-AAAAGAAAATGTTTTGCTATCAATGC |
| PG0893I R | PG0893 (hcp) reverse | 5′-TCAGCGTCCGAATATCTTCTTC |
| PG0893 F | PG0893 (hcp and upstream sequence) forward | 5′-ATGTCCCTTGCGACAATTGAT |
| PG0893 R | PG0893 (hcp and downstream sequence) reverse | 5′-GCCAATAACCCCACTACATAC |
| PG2213I F | PG2213 (putative nitrite reductase) forward; also used for real-time PCR | 5′-ATGTCACAACAAGATGAAATT |
| PG2213I R | PG2213 (putative nitrite reductase) reverse; also used for real-time PCR | 5′-ACCGTTGATTTCGTCTAAAATA |
| PG2213 F | PG2213 (putative nitrite reductase and upstream sequence) forward | 5′-TGCCGCCATCGCCAAGCATCG |
| PG2213 R | PG2213 (putative nitrite reductase and downstream sequence) reverse | 5′-AGTACGCGAGATGAGATTTGA |
| PG16s F | 16S forward | 5′-AGGCAGCTTGCCATACTGCG |
| PG16s R | 16S reverse | 5′-ACTGTTAGCAACTACCGATGT |
| PGUstA F | ustA forward; also used for real-time PCR | 5′-ATGCAACGCATTGCCG |
| PGUstA R | ustA reverse; also used for real-time PCR | 5′-TGCATTGGCTTCGCC |
| PG1181 F | PG1181 forward | 5′-ATGGATGCTCGTGCCAGAAT |
| PG1181 R | PG1181 reverse | 5′-TATCTGCATCAGTGCTCGCC |
| PG1019 F | PG1019 forward | 5′-TGAAAAAAAATTTTCTTTTTTTCTCCC |
| PG1019 R | PG1019 reverse | 5′-GAATCTTGCAGGAAAAGAGTTTTGT |
| PG0612 F | PG0612 forward; also used for real-time PCR | 5′-ATGAAACGAATAATTTTATTACTCAGT |
| PG0612 R | PG0612 reverse; also used for real-time PCR | 5′-TAGAAATCTTTACCACAGAGAGCG |
| PG0627 F | PG0627 forward | 5′-GATGAGTATGAACATCTACGTAGGGAA |
| PG0627 R | PG0627 reverse | 5′-AATAGCGATCTTCGTGTCGGA |
| PG1467 F | PG1467 forward | 5′-GATGAGACTGTTCAAGAAGTTCATAAG |
| PG1467 R | PG1467 reverse | 5′-ATCGCCCGATAACACAGATAC |
| PG0893 RltPCR F | PG0893 forward for real-time PCR | 5′-CGGCTACGTGGATGAAGAGATCAA |
| PG0893 RltPCR R | PG0893 reverse for real-time PCR | 5′-CGACACCTATATTTACTTCCGTGATTT |
| PG16s RltPCR F | 16S forward for real-time PCR | 5′-CGATGATTACTAGGAGTTTGCGAT |
| PG16s RltPCR R | 16S reverse for real-time PCR | 5′-CACCATCCGTCATCTACATTTCAA |
| PG1181 RltPCR F | PG1181 forward for real-time PCR | 5′-ATCTGCAGCTCTTCAAAGAAAAATACC |
| PG1181 RltPCR R | PG1181 reverse for real-time PCR | 5′-TATCTGCATCAGTGCTCGCC |
| PG1019 RltPCR F | PG1019 forward for real-time PCR | 5′-TGAAAAAAAATTTTCTTTTTTTCTCCC |
| PG1019 RltPCR R | PG1019 reverse for real-time PCR | 5′-TCCACTTTCTGTTGGGTACGAAG |
| PG0627 RltPCR F | PG0627 forward for real-time PCR | 5′-GATGAGTATGAACATCTACGTAGGGAA |
| PG0627 RltPCR R | PG0627 reverse for real-time PCR | 5′-GATCGAACAGCTCTTCGATGG |
| PG1467 RltPCR F | PG1467 forward for real-time PCR | 5′-GATGAGACTGTTCAAGAAGTT |
| PG1467 RltPCR R | PG1467 reverse for real-time PCR | 5′-TCAGAAAGTTGGCGATGTTTT |
| PG1080 RltPCR F | PG1080 forward for real-time PCR | 5′-AGCAGTAGTAGGTACCGGAGCC |
| PG1080 RltPCR R | PG1080 reverse for real-time PCR | 5′-GTACGGATACGAGCCATAACG |
| PG1145 RltPCR F | PG1145 forward for real-time PCR | 5′-TGGCGGTCACTATCTATCATCC |
| PG1145 RltPCR R | PG1145 reverse for real-time PCR | 5′-AGATACCGATTCGTTCCCCC |
| PG1829 RltPCR F | PG1829 forward for real-time PCR | 5′-ACAGAATTTCATCCGACTTTATCAA |
| PG1829 RltPCR R | PG1829 reverse for real-time PCR | 5′-CACCATTCGGCAGAGTCTTT |
| PG0071 RltPCR F | PG0071 forward for real-time PCR | 5′-AAACAACAAACACTGAAAGATAAGTTC |
| PG0071 RltPCR R | PG0071 reverse for real-time PCR | 5′-GCCTTTTTTCAGCACCGTA |
| PG0075 RltPCR F | PG0075 forward for real-time PCR | 5′-AATTATTATACAGACACCCCGCA |
| PG0075 RltPCR R | PG0075 reverse for real-time PCR | 5′-GTTGGGTGCGATGATATCG |
| BamHI 0893 F | PG0893 forward (hcp and promoter region with attached BamHI sequence) for complementation | 5′-GGATCCTGATTTTTCTCTGAAT |
| BamHI 0893 R | PG0893 reverse (hcp with attached BamHI sequence) for complementation | 5′-GGATCCTGCGATCAGCGTCCG |
Creation of P. gingivalis FLL455 (PG0893∷ermF) and FLL456 (PG2213∷ermF) mutants.
