Abstract
ortho-Nitrophenol 2-monooxygenase (EC 1.14.13.31) from Alcaligenes sp. strain NyZ215 catalyzes monooxygenation of ortho-nitrophenol to form catechol via ortho-benzoquinone. Sequence analysis of this onpA-encoded enzyme revealed that it contained a flavin-binding monooxygenase domain and a heme-binding cytochrome b5 domain. OnpA was purified to homogeneity as a His-tagged protein and was considered a monomer, as determined by gel filtration. FAD and heme were identified by high-performance liquid chromatography (HPLC) and HPLC-mass spectrometry (HPLC-MS) as cofactors in this enzyme, and quantitative analysis indicated that 1 mol of the purified recombinant OnpA contained 0.66 mol of FAD and 0.20 mol of heme. However, the enzyme activity of OnpA was increased by 60% and 450% after addition of FAD and hemin, respectively, suggesting that the optimal stoichiometry was 1:1:1. In addition, site-directed mutagenesis experiments confirmed that two highly conserved histidines located in the cytochrome b5 domain were associated with binding of the heme, and the cytochrome b5 domain was involved in the OnpA activity. These results indicate that OnpA is an unusual FAD-dependent monooxygenase containing a fused cytochrome b5 domain that is essential for its activity. Therefore, we here demonstrate a link between cytochrome b5 and flavin-dependent monooxygenases.
INTRODUCTION
Heme-binding proteins are ubiquitous and involved in many bioprocesses. Cytochrome b5 (Cyt b5) is a small heme-binding protein which is widespread in eukaryotes. Its ligand heme is buried in a hydrophobic pocket and connected to two highly conserved histidine residues (28). The physiological functions of Cyt b5 are associated with fatty acid desaturation, biosynthesis of the N-glycolylneuraminic acid, plasmalogen, and cholesterol, and reduction of cytochrome P450 (35). In some cases, Cyt b5 appears as a domain fused onto proteins with other functional domains (22, 28), such as human sulfite oxidase (15), human fatty acid desaturase (8, 9), plant nitrate reductase (18, 23), yeast flavocytochrome b2 (19), and mammalian cytochrome b-type NAD(P)H oxidoreductase (44). Interestingly, all these Cyt b5 fusion proteins are found exclusively in eukaryotic lineages (22, 25, 28), not prokaryotic ones (17, 22).
Cytochrome P450 (Cyt P450) is one of the best characterized monooxygenases. It is involved in a variety of cellular reactions, such as biotransformation of drugs, bioconversion of xenobiotics, and biosynthesis of physiological compounds (3). It is also part of a superfamily of heme-binding proteins but distinct from Cyt b5; for all Cyt P450, an absolutely conserved cysteine is the fifth ligand of the heme iron (16). This differs from Cyt b5, in which the fifth and sixth ligands of the heme iron are two highly conserved histidines, which prevents its direct interaction with molecular oxygen (28).
Flavin-dependent monooxygenase is another important class of monooxygenases. In contrast to Cyt P450, which is abundant in eukaryotes, flavin-dependent monooxygenase appears to be most prevalent among prokaryotic organisms (33, 34). It plays an important role in biodegradation of aromatic compounds, drug detoxification, and biosynthesis of sterols, antibiotics, and plant hormones (1, 14). While Cyt P450 uses heme, flavin-dependent monooxygenase uses flavins, either FAD or FMN, to bind and activate oxygen for catalyzing monooxygenation. Cyt P450 gets electrons from Cyt P450 reductase or from Cyt b5 in some cases, whereas flavin-dependent monooxygenase receives electrons directly from NAD(P)H. The relationship between Cyt P450 and Cyt b5 has been broadly studied (11, 28, 42). It appears that Cyt b5 has a rather complex interaction with different Cyt P450 and can stimulate, inhibit, or have no effect on the activity of Cyt P450 through diverse strategies. However, there are no reports on interactions between Cyt b5 and flavin-dependent monooxygenase.
ortho-Nitrophenol 2-monooxygenase (EC 1.14.13.31) catalyzes monooxygenation of ortho-nitrophenol (ONP), which is known to be an aromatic pollutant as well as an indicator in enzyme assays. This enzyme was purified and characterized from the ONP-utilizing bacterium Pseudomonas putida B2 two decades ago (40), but its encoding gene, onpA (GenBank accession no. EF547253), was only recently identified in a newly isolated ONP utilizer, Alcaligenes sp. strain NyZ215 (38). OnpA converts ONP to catechol via ortho-benzoquinone, concomitant with oxygen and NADPH consumption and nitrite release. When NADPH was replaced by NADH, its activity was reduced by 92%. Sequence analysis revealed a Cyt b5 domain in this protein. However, the involvement of Cyt b5 in ONP oxidation has not previously been reported. Here we report that the OnpA from strain NyZ215 is an unusual FAD-dependent monooxygenase containing a cytochrome b5-like heme binding domain and OnpA activity is dependent on the binding of heme to the Cyt b5 domain.
