Abstract
Leptospira interrogans is the primary causative agent of the most widespread zoonotic disease, leptospirosis. An in-depth structural characterization of L. interrogans is needed to understand its biology and pathogenesis. In this study, cryo-electron tomography (cryo-ET) was used to compare pathogenic and saprophytic species and examine the unique morphological features of this group of bacteria. Specifically, our study revealed a structural difference between the cell envelopes of L. interrogans and Leptospira biflexa involving variations in the lipopolysaccharide (LPS) layer. Through cryo-ET and subvolume averaging, we determined the first three-dimensional (3-D) structure of the flagellar motor of leptospira, with novel features in the flagellar C ring, export apparatus, and stator. Together with direct visualization of chemoreceptor arrays, DNA packing, periplasmic filaments, spherical cytoplasmic bodies, and a unique “cap” at the cell end, this report provides structural insights into these fascinating Leptospira species.
INTRODUCTION
The genus Leptospira belongs to the phylum Spirochetes and includes both pathogenic and saprophytic species (37). L. interrogans is the predominant pathogenic species causing leptospirosis, a global reemerging zoonotic disease (for reviews, see references 17 and 20). In nature, L. interrogans colonizes the renal tubules of reservoir hosts (e.g., wild rodents). Humans are infected via contact with infected animals or contaminated soil or water. Outbreaks occur frequently in tropical regions (2, 20), with more than 500,000 reported severe cases of leptospirosis annually with a 10% mortality rate (29). In contrast, Leptospira biflexa is a free-living saprophytic organism that is unable to infect the mammalian host despite its extensive genetic and structural similarities with L. interrogans. Recent comparative genomic analyses of L. interrogans and L. biflexa have identified many unique genes in L. interrogans that may be potentially associated with virulence and pathogenesis (6, 35, 40, 42, 49).
Leptospira spp. share a number of general features with other spirochetes, yet they differ in many ways from other spirochetal pathogens, such as Treponema pallidum and Borrelia burgdorferi (42). As with other spirochetes, Leptospiral motility is driven by the rotation of periplasmic flagella (PF) that are located between the inner membrane (IM) and outer membrane (OM). PF of Leptospira species extend a relatively short distance from each pole, resulting in the characteristic hook-shaped ends of the cells (11). Leptospira also has an abundance of lipopolysaccharide (LPS) that is absent from both T. pallidum and B. burgdorferi (42).
Cryo-electron tomography (cryo-ET) is a powerful three-dimensional (3-D) imaging technique that is capable of visualizing cellular components in living organisms at the molecular level (23, 26, 30). This technique involves collecting a tilt series of images from different angles at high magnification and computationally reconstructing a 3-D density map of the intact organism. Recently, cryo-ET has been extensively utilized to study many spirochetes (1, 7, 14, 15, 19, 28, 32, 33), particularly, in our hands, to study the intact flagellar motor structure of B. burgdorferi (25) as well as the cell envelope architecture of T. pallidum (24).
In this work, we employed cryo-ET for a systematic comparison of cellular structures from pathogenic L. interrogans and saprophytic L. biflexa. Our study revealed cellular features unique to Leptospira spp., as well as structural differences between pathogenic L. interrogans and nonpathogenic L. biflexa. Possible implications of our findings in leptospiral motility, physiology, and pathogenesis are discussed.
MATERIALS AND METHODS
Leptospira culture preparation.
L. interrogans serovar Lai strain Lai 56601 and L. biflexa serovar Patoc strain Patoc I (Paris) were kindly provided by James Matsunaga at the VA Medical Center in Los Angeles, CA. Leptospira spp. were cultured in 50 ml Ellinghausen-McCullough-Johnson-Harris (EMJH) medium at 30°C under aerobic conditions for 100 h to reach exponential growth phase. Leptospiral pellets were harvested by centrifugation at 4,000 × g at 22°C for 20 min and resuspended in EMJH medium.
Cryo-ET data collection and 3-D reconstruction.
Viable bacterial cultures were centrifuged to increase the concentration to ∼2 × 109 cells/ml. four-microliter samples were deposited onto freshly glow-discharged holey carbon grids for 1 min. The grids were blotted with filter paper and rapidly frozen in liquid ethane using a gravity-driven plunger apparatus as previously described (25). The resulting frozen-hydrated specimens were imaged at −170°C using a Polara G2 electron microscope (FEI Company, Hillsboro, OR) equipped with a field emission gun and a 4,000 (4K) × 4K charge-coupled-device (CCD) (16-megapixel) camera (TVIPS; GMBH, Germany). The microscope was operated at 300 kV with a magnification of ×31,000, resulting in an effective pixel size of 5.6 Å after 2 × 2 binning. Using the FEI “batch tomography” program, low-dose single-axis tilt series were collected from each bacterium at −4- to −6-μm defocus with a cumulative dose of ∼100 e−/Å2 distributed over 65 images, covering an angular range from −64° to +64°, with an angular increment of 2°. Tilted images were aligned and reconstructed using the software package Protomo (48). In total, 127 and 420 reconstructions were generated from L. interrogans and L. biflexa, respectively.