Inactivation of the PG0893 and PG2213 genes followed the method of Dou et al. (20). The primers used are listed in Table 2. Briefly, 1-kb upstream and downstream flanking regions for PG0893 and PG2213 were amplified from chromosomal P. gingivalis DNA. The latter were fused to one ermF fragment from pVA2198 plasmid amplification, using the upstream fragment forward primer and the downstream fragment reverse primer. The PCR program included one 5-min cycle at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 56°C and 55°C for PG0893 and PG2213, respectively, and 4 min at 68°C, with a 5-min final extension at 68°C. The fragment obtained was electroporated into P. gingivalis as described by Abaibou et al. (1), and the transformed cells were plated on BHI agar with 10 μg ml−1 of erythromycin. The plates were incubated for 7 days at 37°C. Colony PCR and DNA sequencing were used to confirm appropriate gene replacement in erythromycin-resistant mutant colonies. RT-PCR was also performed on mutant RNA samples to confirm the absence of gene expression for PG0893 and PG2213.
Complementation of the P. gingivalis FLL455 (PG0893∷ermF) mutant.
A DNA fragment containing the PG0893 open reading frame and its upstream promoter region was amplified from P. gingivalis W83 chromosomal DNA using oligonucleotide primers engineered with BamHI restriction sites (Table 2). The BamHI restriction site was created at the ends of both primers to facilitate the subcloning of the PCR fragment. BamHI-digested pTCOW (26) and the BamHI-digested PCR fragment were ligated and used to transform E. coli DH5α cells. The purified recombined plasmid, designated pFLL455a, was used to transform the P. gingivalis FLL455 (PG0893∷ermF) mutant by electroporation. The transformed cells were grown on BHI agar plates in the presence of erythromycin and tetracycline for 7 to 10 days at 37°C.
In silico analysis.
In silico analysis of microarray data and data interpretation for the metabolome analysis were carried out using the ArrayStar software package, version 3. Metabolome analysis was done using the KEGG pathway modules and pathway mapping modes (http://www.genome.jp/kegg/pathway.html) (4). The KEGG expression database (40) and the NCBI Gene Expression Omnibus database (45) were used for reference and analysis.
Glutamate concentration in P. gingivalis W83 and FLL455 (PG0893∷ermF) strains.
P. gingivalis strains were grown anaerobically to early exponential phase (OD600 ∼ 0.3) in BHI broth at 37°C. DEA NONOate (0.66 μmol for P. gingivalis W83 and 0.36 μmol for P. gingivalis FLL455 [PG0893∷ermF] for NO sensitivity reasons) was added to the cultures, which were further incubated at 37°C anaerobically. Culture samples were taken 15 min posttreatment and centrifuged (RC5C; Sorvall Instruments, DuPont) at 13,000 × g for 10 min. Cell pellets were washed and resuspended in phosphate-buffered saline (PBS; pH 7.4, 10 mM) and then lysed by sonication (Sonic Dismembrator, Fischer Scientific, NJ) at 30% amplitude for 15 min. Lysates were centrifuged (RC5C; Sorvall Instruments, DuPont) at 4,000 × g for 10 min. Supernatants were used for glutamate concentration determination with a glutamate assay kit (BioVision, Mountain View, CA), and the endpoint OD was measured at 405 nm using a microplate reader (Bio-Rad, CA). Untreated cell cultures grown at 37°C were similarly processed and used as controls. Each experiment was done in triplicate.
RESULTS
DEA NONOate dosage to mimic NO physiological range.
NO has a half life of a few seconds, so the study of its effects on P. gingivalis required the use of a NO donor (12) to maintain exposure of the bacterial cells to the compound. Among many methods (30, 31, 43, 44, 48, 56), DEA NONOate had been successfully used in studies involving bacteria (42) and had a short enough half life to avoid unnecessary prolonged bacterial exposure to NO and allow us to determine easily the dose needed to replicate NO levels found in the periodontal pocket (47). Our studies revealed that, to mimic the average NO physiological range of periodontal pockets, the ideal DEA NONOate dose in hypoxic BHI was 10 μl of 36 mM, or 0.36 μmol. This dose yielded an average NO peak concentration of 20.6 μM ± 7.7 μM, followed by a much slower gradual decline to an average of 8.10 μM ± 3 μM after 15 min, which covered our desired average NO physiological range in the microenvironment of the periodontal pocket. To maintain 1-h NO stress exposure of the bacteria within the same NO range, one injection of DEA NONOate (10 μl; 36 mM) followed by another one (8 μl; 36 mM) every 15 min was found to give good consistency.
The growth of P. gingivalis W83 was inhibited under NO stress.
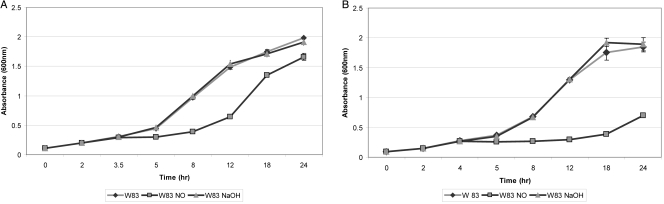
P. gingivalis was grown in the presence of 0.36 μmol DEA NONOate to evaluate its effect on growth. As shown in Fig. 1, the growth of P. gingivalis in the absence of NO had a 3-h generation time, as previously reported (24). The growth rate was similar in the presence of NaOH, which was used as the solvent for our NO donor. In the presence of a single exposure to NO, the generation time of P. gingivalis was significantly increased (P ≤ 0.05) by approximately 40% up to 5 h posttreatment (Fig. 1A). Upon chronic exposure to NO for 1 h, the growth of P. gingivalis was significantly inhibited (P ≤ 0.05) compared to that of the control (Fig. 1B). These results suggest that NO-induced stress can inhibit the growth of P. gingivalis.
Fig 1.
(A) Sensitivity of P. gingivalis W83 to single NO stress. P. gingivalis W83 strain was grown anaerobically to early exponential phase in BHI broth at 37°C, DEA NONOate (NO) or NaOH was added to the cultures, and the cultures were further incubated for 24 h. Each experiment was done in triplicate. The error bars show standard deviations. (B) Sensitivity of P. gingivalis W83 to multiple NO stress. P. gingivalis W83 strain was grown anaerobically to early exponential phase in BHI broth at 37°C, and DEA NONOate (NO) or NaOH was added to the cultures, followed by repeated DEA NONOate or NaOH additions for 1 h. The cultures were further incubated for 24 h. Each experiment was done in triplicate. The error bars show standard deviations.