MATERIALS AND METHODS
Expression and purification of OnpA in E. coli.
The gene onpA was PCR amplified from Alcaligenes sp. strain NyZ215 (38) with the high-fidelity DNA polymerase Pyrobest (Takara) using a pair of primers (forward, 5′-GAGGGATCCATGCGAGCTGTCATTATCG-3′; reverse, 5′-GGCAAGCTTAATATCAAGCCGTATGAGGC-3′). The PCR product containing onpA was double-digested with BamHI and HindIII and then ligated into similarly treated pET28a to form the expression construct pZWX28A, in which an N-terminal six-His-tag-encoding sequence was fused into onpA. Its sequence was verified by DNA sequencing to ensure that no mutations were incorporated. Escherichia coli BL21(DE3) carrying pZWX28A was grown on lysogeny broth (LB) at 37°C to an A600 of around 0.7 and induced by the addition of 0.1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) for 3 to 4 h at 20°C. The cells were harvested and stored at −80°C until use.
All steps for purification were performed at 4°C. After the cells were disrupted by sonication in the binding buffer (300 mM NaCl, 50 mM sodium phosphate buffer [pH 8.0]), the cellular lysate was centrifuged at 13,500 × g for 30 min, and the supernatant was taken for protein purification. OnpA was then purified via affinity chromatography using nickel-nitrilotriacetic acid agarose (Merck Biosciences) according to the supplier's recommendations. Finally, imidazole and salts were removed from the eluted fractions by overnight dialysis against 20 mM sodium phosphate buffer (pH 7.5).
Determination of molecular mass of OnpA.
The molecular mass of OnpA was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gel filtration chromatography was used to determine the native molecular mass of OnpA. The experiment was performed on an Äkta fast-performance liquid chromatography system (Amersham Pharmacia Biotech) using a Superdex 200 10/300 GL column (Amersham Pharmacia Biotech). The 50 mM phosphate buffer (pH 7.2) contained 0.15 M NaCl, and the flow rate was 0.25 ml/min. The native molecular mass was estimated from a calibration curve plotted using standard proteins (Sigma): carbonic anhydrase (29 kDa), bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), β-amylase (200 kDa), apoferritin (443 kDa), and thyroglobulin (669 kDa).
Enzyme assay and kinetic measurement.
ortho-Nitrophenol 2-monooxygenase activity was determined spectrophotometrically by measuring the decrease in ONP absorbance at 410 nm due to substrate consumption (40). The molar extinction coefficients for ONP and NADPH were 3.47 mM−1 cm−1 at 410 nm and 6.22 mM−1 cm−1 at 340 nm, respectively. The standard enzyme assay mixture contained 40 nM purified OnpA, 0.05 mM ONP, 0.2 mM NADPH, and 4 mM Mg2+ in 500 μl (final volume) of phosphate buffer (20 mM, pH 7.5). The assay was initiated through addition of substrate ONP. The halogen-containing salts, hemin, protoporphyrin IX, FAD, and FMN were used to test their impact on OnpA activity. The kinetic analysis was carried out following the method described (40). ONP was varied from 1 to 100 μM when NADPH was constant at 0.5 mM; NADPH was varied from 20 to 400 μM when ONP was constant at 0.03 mM. One unit of enzyme activity was defined as the amount required for the disappearance of 1 μmol of substrate per minute at room temperature. Specific activities are expressed as units per gram of protein. Protein concentration was determined by the Bradford method with bovine serum albumin as the standard.
Isolation and determination of the cofactors from OnpA.
The flavin cofactor was released by treating 400 μl of OnpA (0.3 mg/ml) with 1 mg/ml proteinase K for 2 h at room temperature in the dark (30), followed by centrifugation at 13,500 × g for 10 min. The supernatant was subjected to HPLC analysis.