Subvolume averaging of flagellar motor.
A total of 496 subvolumes (320 × 320 × 320 voxels) containing entire flagellar motors and their surrounding membranes were extracted from the original 547 tomograms. The initial orientation of each flagellar motor in each organism was determined manually. Subvolume analysis of flagellar motors was carried out as described before (24, 25).
Visualization of 3-D reconstruction.
Tomographic reconstructions were visualized using IMOD software (18), and surface rendering of flagellar structures was carried out with the software package UCSF Chimera (38).
Reconstructive microscopy.
L. biflexa and L. interrogans at a concentration of ∼1 × 109 cells/ml were mounted with Hanks ' buffered salt solution (HBSS) on pads in preparation for imaging. 4′,6-Diamidino-2-phenylindole (DAPI) and FM 1-43FX (Invitrogen) were added to the cells at concentrations of 2.5 μg/ml and 5 μg/ml, respectively, and simultaneously incubated for 15 min. Specimens were scanned with an Applied Precision DeltaVision (Issaquah, WA) system fitted with an Olympus IX 70 inverted microscope employing a 100-W mercury arc lamp for illumination (Olympus America, Melville, NY) and excitation/emission filter sets (Chroma Technology Corp, Brattleboro, VT) specific for each of the fluorescent dyes. The filter set combination for DAPI (blue membrane-permeating DNA-specific dye) was a 340-nm excitation filter with a band pass of 20 nm and an emission filter of 390 nm with a band pass of 20 nm. FM 1-43FX (cell envelope stain for cell localization) was visualized with an excitation filter of 555 nm, band pass of 28 nm, and an emission filter of 617 nm, band pass of 73 nm. Image scans for each dye were acquired in series of Z sections at a step size of 0.02 μm with a Sony Interline CCD camera—16 total images for the combined reconstruction of the two dyes used. Objective magnification was ×100. Deconvolution and image analysis were performed on a Linux/RedHat workstation employing the SoftWoRx software program (Applied Precision).
RESULTS AND DISCUSSION
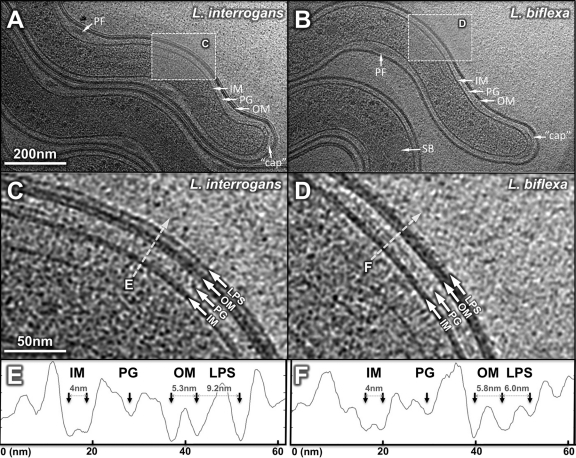
Three-dimensional (3-D) reconstructions of 547 different organisms were generated using cryo-ET to examine the cellular characteristics of pathogenic and saprophytic Leptospira (Fig. 1). The data presented are organized from the exterior to the center of the organisms as follows: cell envelope, cell end “cap,” periplasmic filaments, flagellar motor, chemoreceptor receptors, spherical bodies, and DNA.
Fig 1.
Ultrastructure from 3-D reconstruction of intact L. interrogans (A) or L. biflexa (B). The outer membrane (OM), inner membrane (IM), peptidoglycan layer (PG), and periplasmic flagellum (PF), the “cap” at the cell end, and a spherical body (SB) can be discerned from the picture. Zoom-in views reveal the structural details of the cell envelope of L. interrogans (C) or L. biflexa (D), respectively. LPS extends from the outer leaflet to a strong continuous density layer. Density profiles across the membranes of L. interrogans and L. biflexa (dashed line) are shown in panels E and F, respectively. The LPS layer was considerably thicker in L. interrogans than that in L. biflexa.
Ultrastructure of leptospiral cell envelope.