Role of P. gingivalis FLL455 (PG0893∷ermF) and FLL456 (PG2213∷ermF) strains in NO-induced stress.
In several bacteria, HCP and nitrate reductases are involved in the metabolism and detoxification of NO (15, 35, 49). In P. gingivalis, PG0893 and PG2213 are predicted to be an HCP and a possible nitrite reductase-related protein, respectively (www.oralgen.lanl.gov). In addition, PG2213 may have hydroxylamine oxidoreductase properties that would imply a NO detoxification function. To evaluate their role in NO stress, the genes encoding these proteins were inactivated by allelic-exchange mutagenesis. Following electroporation and plating on selective medium, we obtained several erythromycin-resistant mutant colonies after a 5- to 7-day incubation period. The mutants were confirmed by PCR and DNA sequencing (data not shown). Similar to the wild-type strain, the erythromycin-resistant mutants were black pigmented and showed beta-hemolysis on brucella blood agar plates. One mutant for each gene deletion was randomly chosen for further study. In BHI broth culture, P. gingivalis W83, FLL455 (PG0893∷ermF), and FLL456 (PG2213∷ermF) displayed similar growth rates when incubated at 37°C (data not shown). Additionally, no differences in gingipain activity were observed between the wild-type and the isogenic mutant strains (data not shown).
The P. gingivalis FLL455 (PG0893∷ermF) and FLL456 (PG2213∷ermF) strains had growth comparable to that of the wild-type W83 under increased temperature.
In the inflammatory environment of the periodontal pocket, P. gingivalis successfully survives increased temperature and NO and H2O2 stress and keeps its virulent properties (16, 25, 58). To investigate the possible relevance of PG0893 and PG2213 to increased temperature survival, the growth of P. gingivalis FLL455 (PG0893∷ermF), FLL456 (PG2213∷ermF), and wild-type strains was studied at 42°C. P. gingivalis W83 and the mutant strains grew similarly at 42°C (P ≥ 0.05) (data not shown).
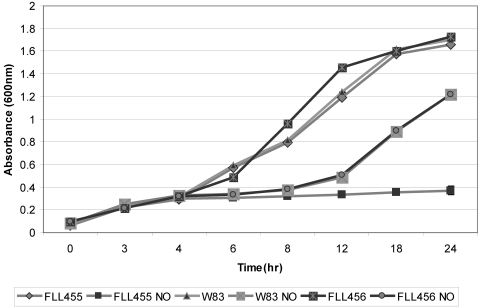
The P. gingivalis FLL455 (PG0893∷ermF) strain is sensitive to NO stress.
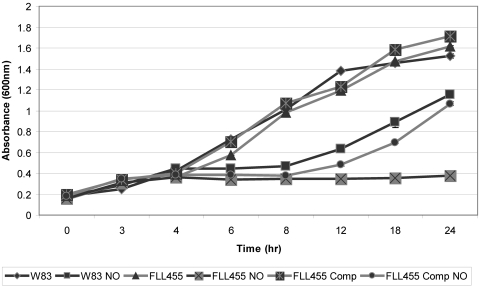
The relative significance of the PG0893 and PG2213 genes in NO stress resistance was evaluated by the exposure of P. gingivalis FLL455 (PG0893∷ermF) and FLL456 (PG2213∷ermF) strains to NO. As shown in Fig. 2, P. gingivalis FLL456 and the wild-type strain had similar sensitivities to NO, in contrast to P. gingivalis FLL455, which showed significantly increased sensitivity (P ≤ 0.05). Complementation of P. gingivalis FLL455 (PG0893∷ermF) with the wild-type gene significantly (P ≤ 0.05) restored NO stress sensitivity to a level similar to that of the wild-type strain (Fig. 3).
Fig 2.
Sensitivity of P. gingivalis W83, FLL455 and FLL456 to single NO stress. P. gingivalis strains were grown anaerobically to early exponential phase in BHI broth at 37°C, DEA NONOate (NO) was added to the cultures, and the cultures were further incubated for 24 h. Each experiment was done in triplicate. The error bars show standard deviations.
Fig 3.
Sensitivity of P. gingivalis W83, FLL455, and complemented (comp) FLL455 to NO stress. P. gingivalis strains were grown anaerobically to early exponential phase in BHI broth at 37°C, DEA NONOate (NO) was added to the cultures, and the cultures further incubated for 24 h. Each experiment was done in triplicate. The error bars show standard deviations.
H2O2 sensitivity of P. gingivalis FLL455 (PG0893∷ermF) and FLL456 (PG2213∷ermF) isogenic mutants.
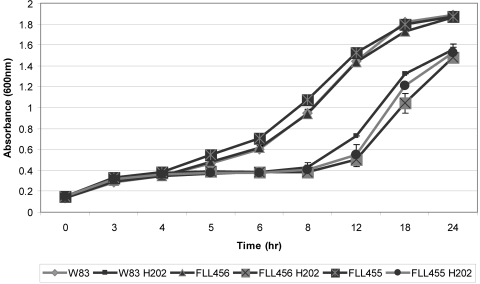
PG2213 was previously found to be upregulated upon microaerophilic exposure (37), and HCP from other bacteria was shown to have a protective role in oxidative stress (3, 5). The importance of the PG2213 and PG0893 genes in oxidative stress was explored by exposing P. gingivalis W83 and isogenic mutants defective in those genes to H2O2. As shown in Fig. 4, there was no significant difference (P ≥ 0.05) in sensitivity to H2O2 between the mutants and the wild-type strain.
Fig 4.
Sensitivity of P. gingivalis W83, FLL455, and FLL456 to H2O2 stress. P. gingivalis strains were grown anaerobically to early exponential phase in BHI broth at 37°C, H2O2 was added to the cultures, and the cultures further incubated for 24 h. Each experiment was done in triplicate. The error bars show standard deviations.
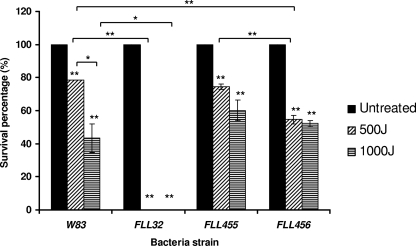
The P. gingivalis FLL455 (PG0893∷ermF) and FLL456 (PG2213∷ermF) strains were not sensitive to UV irradiation.