The method for heme extraction was described previously (24). Briefly, 50 μl of solution containing OnpA was mixed with 450 μl of acetone-HCl (19:1, vol/vol) by gentle shaking at room temperature for 20 min. After centrifugation at 13,500 × g for 5 min, 1 ml of ice-cold water and 0.3 ml of ethyl acetate were added to the supernatant, followed by vortexing and centrifugation. The ethyl acetate phase was recovered, and the solvent was removed with a vacuum concentrator. The residue was dissolved in 50 μl of acetonitrile and analyzed immediately. Myoglobin (Sigma) was used as a positive control for heme extraction and identification.
Analytical methods.
To analyze the flavin extracted from OnpA, high-performance liquid chromatography (HPLC) analysis was performed using a Gilson 715 system equipped with a C18 reversed-phase column from Supelco (250 by 4.6 mm, 5 μm) with a column temperature of 30°C. The mobile phase consisted of methanol (40%) and 10 mM H3PO4 (60%) at a flow rate of 1.0 ml/min. The flavins were monitored at 450 nm with a Gilson 119 UV/Vis detector. The retention times of the authentic FAD and FMN (Sigma) were 3.53 and 4.14 min, respectively. FAD concentration was determined by reference to a standard of known concentration.
To identify the heme extracted from OnpA and myoglobin, HPLC-MS (mass spectrometry) analysis was carried out on an Agilent 1200 series HPLC system (Agilent Technologies) consisting of a quaternary solvent delivery system, an on-line degasser, an autosampler, a column temperature controller, and a diode array detector. The mass spectra were acquired using a micro-QTOF mass spectrometer (Bruker Daltonics) equipped with an ESI (electrospray ionization) interface. Five percent of the eluent was directed to MS using a Bruker nuclear magnetic resonance-MS interface unit (Bruker BioSpin). Chromatographic separation was carried out on an ACE C18-HL (Hi-Load) column (5 μm, 250 by 4.6 mm; Advanced Chromatography Technologies) with a C18 guard column at 25°C. The detection wavelength was set at 405 nm. The mobile phase consisted of water (A) and acetonitrile (B), both containing 0.1% (vol/vol) formic acid. A gradient program was used as follows: 0 to 5 min, 20% B; 5 to 15 min, 20 to 80% B; 15 to 20 min, 80 to 100% B. The flow rate was 1.0 ml/min. The optimized mass spectrometric parameters were as follows: positive-ion mode; capillary voltage, 4,500 V; nebulizer gas pressure, 0.8 × 105 Pa; drying gas flow rate, 8 liter/min; gas temperature, 180°C. The spectra were recorded in the range of m/z 50 to 1,000.
UV-visible absorption spectra were recorded with a Perkin Elmer Lambda 25 UV/Vis spectrometer. The concentration of heme was determined using an absorption coefficient of 130 mM−1 cm−1 at 413 nm (2).
Site-directed mutagenesis.
To confirm coordination of the two highly conserved histidines with the heme in the cytochrome b5 domain, both of them were individually replaced by aspartic acids. Overlapping PCR was adopted to construct the site-directed mutants. The desired mutations were identified by DNA sequencing. Two mutant proteins, OnpA(H515D) and OnpA(H538D), were expressed and purified by the methods described above.
RESULTS
Sequence analysis.
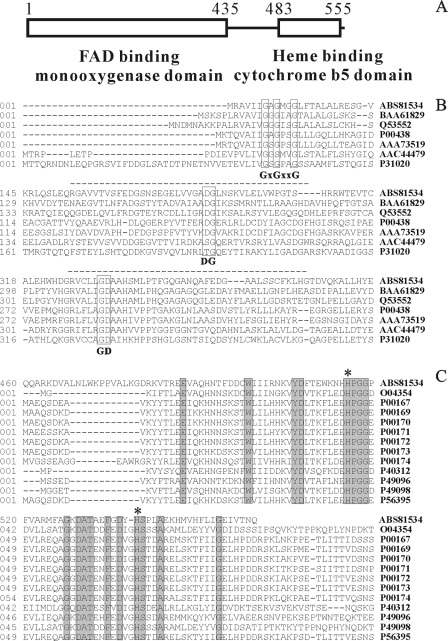
Bioinformatic analysis of OnpA using the software protein blast (available at http://blast.ncbi.nlm.nih.gov/) revealed that this 558-amino-acid protein has a salicylate 1-monooxygenase domain (bit score, 121.1; E-value, 5.34e−28) located between positions 1 and 435 in the N terminus and a cytochrome b5-like heme binding domain (bit score, 99.59; E-value, 1.47e−21) between positions 483 and 555 in the C terminus (Fig. 1A). The salicylate 1-monooxygenase is characteristic of a flavin-dependent monooxygenase (39). Thus, OnpA and other known flavin-dependent monooxygenases (14), including phenol hydroxylase, 4-methyl-5-nitrocatechol oxygenase, 4-hydroxybenzoate hydroxylase, and salicylate 1-monooxygenase, were selected for sequence alignment (Fig. 1B) using Clustal W software. The result showed that the motifs (GXGXXG, DG, and GD) associated with