Leptospira has a dual-membrane architecture similar to that of Gram-negative bacteria. Leptospiral LPS and outer membrane proteins are major components of the outer membrane and play important roles in pathogenesis during infection (12, 34, 47). Cryo-ET of intact organisms provided a detailed view of the leptospiral cell envelope (Fig. 1). The envelope is composed of an IM, OM, and peptidoglycan (PG) layer. The IM contains two density layers corresponding to the phospholipid bilayer with a 4.0-nm spacing (Fig. 1E and F). There are three distinct density layers associated with the Leptospira OM, which is considerably thicker than the OMs of B. burgdorferi and T. pallidum (24, 25). The first two density layers likely correspond to the inner leaflet and the outer leaflet of the OM, with a spacing of 5.3 nm and 5.8 nm in L. interrogans and L. biflexa, respectively. A major structural difference between L. interrogans and L. biflexa was the third layer of the OM, which appears more pronounced in L. interrogans than in L. biflexa (Fig. 1C, D, E, and F). This third layer was 9.2 nm away from the outer leaflet of the OM in L. interrogans, compared to 6.0 nm in L. biflexa. Proteinase K treatment was ineffective in eliminating any of the OM layers (see Fig. S1 in the supplemental material), suggesting that none of the three density layers is formed solely by outer membrane proteins (which is unlike what has been observed in B. burgdorferi [25]). In addition, the outermost density layer is absent in T. pallidum, a spirochete that lacks both LPS and major outer surface proteins (24), suggesting that LPS likely extends from the outer leaflet of the OM to the third density layer on the envelope of Leptospira. LPS appears more abundant and longer (9.2 nm) in L. interrogans than that in L. biflexa (6.0 nm) (Fig. 1E and F). This finding supports the model that LPS is highly variable on cell surfaces of pathogenic and saprophytic species and plays an essential role in leptospiral virulence (34, 47).
Cryo-ET revealed the most detailed images of Leptospira species to date (Fig. 1C and D). Multiple density layers were observed in the periplasmic space, in an arrangement similar to that observed in T. pallidum (24). Therefore, we hypothesize that spirochetes share a similar envelope architecture (24), with a thin PG layer lying between two putative lipoprotein layers that are associated with either the IM or the OM. In addition, we observed that membrane vesicles are commonly formed by the OM without obvious involvement of the PG/IM layer (see Fig. S2 in the supplemental material), suggesting that despite the presence of LPS, the OM is fluid and loosely connected to the cell body.
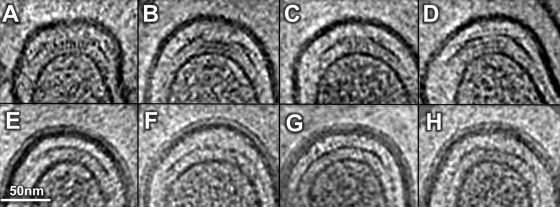
Cryo-ET also revealed a “cap”-like structure at the polar ends in both L. biflexa (Fig. 2A to D) and L. interrogans (Fig. 2E to H). Multiple density layers are located between the tips of OM and IM. The “cap” structure is distinct from the cone-shaped structures reported at the cell ends of Treponema primitia (33) and T. pallidum (15, 24). The leptospiral cell end is known for its attachment to the host cells (17, 22, 35), suggesting that this “cap” structure might play a role in mediating such interactions. The nature of the polar “cap,” as well as its physiological functions, remains to be explored.
Fig 2.
A gallery of the cell end reconstructions reveals a “cap”-like feature of L. biflexa (A to D) and L. interrogans (E to H).
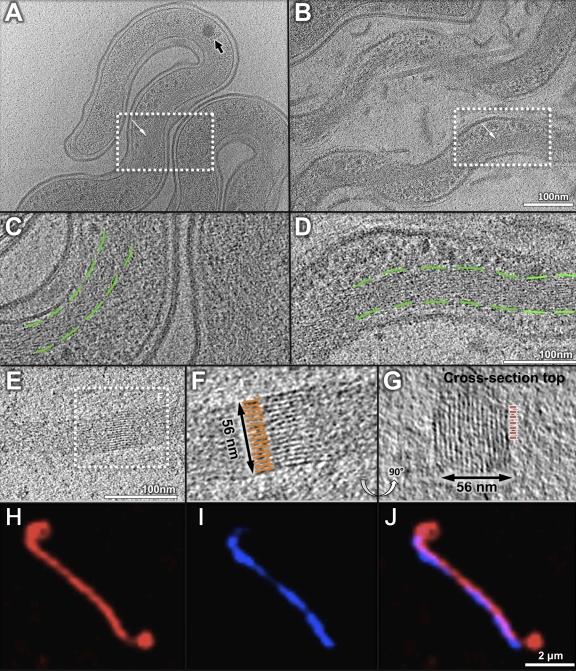
Novel filaments in the periplasm.