The persistence of P. gingivalis in the inflammatory environment of the periodontal pocket requires an ability to overcome oxidative and NO stress, including the damage they cause to the bacterial DNA. UV stress causes DNA mutations similar to the ones found in NO- and H2O2-induced stress and has been widely used for DNA repair mechanism studies. Unlike other bacteria, P. gingivalis has been shown to use a yet-unknown repair mechanism for the removal of 8-oxo-7,8-dihydroguanine (8-oxo-G) lesions caused by NO- and H2O2-induced stress (11, 29, 53). To investigate the potential effect of PG0893 and PG2213 in the cascade of events for NO detoxification, cell repair (including DNA repair) and survival, we evaluated the sensitivity of the P. gingivalis FLL455 (PG0893∷ermF) and FLL456 (PG2213∷ermF) mutants to UV stress compared to the wild-type strain W83. FLL32, a recA-deficient and UV stress-sensitive mutant (2), was used as a control. As demonstrated in Fig. 5, P. gingivalis W83 and the P. gingivalis mutants FLL455 (PG0893∷ermF) and FLL456 (PG2213∷ermF) displayed no major differences in their sensitivity to UV stress.
Fig 5.
Sensitivity of P. gingivalis W83, FLL32, FLL455, and FLL456 to UV irradiation. Dilutions of fresh cultures of the P. gingivalis strains were grown anaerobically to exponential phase in BHI broth from overnight cultures and spread on BHI agar. The cells were irradiated with the indicated UV radiation. Plates were incubated at 37°C for 7 to 10 days, and CFU were counted. Each experiment was done in triplicate. The error bars show standard deviations. *, P ≤ 0.05; **, P ≤ 0.01; asterisks without brackets represent comparison to the value for untreated samples.
The transcriptome response of P. gingivalis to NO exposure.
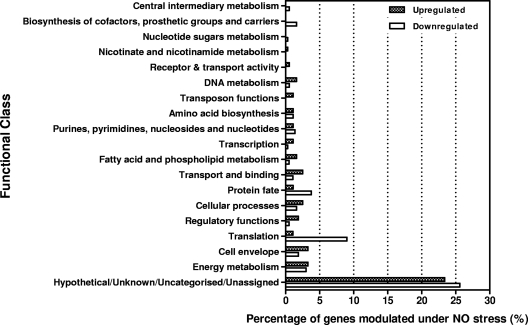
To further determine the role of other P. gingivalis genes in NO stress, we performed whole-genome profiling by DNA microarray analysis. P. gingivalis W83 in exponential growth phase was exposed to 0.36 μmol of DEA NONOate for 15 min. The results summarized in Fig. 6 and Table S1 in the supplemental material are derived from three independent experiments performed in triplicate. Approximately 19% of the P. gingivalis genome was modulated up and down (≥2-fold) in response to NO exposure. Analysis of these data revealed that 179 genes were upregulated and 201 genes downregulated. It is noteworthy that PG0893 and PG2213 gene expression was upregulated 1.4- and 2.2-fold, respectively. The pattern of expression of selected genes using real-time PCR confirmed those observed in the DNA microarray analysis (Table 3). The modulated genes, when classified into functional groups according to the annotation in the Oral Pathogen Sequence Databases at the Los Alamos National Laboratory (www.oralgen.lanl.gov), showed that the majority of those genes were hypothetical or of unknown function (Fig. 6), including PG1181, which was upregulated 76-fold.
Fig 6.
Distribution of functions for genes affected by exposure of P. gingivalis to NO for 15 min. The genes that were differentially expressed (≤2-fold up- or downregulation and a P value of ≤0.05) were grouped by functional class according to the Los Alamos P. gingivalis genome database (www.oralgen.lanl.gov).
Table 3.
Real-time PCR analysis of selected genes modulated in the NO microarray study
| Gene | Avg change (fold)a |
|---|---|
| PG0893 | 17 ± 7 |
| PG2213 | 3.8 ± 0.6 |
| PG1181 | 84 ± 3 |
| PG0612 | 23 ± 9 |
| PG0246 | 3 ± 0.6 |
| PG1019 | 5.4 ± 1.3 |
| PG1467 | 1.9 ± 0.6 |
| PG0627 | −29 ± 7 |
| PG1080 | 9.5 ± 0 |
| PG1145 | 2 ± 0.2 |
| PG1829 | 5 ± 0.1 |
| PG0071 | 2 ± 0.4 |
| PG0075 | 1.2 ± 0.02 |
Values are means ± standard deviations.
Metabolome variations during NO stress.
An in silico transcriptome interrogation of P. gingivalis during nitric oxide stress revealed the modulation of genes that can affect several metabolic pathways (Table 4). An upregulation of PG1829 predicts an increase in fatty acid synthesis and metabolism. There is also a predicted increase in fatty acid elongation. The initiation of these pathways, which usually occurs through the acetyl coenzyme A (acetyl-CoA) acylation process, is also predicted to increase with the upregulation of the PG0775 and PG1080 genes. There is also an upregulation of PG0071, which encodes the hydroxyl acyl carrier protein, which can enhance palmitic acid biosynthesis. The PG0071 gene can also enhance the fatty acid elongation process in which PG1145 and PG1829 are the key genes and are upregulated more than 2-fold. Other pathways predicted to be upregulated include isoleucine and arginine degradation and cyanocobalamin synthesis.
Table 4.