monooxygenase activity and that FAD and NAD(P)H binding (14) are conserved in OnpA. However, OnpA is about 100 amino acids longer than all the other flavin-dependent monooxygenases examined. Sequence alignment (Fig. 1C) of the Cyt b5 domain from OnpA against various independent b5 cytochromes (28) from discrete eukaryotic origins suggested that as many as 18 amino acid residues were conserved. Most importantly, the two highly conserved histidines which form the fifth and sixth ligands of the heme iron (22, 25, 28) were found in OnpA (H515 and H538). However, the carboxyl-terminal membrane anchor domain (11, 12, 28) was not observed in OnpA or in a bacterial Cyt b5 of unknown function (21, 28). Interestingly, the Cyt b5 in OnpA has moderate identity (40 to 50%) to the listed eukaryotic Cyt b5 but a lower identity (37%) to the bacterial Cyt b5. A phylogenetic tree (Fig. 2) for Cyt b5 from different sources was constructed with Mega 5.0 (32) using the neighbor-joining method. This shows that the Cyt b5 in OnpA has a closer phylogenetic relationship to independent eukaryotic Cyt b5 than those originating from bacteria or other fusion Cyt b5, implying that the Cyt b5 domain of onpA is of eukaryotic origin.
Fig 1.
(A) Outline of OnpA primary structure. (B) Multiple-sequence alignment of flavin-dependent monooxygenases and the N terminus of OnpA. (C) Multiple-sequence alignment of b5 cytochromes and the C terminus of OnpA. GenBank accession numbers are provided on the right. OnpA is available under accession no. ABS81534. The selected flavin-dependent monooxygenases (14) include phenol hydroxylases (P31020), 4-methyl-5-nitrocatechol oxygenase (AAC44479), 4-hydroxybenzoate hydroxylases (AAA73519 and P00438), salicylate monooxygenase (BAA61829 and Q53552). The conserved motifs GXGXXG, DG, and GD are boxed and labeled. The sequences of b5 cytochromes obtained from mouse (P56395), tobacco (P49098), housefly (P49096), yeast (P40312), chicken (P00174), rat (P00173), pig (P00172), bovine (P00171), horse (P00170), rabbit (P00169), human (P00167), and borage (O04354) were all collected from a published review (28). Gray shading indicates conserved residues, and asterisks indicate two histidines coordinating with the heme.
Fig 2.
Phylogenetic tree of cytochrome b5 (Cyt b5) from OnpA, independent Cyt b5 from eukaryotic and prokaryotic origins, and fused Cyt b5. The phylogenetic tree was constructed (using the neighbor-joining method) with Mega 5 software. The scale at the bottom indicates sequence divergence. OnpA is indicated by a plus sign.
It is very unusual to find a combination of a flavin-dependent monooxygenase and a cytochrome b5 in a single polypeptide. Besides OnpA, Blast analysis of the current entire GenBank database showed that only an unidentified open reading frame (accession no. AAV96388) from the strain Ruegeria (formerly Silicibacter) pomeroyi DSS-3 encoded a protein with the same domain organization. This 563-amino-acid putative monooxygenase exhibits 63% identity to OnpA, but its function is unknown, and no genes encoding enzymes involved in ONP catabolism were found in close proximity to it.
Properties of OnpA.
OnpA was overexpressed in E. coli BL21(DE3) and purified by Ni2+-nitrilotriacetic acid affinity chromatography with a specific activity of 450 U g−1. The purified protein was tinged brown, a color clearly distinct from the typical red hue of cytochrome b5 and the yellow of flavin-containing proteins. A single band of OnpA with an apparent molecular mass of 65 kDa was detected by SDS-PAGE (data not shown), consistent with the molecular mass of OnpA as deduced from its amino acid sequence. The native molecular mass of OnpA was calculated to be 71.8 ± 4.0 kDa by gel filtration chromatography, indicating that OnpA was a monomer. In 20 mM phosphate buffer (pH 7.5), OnpA was stable at 4°C for 1 week without any apparent loss of activity. If kept in binding buffer at 4°C, the purified OnpA could be stored for 1 month with more than 75% of the initial activity. When OnpA was incubated at 40°C for 5 min, complete loss of enzyme activity was observed.