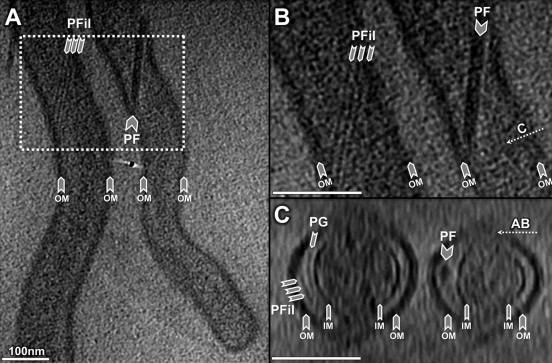
Cryo-ET revealed two types of filaments in the periplasmic space: flagella and novel periplasmic filaments (Fig. 3; see also Fig. S3 in the supplemental material). They differ in their diameters, locations, and possible functions. Flagella of Leptospira are located near the cell termini. They are responsible for bacterial motility and for the unique hook shape of the cell end(s) (5), but they do not determine the spiral shape of the cell (39). In this respect, Leptospira spp. are very different from B. burgdorferi, in which the flagella define the flat wave shape of the cell, since inactivation of flagellar genes results in long, rod-shaped bacteria (31). Additional filaments were found in the periplasmic space of both Leptospira species. The diameter of these filaments is considerably smaller than that of flagellar filaments (8 nm versus 22 nm). They wrap around the cell body in a right-handed fashion, particularly in the middle of the elongated organisms (Fig. 3), where PF is absent. Given that PF of Leptospira does not play a major role in defining the spiral shape of the cell body (39), it has been postulated that the cell morphology of Leptospira is determined by the cytoskeleton, the PG layer, or another undefined structure (17). The presence of these additional filaments in the periplasmic space is intriguing, since they represent novel candidates for shape determination in Leptospira. Further studies are required to provide direct evidence for this hypothesis.
Fig 3.
Periplasmic filaments in Leptospira spp. (A) Periplasmic filaments (PFil) are visible in the middle of the elongated organisms, while PF of Leptospira is located near the termini. (B) An enlarged section from panel A of PFil in L. interrogans. Three filaments are observed within the boundary of the outer membrane (OM). (C) A cross-sectional view indicates that PFil are located between the outer membrane (OM) and the inner membrane (IM).
Molecular architecture of leptospiral flagellar motor.
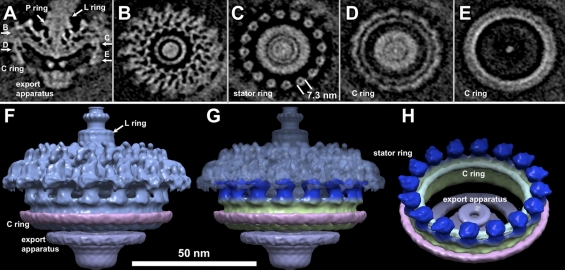
Flagella play important roles in spirochetal motility and host tissue invasion (16, 43). Unlike many other spirochetes (e.g., T. pallidum and B. burgdorferi) that have multiple flagellar motors, Leptospira species have a single flagellar motor at each end, and yet the organism is highly motile. To better understand this fascinating nanomachine, a total of 496 motors were extracted from 3-D reconstructions of intact organisms, and the molecular architecture of the intact flagellar motor was determined by subvolume averaging (Fig. 4). Leptospiral flagellar motors possess a relatively complex structure, which includes major flagellar components, including the hook, the rod, the MS ring, the C ring, and the export apparatus (Fig. 4). It appears that the leptospiral flagellar motor differs considerably from those in T. primitia (32), B. burgdorferi (25), and T. pallidum (24). Noticeably, the hook on top of the motor is wider (a diameter of ∼21 nm) than that of B. burgdorferi (∼16 nm). The distal rod is associated with two layers of density, identified as the putative L and P rings (Fig. 4A), as the homologous gene products of flgI and flgH in Escherichia coli are responsible for the L and P rings, respectively (27). The presence of flgH in Leptospira species is surprising, since the leptospiral PF is located in periplasmic space. Our map suggests that the L ring in the leptospiral flagellar motor is not embedded in the outer membrane, and therefore it likely lacks the function as a bushing.
Fig 4.