Metabolic variations in P. gingivalis W83 during nitric oxide stress
| Pathway | Gene(s) involved |
|---|---|
| Upregulated | |
| Fatty acid elongation | PG1145 |
| Fatty acid biosynthesis | PG1829 |
| Acyl-CoA acylation | PG0775, PG1080 |
| GDP mannose biosynthesis | PG2010 |
| Cyanocobalamine synthesis | PG1124, PG1858, PG0703 |
| Palmitic acid biosynthesis | PG0071 |
| Isoleucine degradation | PG1079 |
| Arginine degradation | PG1271 |
| Downregulated | |
| N-Acetylglucosamine degradation | PG1286 |
| G protein biosynthesis | PG1808 |
| Flavin biosynthesis | PG0155 |
| Folate transformation | PG1854 |
| tRNA precursor synthesis | PG1596 |
| Pyruvate fermentation | PG0429 |
| Glyoxal degradation | PG0745 |
| Mixed acid fermentation | PG0429 |
| Nucleotide synthesis salvage pathway | PG1260, PG0558 |
| Beta oxidation | PG1829, PG0775, PG1145, PG1080 |
| Anaerobic respiration | PG1576, PG1824 |
| Glycolysis | PG1755 |
| Glycogen biosynthesis | PG1834, PG1042 |
| Calvin-Benson-Bassham cycle | PG1747, PG1515 |
| Gluconeogenesis pathway | PG1677 |
| Arginine biosynthesis | PG1271 |
| Acyl carrier protein synthesis and assimilation | PG0273 |
| Heme biosynthesis | PG0475, PG0195 |
| Thiamine biosynthesis | PG1693 |
| Lipid A synthesis | PG0072, PG0638 |
The predicted downregulated pathways include arginine biosynthesis, heme biosynthesis, N-acetylglucosamine degradation, folate transformation, and glyoxal degradation. Energy pathways such as glycolytic pathway, glycogen biosynthesis, gluconeogenesis, anaerobic respiration, and the Calvin-Benson-Bassham cycle were downregulated. The heme biosynthesis pathway was downregulated due to PG0475 (oxygen independent coproporphyrinogen III oxidase).
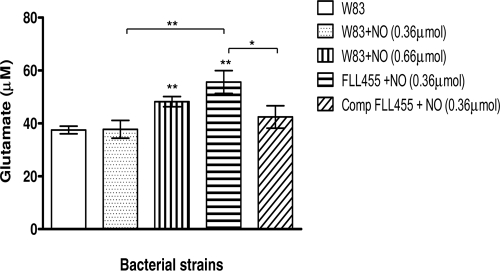
Glutamate concentration was increased in P. gingivalis W83 and FLL455 (PG0893∷ermF) strains under conditions of NO stress.
The arginine degradation pathway can lead to glutamate, which can be a substrate for energy metabolism and would be vital for survival (10). To confirm the hypothesis of an increase in bacterial intracellular glutamate levels following exposure to NO, the glutamate concentration of lysed NO-treated cells was determined for the P. gingivalis W83 and FLL455 (PG0893∷ermF) strains. As shown in Fig. 7, the glutamate concentrations were significantly increased (P ≤ 0.05) in cells exposed to NO compared to the untreated W83 control. Although P. gingivalis FLL455 (PG0893∷ermF) was exposed to a lower dose of NO than the parent strain, the highest glutamate level was observed in this strain. Complementation of this strain with a wild-type copy of the PG0893 gene significantly (P ≤ 0.05) restored the glutamate levels to those of the parent strain exposed to NO.
Fig 7.
Glutamate concentration in P. gingivalis W83, FLL455, and complemented FLL455. P. gingivalis strains were grown anaerobically to early exponential phase in BHI broth at 37°C, and cultures were either left untreated or treated with the indicated amount of DEA NONOate. Samples were taken after 15 min and centrifuged. Cell pellets were washed, resuspended in PBS (pH 7.4), and sonicated. The glutamate concentration of the samples was determined. Each experiment was done in triplicate. The error bars show standard deviations.*, P ≤ 0.05; **, P ≤ 0.01; asterisks without brackets represent comparison to untreated W83 samples.
Data and statistical analysis.
All studies were performed as three independent experiments. Statistical analysis was done using the Microsoft Office Excel 2003 software package (Microsoft, Mountain View, CA). The t test was used for one-to-one comparisons, and ANOVA was used for multiple comparisons.
DISCUSSION
Survival in the periodontal pocket requires P. gingivalis to withstand NO stress generated by immune cells, such as macrophages and neutrophils. When the bacterium is exposed to NO stress within the physiological range found in the periodontal pocket, PG0893 appears to be essential for NO stress resistance but shows little importance in oxidative stress resistance (47). These results are consistent with other studies, in which HCP has been shown to be involved in NO and hydroxylamine reduction and detoxification in other strictly or facultatively anaerobic bacteria (15, 27, 36, 49). In other bacteria (3, 5, 39), HCP has been implicated in protection against H2O2-induced stress.
Conversely, PG2213 did not appear to play any major role in NO detoxification. Under external NO stress, controlling internal NO bacterial levels would be of critical importance for survival. The ammonification pathway that involves a nitrite reductase and hydroxylamine reductase is induced under conditions of nitrite or nitrate stress (49, 62). The iron-sulfur cluster and residue similarities of PG2213 with NirB and with other nitrite reductases from S. aureus and B. subtilis (www.oralgen.lanl.gov) would imply a role in nitrite reduction to NO, affecting bacterial intracellular NO levels and thus HCP activity. The gene encoding PG2213 was found to be induced under conditions of NO stress; however, when the gene was inactivated, the mutant strain showed a level of NO sensitivity similar to that of the wild-type strain, which suggests the possibility of multiple mechanisms for NO detoxification. Additionally, the PG2213-defective mutant showed sensitivity to oxidative stress similar to that of the wild-type, which implied a reductase function that could possibly exclude the use of H2O2 as a substrate.
DNA microarray analysis revealed that multiple genes are modulated in response to NO stress in P. gingivalis. Because the majority of these genes were hypothetical or of unknown function, it is likely that they could represent unique nitric oxide stress resistance pathways. Several putative transcriptional regulators (PG1181, PG1237, PG0997, PG1000, PG0004, and PG1240) were highly upregulated. PG1181 was most highly upregulated and may be one of the key regulators in NO stress resistance in P. gingivalis. This gene is predicted to be part of a seven-gene transcriptional unit that includes PG1775, PG1176, PG1177, PG1178, PG1179, and PG1180, all of which were upregulated except PG1177 when P. gingivalis was exposed to NO. These genes all encode hypothetical proteins of unknown function except for PG1175 and PG1176, whose products may be ABC transporter proteins.
PG1181 and PG1240 are predicted to belong to the TetR family. Many bacteria have been reported to use mechanisms involving TetR regulators in resistance to stress, multidrug resistance, biofilm formation, control of metabolic pathways, and virulence (46). The exact roles the other putative transcriptional regulators play in NO stress resistance and virulence in P. gingivalis are still unclear, and this is under investigation.