Molecular architecture of the intact flagellar motor of Leptospira spp. (A) Centered section parallel to the direction of the filament of an asymmetric reconstruction of the Leptospiral motors. Panels B, C, D, and E are horizontal cross sections. The locations of the sections are indicated in panel A. Panel B shows a section that lies inside the socket formed by the collar in periplasmic space. Section C transects the cytoplasmic side of the MS ring, just above the C ring. Sections D and E are located on the top and bottom of the C ring, respectively. Surface rendering of the Leptospira flagellar motor is presented in panels F and G. The side view (F and G) shows a novel C-ring structure. Extra density (in pink) is found at the bottom of the C ring. In panel G, 16 ring-like structures (colored in blue) are located on top of the C ring. Each ring is about 7.3 nm in diameter and 7.5 nm in height. The top portion of the ring is embedded in the cytoplasmic membrane, while the bottom portion is likely interacting with the C ring. Panel H is a view 45° rotated from panel G, revealing the interaction between the C ring and the stators.
The C ring and export apparatus in leptospiral flagellar motors are more complex than those of any recently determined motor structures (8). Additional density (colored in pink), which has not been shown in other motor structures, is visible around the bottom part of the C ring. The export apparatus is also different from other motor structures. It appears that the cytoplasmic part of the export apparatus is bigger than those from other spirochetes and bacteria. On the top of the C ring, the putative stators embedded in the cytoplasmic membrane were visualized in sufficient detail to distinguish the ring shape structures with a diameter of 7.3 nm (Fig. 4C and H). The shape and diameter of individual rings are similar to those of a stator complex purified from Vibrio alginolyticus (50). However, it remains to be answered if these rings correspond to the individual torque generators.
Recently, analysis of a large number of spirochete genome sequences uncovered remarkable genetic diversity among flagellar systems, even though many flagellar genes have been conserved during the evolutionary process (36). Our studies indicate that the spirochetal motors (including those of L. interrogans, T. pallidum, and B. burgdorferi) share overall similar sizes and shapes, yet there are considerable differences in the C ring, P ring, and export apparatus. This is consistent with the recent finding that the flagellar motors exhibit a common core architecture, despite the striking differences in their overall appearance (8). The novel structures of the C ring, export apparatus, and stator provide new insight into the diversity of bacterial flagellar motors.
Morphology and distribution of chemoreceptor arrays.
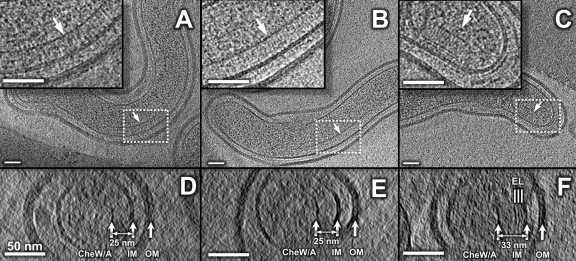
Chemotaxis is an important part of the bacterial signal transduction system and is closely associated with bacterial motility (46). Cryo-ET analysis allows direct visualization of the putative chemoreceptor arrays based on the fact that they form a characteristic pattern extending from the inner membrane to a concave “basal plate” (4, 28). The locations of these putative chemoreceptor arrays are variable, typically from 500 nm to 1,000 nm away from the cell ends. The width of arrays is also variable; however, the length of the receptor (from a prominent line “base plate” to the cell membrane) is relatively consistent at ∼25 nm (Fig. 5). Another type of array was visible in ∼20% of the L. biflexa organisms and was consistently located at the cell end (Fig. 5C). This array is structurally different from the chemoreceptor arrays because of the presence of additional domains. It is slightly longer (∼33 nm) yet narrower in width and is commonly located in close proximity to the only flagellar motor at the polar end (Fig. 5B). Overall, our cryo-ET observations indicate that the chemoreceptor arrays in L. biflexa are more readily visible than in L. interrogans under the current experimental conditions.
Fig 5.
Visualization of chemoreceptor arrays of Leptospira spp. Putative chemoreceptor arrays were observed in L. interrogans (A) and L. biflexa (B). The insets are the corresponding zoom-in views of the arrays outlined with white dashed lines. The arrays appear as clusters of pillar-like densities that extend from the IM and connect with a layer of high electron density at the membrane-distal ends. These are likely formed by CheA/W, which is known to form a continuous layer at the bottom of the chemoreceptor arrays. An additional array, unique to the saprophytic species, is found at the cell end (C). The inset is the zoom-in image of the array, indicating the presence of extra density layers (EL). The distance between the IM and the basal layer of CheA/W is relatively constant (25 nm) for most of the arrays, as shown in the cross sections (D and E). However, the novel array is longer (33 nm), as shown in the cross sections (F). The scale bar is 50 nm.
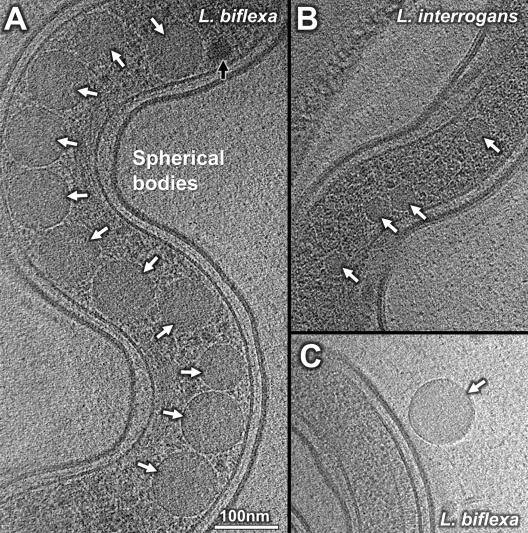
Spherical cytoplasmic bodies.
Two distinct morphological types of spherical cytoplasmic bodies were frequently observed along the cell bodies of L. biflexa and L. interrogans. One type is of high density (dark arrows in Fig. 6 and 7). The other type is of low density with a uniform internal texture (white arrows in Fig. 6A and B). These distinct spherical cytoplasmic bodies were frequently observed along the cell body (Fig. 6) and occasionally outside the cell body (Fig. 6C). No membrane boundary was found between spherical bodies and the cytoplasm (Fig. 6). Furthermore, the spherical bodies were larger in L. biflexa (∼100 nm) than in L. interrogans (∼30 nm). The spherical bodies did not contain molecular complexes, such as ribosomes, and appeared similar to the phosphate- and carbon-rich bodies in Caulobacter crescentus (9), suggesting a possible role in nutrient storage.
Fig 6.
Two types of spherical bodies in Leptospira spp. High- and low-density spherical bodies were labeled with dark and white arrows, respectively. (A) Cryo-ET tomogram slice of an L. biflexa cell, with spherical bodies (∼100 nm in size) located in the cell center. (B) Cryo-ET of an L. interrogans cell shows that the spherical bodies are randomly positioned along the cell body, with a diameter of ∼30 nm. (C) High-magnification view of a free-in-solution spherical body showing the absence of a phospholipid bilayer.
Fig 7.
Putative DNA bundles in intact and lysed cells and in cell medium. (A) Cryo-ET of L. biflexa cell reveals putative DNA bundles in the cell body. (B) Image of a partially lysed L. interrogans cell, showing that the putative DNA maintains a filamentous structure. Panels C and D show the zoom-in views of the areas highlighted in panels A and B, respectively. The contour lines outline the boundary along the putative DNA structure. (E) Top view of the putative DNA bundle in cell medium. (F) Zoom-in view of area indicated in panel E. Orange arrows depict putative DNA strands. (G) Cross-section view of the putative DNA bundle shown in panel F. The bundle is tightly packed by filaments with a spacing of ∼4 nm and a diameter of ∼2 nm. Fluorescence microscopy shows continuous dyed DNA in blue (I), cell envelope in red (H), and a composite image (J).
Visualization of DNA in situ.
Fluorescence microscopy analysis of both L. biflexa and L. interrogans cells indicated that the DAPI-stained nuclei, containing DNA (in blue, Fig. 7I), were homogeneously distributed along the cell body but not present at the cell ends (the cell envelope was stained in red; Fig. 7H). Unlike E. coli, where the genomic DNA is contained within the central area called the nucleoid (45), DNA inside Leptospira is distributed along the cell body (Fig. 7). Cryo-ET analysis of intact leptospiral cells revealed extensive ribosome-excluding regions, packed with long filamentous structures. These filamentous bundles were continuous along the cell body except at the cell ends, suggesting that they are likely related to DAPI-stained nuclei.
For better visualization of the leptospiral nuclei, we focused on a small percentage of cells that partially lysed during preparation. The putative DNA-containing bundles remained mostly trapped inside the cells (Fig. 7B and D). To our surprise, this type of bundle also existed by itself outside the cell, which allowed a more detailed structural analysis of the bundle itself, both in a horizontal (Fig. 7E and F) and a perpendicular cross-section (Fig. 7G) view. The close packing was evident in the cross-section view (Fig. 7G), with filaments spaced ∼4 nm apart. The filaments could be separated as shown in the arrows in panel G, despite the poor resolution in the Z direction. This tightly packed filamentous bundle is different from the cytoplasmic filaments found in other bacteria. For example, MreB is known to form filaments, yet the reported interfilament distance is significantly larger (44). The structural characteristics of the bundle were consistent with the molecular arrangement of DNA observed in Deinococcus radiodurans nucleoids, in which cholesteric liquid crystalline order was observed and the average interfilament distance was 4 nm (10). Therefore, we suggest that the DNA in the leptospiral nucleoids forms a long filamentous bundle along the cell body. The appearance of a liquid crystalline organization of the putative DNA is consistent with the notion that formation of a liquid crystalline arrays is required for the intracellular packaging of high-density DNA materials (similar to the situation for packaging high-copy-number supercoiled plasmids) (41).