Several genes, most of which are hypothetical or of unknown function and are part of multiple transcriptional units (e.g., PG1175 to -81, PG0612 to -14, PG1236 and -37, PG1239 and -40, and PG0409 to -11), were modulated in P. gingivalis exposed to NO stress. This may suggest that the response of P. gingivalis to NO stress is tightly regulated and/or coordinated. Four genes usually induced by stress or involved in mechanisms of adaptation to atypical conditions [uspA (PG0245), htpG (PG0045), ustA (PG0246), and dps (PG0090)] were also noticeably upregulated. Additionally, 12 upregulated genes (PG1858, dps, PG0777, PG0776, PG1172, PG1171, PG1239, hcp, PG2034, PG0108, PG0616, and PG0195) that were differentially expressed following NO stress were found to encode proteins with electron carrier and oxidoreductases functions. A gene encoding rubrerythrin (PG0195) and a putative thioredoxin gene (PG0616), known to be involved in oxidative stress, were highly downregulated.
Rubrerythrin has been reported to play a protective role against reactive nitrogen species in P. gingivalis in vivo (41); thus, its downregulation in our studies is unclear. This difference may be partially due to the multiple signals that can be induced by the host-microbe response and further underscores the complex nature of that interaction. The gene encoding PG0616, a hypothetical protein with a thioredoxin-like motif, may be involved in stabilizing the redox status of the cell surface, an important mechanism for maintaining cellular integrity (51). Its downregulation differed from the E. coli thioredoxin Trx patterns in oxidative stress, where it was shown to be upregulated and played a role in detoxification to promote bacterial survival (13, 14). Our results indicate that it may not contribute to NO stress resistance in P. gingivalis.
The expression of several genes encoding proteins involved with protein fate, degradation, and stabilization (e.g., PG0521 [GroES] and PG0778) was upregulated during NO stress. GroES is a 10-kDa chaperonin heat shock protein that is important in protein repair under conditions of environmental stress (33, 61), while PG0778, encoding an O-sialoglycoprotease in P. gingivalis, was earlier reported to be important in gingipain biogenesis and virulence modulation through the release of sialic acid (6), which is known to be an important scavenger of reactive oxygen species and plays a role in oxidative stress resistance. However, it is unclear if a similar strategy is functional during NO stress in P. gingivalis.
Changes in genetic expression relevant to metabolism might also give an indication as to the pathways and the compounds that would be most beneficial to the bacteria under conditions of NO stress. Many of the intermediary metabolites in the metabolome of NO-stressed P. gingivalis may be important in its growth and survival. Though we did not find the involvement of major nitrate regulating genes in our transcriptome analysis, we could identify two major metabolomic variations in key pathways: (i) there was increased fatty acid synthesis or fatty acid elongation; and (ii) there was a decrease in common energy metabolic pathways.
Multiple pathways can facilitate the energy requirements of the cells. Under NO stress, a shift from the major energy pathways, including glycolysis, gluconeogenesis, and glycogen biosynthesis, was observed. It is predicted that to overcome this deficit, substrates from other catabolic processes provide appropriate energy metabolites. NO stress showed increased isoleucine degradation, which forms acetyl-CoA, which in turn can be used by the energy pathways in P. gingivalis. There is also degradation of several other amino acids, such as isoleucine and arginine, which can eventually be converted into glutamate, which can then enter the tricarboxylic acid (TCA) cycle. In addition, with the downregulation of pyruvate fermentation, there would be a shift in the substrate to glutamate. The glutamate can be degraded further into aspartate and fumarate, which can also enter the TCA cycle.
NO generated from arginine via the NO synthase enzyme is involved in homeostasis and is vital to cell survival (34, 38, 59, 63). This study showed a predicted downregulation of arginine synthesis and upregulation of arginine degradation. It is likely that downregulation of arginine synthesis could reduce the inflow of nitrogen to the ammonia cycle during NO stress. Upregulation of arginine degradation could cause a shift in the use of the common intermediary metabolites like pyruvate and aspartate to glutamate generated by arginine degradation. Additionally, the formate which can be formed from glyoxylate metabolism and folate transformation was also observed to be reduced.
Furthermore, N-acetylglucosamine, an important cell wall component, was found to be protected from degradation during NO stress. A balance in the formation of fructose-6-phosphate from the TCA cycle leading to mannose synthesis is maintained, and this is evident from the upregulation of the mannose pathway. There was an upregulation of mannose biosynthesis due to the enzyme phosphomannose mutase (PG2010). Together, these observations indicate that it is likely that this system helps to maintain cell wall integrity. Overall, the energy requirements for P. gingivalis under NO stress may be provided by fatty acid and amino acid catabolism. In addition, an increase in fatty acid synthesis and acyl-CoA acylation could play a role in an antioxidant mechanism against NO (19). This is under further investigation in our laboratory.
In conclusion, our results suggest that NO stress resistance in P. gingivalis is facilitated by a complex and tightly regulated network of genes involved in multiple pathways, including energy metabolism, gene regulation, detoxification, and virulence. Seemingly, these genes may play important roles in the ability of this bacterium to survive in the inflammatory microenvironment of the periodontal pocket. Further, there may be other unique mechanisms that are important in the pathogenesis of P. gingivalis. Collectively, our observations suggest that further characterization of the NO stress response in P. gingivalis could reveal important therapeutic targets that could have important implications for the eradication of this organism.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Loma Linda University and Public Health grants DE-13664 and DE-019730 from NIDCR (to H.M.F.).