DNA toroids have served as a model for maximally condensed packing in bacteriophages and viruses (3, 13). However, the morphology and the packing of the condensed DNA in bacteria are poorly understood. In addition, the arrangement of the genomic DNA within a long, helical bacterium has rarely been investigated. Our study provides the first structural evidence that the putative DNA forms a rod-like structure spanning the cell body. It does not organize into a toroidal shape (21) or diffuse randomly as in Deinococcus radiodurans (10). Rather, leptospiral DNA is likely packed into a filamentous bundle (41). Nevertheless, these findings raise important questions. How do spirochetes package DNA into this morphology? Why is DNA restricted from the cell end? How is the DNA bundle changed during different growth phases of Leptospira and other spirochetes? Much work will be required to answer these questions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steven Norris, Angel Paredes, Feng Xue, Dinara Yangirova, and Brian Poindexter for their comments and suggestions. We are grateful to Feng Xue for bacterial cultures. We particularly thank the reviewers for numerous critical comments and suggestions.
This work was supported in part by grants R01AI087946 (to J. Liu) and 5R01AI034431 (to D. A. Haake) from the National Institute of Allergy and Infectious Diseases (NIAID), by grant AU-1714 from the Welch Foundation (to J. Liu), and by VA Medical Research funds (to D. A. Haake).
Footnotes
Published ahead of print 6 January 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Beck M, et al. 2009. Visual proteomics of the human pathogen Leptospira interrogans. Nat. Methods 6:817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bharti AR, et al. 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 3:757–771 [DOI] [PubMed] [Google Scholar]
- 3. Bloomfield VA. 1996. DNA condensation. Curr. Opin. Struct. Biol. 6:334–341 [DOI] [PubMed] [Google Scholar]
- 4. Briegel A, et al. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. U. S. A. 106:17181–17186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bromley DB, Charon NW. 1979. Axial filament involvement in the motility of Leptospira interrogans. J. Bacteriol. 137:1406–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bulach DM, et al. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. U. S. A. 103:14560–14565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charon NW, et al. 2009. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J. Bacteriol. 191:600–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen S, et al. 2011. Structural diversity of bacterial flagellar motors. EMBO J. 30:2972–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Comolli LR, Kundmann M, Downing KH. 2006. Characterization of intact subcellular bodies in whole bacteria by cryo-electron tomography and spectroscopic imaging. J. Microsc. 223:40–52 [DOI] [PubMed] [Google Scholar]
- 10. Eltsov M, Dubochet J. 2005. Fine structure of the Deinococcus radiodurans nucleoid revealed by cryoelectron microscopy of vitreous sections. J. Bacteriol. 187:8047–8054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goldstein SF, Charon NW. 1988. Motility of the spirochete Leptospira. Cell Motil. Cytoskeleton 9:101–110 [DOI] [PubMed] [Google Scholar]
- 12. Haake DA, Matsunaga J. 2010. Leptospira: a spirochete with a hybrid outer membrane. Mol. Microbiol. 77:805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hud NV, Vilfan ID. 2005. Toroidal DNA condensates: unraveling the fine structure and the role of nucleation in determining size. Annu. Rev. Biophys. Biomol. Struct. 34:295–318 [DOI] [PubMed] [Google Scholar]
- 14. Izard J, Hsieh CE, Limberger RJ, Mannella CA, Marko M. 2008. Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. J. Struct. Biol. 163:10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Izard J, et al. 2009. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J. Bacteriol. 191:7566–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kennedy MJ, Rosey EL, Yancey RJ., Jr 1997. Characterization of flaA- and flaB- mutants of Serpulina hyodysenteriae: both flagellin subunits, FlaA and FlaB, are necessary for full motility and intestinal colonization. FEMS Microbiol. Lett. 153:119–128 [DOI] [PubMed] [Google Scholar]
- 17. Ko AI, Goarant C, Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 7:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 19. Kudryashev M, et al. 2009. Comparative cryo-electron tomography of pathogenic Lyme disease spirochetes. Mol. Microbiol. 71:1415–1434 [DOI] [PubMed] [Google Scholar]
- 20. Levett PN. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levin-Zaidman S, et al. 2003. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299:254–256 [DOI] [PubMed] [Google Scholar]