Footnotes
Published ahead of print 13 January 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abaibou H, et al. 2001. vimA gene downstream of recA is involved in virulence modulation in Porphyromonas gingivalis W83. Infect. Immun. 69:325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abaibou H, et al. 2000. Unaltered expression of the major protease genes in a non-virulent recA-defective mutant of Porphyromonas gingivalis W83. Oral Microbiol. Immunol. 15:40–47 [DOI] [PubMed] [Google Scholar]
- 3. Almeida CC, Romao CV, Lindley PF, Teixeira M, Saraiva LM. 2006. The role of the hybrid cluster protein in oxidative stress defense. J. Biol. Chem. 281:32445–32450 [DOI] [PubMed] [Google Scholar]
- 4. Aoki-Kinoshita KF, Kanehisa M. 2007. Gene annotation and pathway mapping in KEGG. Methods Mol. Biol. 396:71–91 [DOI] [PubMed] [Google Scholar]
- 5. Aragao D, et al. 2003. Reduced hybrid cluster proteins (HCP) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at high resolution using synchrotron radiation. J. Biol. Inorg. Chem. 8:540–548 [DOI] [PubMed] [Google Scholar]
- 6. Aruni W, et al. 2011. Sialidase and sialoglycoproteases can modulate virulence in Porphyromonas gingivalis. Infect. Immun. 79:2779–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Batista AC, Silva TA, Chun JH, Lara VS. 2002. Nitric oxide synthesis and severity of human periodontal disease. Oral Dis. 8:254–260 [DOI] [PubMed] [Google Scholar]
- 8. Beliaev AS, et al. 2005. Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J. Bacteriol. 187:7138–7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brennan PA, Thomas GJ, Langdon JD. 2003. The role of nitric oxide in oral diseases. Arch. Oral Biol. 48:93–100 [DOI] [PubMed] [Google Scholar]
- 10. Buckel W, Barker HA. 1974. Two pathways of glutamate fermentation by anaerobic bacteria. J. Bacteriol. 117:1248–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cadet J, Douki T, Ravanat JL. 2006. One-electron oxidation of DNA and inflammation processes. Nat. Chem. Biol. 2:348–349 [DOI] [PubMed] [Google Scholar]
- 12. Cai TB, Wang PG, Holder AA. 2005. NO and NO donors, p 3–31. In Wang PG, Cai TB, Taniguchi N. (ed), Nitric oxide donors. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany [Google Scholar]
- 13. Cha MK, Kim HK, Kim IH. 1995. Thioredoxin-linked “thiol peroxidase” from periplasmic space of Escherichia coli. J. Biol. Chem. 270:28635–28641 [DOI] [PubMed] [Google Scholar]
- 14. Cha MK, Kim HK, Kim IH. 1996. Mutation and mutagenesis of thiol peroxidase of Escherichia coli and a new type of thiol peroxidase family. J. Bacteriol. 178:5610–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chismon DL, Browning DF, Farrant GK, Busby SJ. 2010. Unusual organization, complexity and redundancy at the Escherichia coli hcp-hcr operon promoter. Biochem. J. 430:61–68 [DOI] [PubMed] [Google Scholar]
- 16. Cochran DL. 2008. Inflammation and bone loss in periodontal disease. J. Periodontol. 79:1569–1576 [DOI] [PubMed] [Google Scholar]
- 17. Cutruzzola F. 1999. Bacterial nitric oxide synthesis. Biochim. Biophys. Acta 1411:231–249 [DOI] [PubMed] [Google Scholar]
- 18. de Sa Siqueira MA, Fischer RG, da Silva Figueredo CM, Brunini TM, Mendes-Ribeiro AC. 2010. Nitric oxide and oral diseases: can we talk about it? Cardiovasc. Hematol. Agents Med. Chem. 8:104–112 [DOI] [PubMed] [Google Scholar]
- 19. d'Ischia M, Napolitano A, Manini P, Panzella L. 2011. Secondary targets of nitrite-derived reactive nitrogen species: nitrosation/nitration pathways, antioxidant defense mechanisms and toxicological implications. Chem. Res. Toxicol. 24:2071–2092 [DOI] [PubMed] [Google Scholar]
- 20. Dou Y, Osbourne D, McKenzie R, Fletcher HM. 2010. Involvement of extracytoplasmic function sigma factors in virulence regulation in Porphyromonas gingivalis W83. FEMS Microbiol. Lett. 312:24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Einsle O, et al. 2000. Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs. J. Biol. Chem. 275:39608–39616 [DOI] [PubMed] [Google Scholar]
- 22. Fang FC. 1997. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Invest. 99:2818–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fang FC. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2:820–832 [DOI] [PubMed] [Google Scholar]
- 24. Fletcher HM, Morgan RM, Macrina FL. 1997. Nucleotide sequence of the Porphyromonas gingivalis W83 recA homolog and construction of a recA-deficient mutant. Infect. Immun. 65:4592–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frazer LT, et al. 2006. Vaccination with recombinant adhesins from the RgpA-Kgp proteinase-adhesin complex protects against Porphyromonas gingivalis infection. Vaccine 24:6542–6554 [DOI] [PubMed] [Google Scholar]
- 26. Gardner RG, Russell JB, Wilson DB, Wang GR, Shoemaker NB. 1996. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed beta-1,4-d-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Appl. Environ. Microbiol. 62:196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haveman SA, Greene EA, Stilwell CP, Voordouw JK, Voordouw G. 2004. Physiological and gene expression analysis of inhibition of Desulfovibrio vulgaris Hildenborough by nitrite. J. Bacteriol. 186:7944–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He Q, et al. 2006. Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough, inferred from global transcriptional analysis. Appl. Environ. Microbiol. 72:4370–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henry LG, Sandberg L, Zhang K, Fletcher HM. 2008. DNA repair of 8-oxo-7,8-dihydroguanine lesions in Porphyromonas gingivalis. J. Bacteriol. 190:7985–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hetrick EM, Schoenfisch MH. 2007. Antibacterial nitric oxide-releasing xerogels: cell viability and parallel plate flow cell adhesion studies. Biomaterials 28:1948–1956 [DOI] [PubMed] [Google Scholar]
- 31. Heurlier K, Thomson MJ, Aziz N, Moir JW. 2008. The nitric oxide (NO)-sensing repressor NsrR of Neisseria meningitidis has a compact regulon of genes involved in NO synthesis and detoxification. J. Bacteriol. 190:2488–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hochgrafe F, et al. 2008. Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J. Bacteriol. 190:4997–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hotokezaka H, Ohara N, Hayashida H, Matsumoto S, Matsuo T, Naito M, Kobayashi K, Yamada T. 1997. Transcriptional analysis of the groESL operon from Porphyromonas gingivalis. Oral Microbiol. Immunol. 12:236–239 [DOI] [PubMed] [Google Scholar]