- 22. Li L, Ojcius DM, Yan J. 2007. Comparison of invasion of fibroblasts and macrophages by high- and low-virulence Leptospira strains: colonization of the host-cell nucleus and induction of necrosis by the virulent strain. Arch. Microbiol. 188:591–598 [DOI] [PubMed] [Google Scholar]
- 23. Li Z, Jensen GJ. 2009. Electron cryotomography: a new view into microbial ultrastructure. Curr. Opin. Microbiol. 12:333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu J, et al. 2010. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J. Mol. Biol. 403:546–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu J, et al. 2009. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J. Bacteriol. 191:5026–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lucic V, Forster F, Baumeister W. 2005. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 74:833–865 [DOI] [PubMed] [Google Scholar]
- 27. Macnab RM. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77–100 [DOI] [PubMed] [Google Scholar]
- 28. Malmstrom J, et al. 2009. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460:762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McBride AJ, Athanazio DA, Reis MG, Ko AI. 2005. Leptospirosis. Curr. Opin. Infect. Dis. 18:376–386 [DOI] [PubMed] [Google Scholar]
- 30. Milne JL, Subramaniam S. 2009. Cryo-electron tomography of bacteria: progress, challenges and future prospects. Nat. Rev. Microbiol. 7:666–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Motaleb MA, et al. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. U. S. A. 97:10899–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murphy GE, Leadbetter JR, Jensen GJ. 2006. In situ structure of the complete Treponema primitia flagellar motor. Nature 442:1062–1064 [DOI] [PubMed] [Google Scholar]
- 33. Murphy GE, Matson EG, Leadbetter JR, Berg HC, Jensen GJ. 2008. Novel ultrastructures of Treponema primitia and their implications for motility. Mol. Microbiol. 67:1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nahori MA, et al. 2005. Differential TLR recognition of leptospiral lipid A and lipopolysaccharide in murine and human cells. J. Immunol. 175:6022–6031 [DOI] [PubMed] [Google Scholar]
- 35. Nascimento AL, et al. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pallen MJ, Penn CW, Chaudhuri RR. 2005. Bacterial flagellar diversity in the post-genomic era. Trends Microbiol. 13:143–149 [DOI] [PubMed] [Google Scholar]
- 37. Paster BJ, et al. 1991. Phylogenetic analysis of the spirochetes. J. Bacteriol. 173:6101–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pettersen EF, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 39. Picardeau M, Brenot A, Saint Girons I. 2001. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol. Microbiol. 40:189–199 [DOI] [PubMed] [Google Scholar]
- 40. Picardeau M, et al. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3:e1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reich Z, Wachtel EJ, Minsky A. 1994. Liquid-crystalline mesophases of plasmid DNA in bacteria. Science 264:1460–1463 [DOI] [PubMed] [Google Scholar]
- 42. Ren SX, et al. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888–893 [DOI] [PubMed] [Google Scholar]
- 43. Sadziene A, Thomas DD, Bundoc VG, Holt SC, Barbour AG. 1991. A flagella-less mutant of Borrelia burgdorferi. Structural, molecular, and in vitro functional characterization. J. Clin. Invest. 88:82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Swulius MT, et al. 2011. Long helical filaments are not seen encircling cells in electron cryotomograms of rod-shaped bacteria. Biochem. Biophys. Res. Commun. 407:650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tchamedeu Kameni AP, Couture-Tosi E, Saint-Girons I, Picardeau M. 2002. Inactivation of the spirochete recA gene results in a mutant with low viability and irregular nucleoid morphology. J. Bacteriol. 184:452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024–1037 [DOI] [PubMed] [Google Scholar]
- 47. Werts C, et al. 2001. Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat. Immunol. 2:346–352 [DOI] [PubMed] [Google Scholar]
- 48. Winkler H, Taylor KA. 2006. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy 106:240–254 [DOI] [PubMed] [Google Scholar]
- 49. Xue F, Yan J, Picardeau M. 2009. Evolution and pathogenesis of Leptospira spp.: lessons learned from the genomes. Microbes Infect. 11:328–333 [DOI] [PubMed] [Google Scholar]
- 50. Yonekura K, Maki-Yonekura S, Homma M. 2011. Structure of the flagellar motor protein complex PomAB: implications for the torque-generating conformation. J. Bacteriol. 193:3863–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.