- 34. Ignarro LJ. 1990. Nitric oxide. A novel signal transduction mechanism for transcellular communication. Hypertension 16:477–483 [DOI] [PubMed] [Google Scholar]
- 35. Kern M, Volz J, Simon J. 2011. The oxidative and nitrosative stress defence network of Wolinella succinogenes: cytochrome c nitrite reductase mediates the stress response to nitrite, nitric oxide, hydroxylamine and hydrogen peroxide. Environ. Microbiol. 13:2478–2494 [DOI] [PubMed] [Google Scholar]
- 36. Kim CC, Monack D, Falkow S. 2003. Modulation of virulence by two acidified nitrite-responsive loci of Salmonella enterica serovar Typhimurium. Infect. Immun. 71:3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewis JP, Iyer D, Anaya-Bergman C. 2009. Adaptation of Porphyromonas gingivalis to microaerophilic conditions involves increased consumption of formate and reduced utilization of lactate. Microbiology 155:3758–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lundberg JO, Weitzberg E, Gladwin MT. 2008. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 7:156–167 [DOI] [PubMed] [Google Scholar]
- 39. Macedo S, Aragao D, Mitchell EP, Lindley P. 2003. Structure of the hybrid cluster protein (HCP) from Desulfovibrio desulfuricans ATCC 27774 containing molecules in the oxidized and reduced states. Acta Crystallogr. D Biol. Crystallogr. 59:2065–2071 [DOI] [PubMed] [Google Scholar]
- 40. Marti J, Piquemal D, Manchon L, Commes T. 2002. Transcriptomes for serial analysis of gene expression. J. Soc. Biol. 196:303–307 [PubMed] [Google Scholar]
- 41. Mydel P, Takahashi Y, Yumoto H, Sztukowska M, Kubica M, Gibson FC, III, Kurtz DM, Jr, Travis J, Collins LV, Nguyen KA, Genco CA, Potempa J. 2006. Roles of the host oxidative immune response and bacterial antioxidant rubrerythrin during Porphyromonas gingivalis infection. PLoS. Pathog. 2:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ogawa R, et al. 2001. Comparison of control of Listeria by nitric oxide redox chemistry from murine macrophages and NO donors: insights into listeriocidal activity of oxidative and nitrosative stress. Free Radic. Biol. Med. 30:268–276 [DOI] [PubMed] [Google Scholar]
- 43. Pullan ST, et al. 2007. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: unaltered methionine biosynthesis indicates lack of S nitrosation. J. Bacteriol. 189:1845–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pullan ST, Monk CE, Lee L, Poole RK. 2008. Microbial responses to nitric oxide and nitrosative stress: growth, “omic,” and physiological methods. Methods Enzymol. 437:499–519 [DOI] [PubMed] [Google Scholar]
- 45. Quackenbush J. 2001. Computational analysis of microarray data. Nat. Rev. Genet. 2:418–427 [DOI] [PubMed] [Google Scholar]
- 46. Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reher VG, Zenobio EG, Costa FO, Reher P, Soares RV. 2007. Nitric oxide levels in saliva increase with severity of chronic periodontitis. J. Oral Sci. 49:271–276 [DOI] [PubMed] [Google Scholar]
- 48. Rock JD, Thomson MJ, Read RC, Moir JW. 2007. Regulation of denitrification genes in Neisseria meningitidis by nitric oxide and the repressor NsrR. J. Bacteriol. 189:1138–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 1:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roy F, Vanterpool E, Fletcher HM. 2006. HtrA in Porphyromonas gingivalis can regulate growth and gingipain activity under stressful environmental conditions. Microbiology 152:3391–3398 [DOI] [PubMed] [Google Scholar]
- 51. Shoji M, Shibata Y, Shiroza T, Yukitake H, Peng B, Chen YY, Sato K, Naito M, Abiko Y, Reynolds EC, Nakayama K. 2010. Characterization of hemin-binding protein 35 (HBP35) in Porphyromonas gingivalis: its cellular distribution, thioredoxin activity and role in heme utilization. BMC Microbiol. 10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Silvestrini MC, Falcinelli S, Ciabatti I, Cutruzzola F, Brunori M. 1994. Pseudomonas aeruginosa nitrite reductase (or cytochrome oxidase): an overview. Biochimie 76:641–654 [DOI] [PubMed] [Google Scholar]
- 53. Spek EJ, et al. 2001. Recombinational repair is critical for survival of Escherichia coli exposed to nitric oxide. J. Bacteriol. 183:131–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sun W, et al. 2010. Porphyromonas gingivalis stimulates the release of nitric oxide by inducing expression of inducible nitric oxide synthases and inhibiting endothelial nitric oxide synthases. J. Periodontal Res. 45:381–388 [DOI] [PubMed] [Google Scholar]
- 55. Takahashi N. 2005. Microbial ecosystem in the oral cavity: Metabolic diversity in an ecological niche and its relationship with oral diseases. Int. Congr. Ser. 1284:103–112 [Google Scholar]
- 56. Thomson MJ, Stevanin TM, Moir JWB. 2008. Measuring nitric oxide metabolism in the pathogen Neisseria meningitidis. Methods Enzymol. 437:539–560 [DOI] [PubMed] [Google Scholar]
- 57. Ugar-Cankal D, Ozmeric N. 2006. A multifaceted molecule, nitric oxide in oral and periodontal diseases. Clin. Chim. Acta 366:90–100 [DOI] [PubMed] [Google Scholar]
- 58. Van Dyke TE. 2008. The management of inflammation in periodontal disease. J. Periodontol. 79:1601–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu G, Morris SM., Jr 1998. Arginine metabolism: nitric oxide and beyond. Biochem. J. 336:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ye RW, Averill BA, Tiedje JM. 1994. Denitrification: production and consumption of nitric oxide. Appl. Environ. Microbiol. 60:1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yuan L, Hillman JD, Progulske-Fox A. 2005. Microarray analysis of quorum-sensing-regulated genes in Porphyromonas gingivalis. Infect. Immun. 73:4146–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zumft WG. 2002. Nitric oxide signaling and NO dependent transcriptional control in bacterial denitrification by members of the FNR-CRP regulator family. J. Mol. Microbiol. Biotechnol. 4:277–286 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.