The OnpA activity measured spectrophotometrically showed that the purified recombinant OnpA catalyzed degradation of ONP (λmax, 410 nm), together with consumption of NADPH (λmax, 340 nm) and formation of catechol (λmax, 270 nm), as previously observed using cell extracts (38). However, both mutant proteins OnpA(H515D) and OnpA(H538D) completely lost enzyme activity, confirming the importance of these two highly conserved histidines in Cyt b5 and the involvement of the Cyt b5 domain in OnpA activity. In addition, Mg2+, Ca2+, Mn2+, and Co2+ enhanced OnpA activity, whereas Na+, K+, Fe2+, Cu2+, Zn2+, and Ni2+ had no effect or a negative effect. The Km values (mean ± standard error) of OnpA for NADPH and ONP were 35.75 ± 9.85 and 2.11 ± 0.15 μM, respectively; the corresponding kcat values (mean ± standar error) for NADPH and ONP were 52.5 ± 2.5 and 25.0 ± 0.25 min−1, respectively.
In the process of OnpA purification, inhibition of activity by NaCl was observed. Additionally, lower activity was observed in Tris-Cl buffer than in phosphate buffer (38, 40). These observations established that the inhibition factor was chloride ion. Chloride ions are known to compete with aromatic substrate (13) or NAD(P)H (29) in flavin-dependent monooxygenases, causing enzymatic inhibition. Table 1 shows that halogen ions, including F−, Cl−, Br−, and I−, inhibited OnpA activity, but Na2SO4 had no obvious effect. At 75 mM halogen ions, the enzyme lost over 50% of its activity. Salicylate 1-monooxygenase has also been shown to be inhibited by halogen ions (36), suggesting that OnpA shares some characteristics with salicylate 1-monooxygenase. In addition, 75 mM nitrate and nitrite also inhibited OnpA activity by 50% and 25%, respectively.
Table 1.
Effect of halogens on OnpA activity
| Chemical | Relative OnpA activity (%) at concn (mM)a |
|||||
|---|---|---|---|---|---|---|
| 0 | 10 | 25 | 50 | 75 | 100 | |
| Na2SO4 | 100.0 ± 6.9 | 97.9 ± 5.2 | 97.9 ± 5.4 | 87.6 ± 0.9 | 84.2 ± 10.0 | 85.9 ± 3.0 |
| NaCl | 86.3 ± 1.3 | 69.2 ± 2.0 | 58.8 ± 2.1 | 47.8 ± 1.3 | 38.5 ± 1.6 | |
| KCl | 79.5 ± 5.4 | 67.0 ± 5.2 | 41.2 ± 3.5 | 38.9 ± 1.5 | 31.2 ± 3.0 | |
| NaF | 58.4 ± 3.0 | 39.5 ± 5.9 | 30.9 ± 5.2 | 20.6 ± 5.2 | ND | |
| NaBr | 96.2 ± 3.0 | 60.1 ± 2.9 | 43.0 ± 3.0 | 29.2 ± 2.9 | 0 | |
| NaI | 56.7 ± 5.2 | 41.2 ± 8.9 | 24.1 ± 3.0 | 18.9 ± 2.9 | ND | |
The activity determined by the standard enzyme assay of 450 U g−1 was set as 100%. The values are means ± standard deviations calculated from triplicate assays. ND, no determination.
Identification of FAD and heme as cofactors in OnpA.
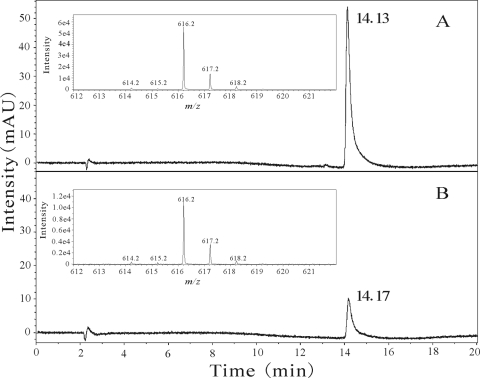
The flavin isolated from OnpA was confirmed to be FAD, not FMN, by HPLC analysis, since it had the same retention time as authentic FAD. Quantitative analysis via HPLC indicated that 1 mol of OnpA contained 0.66 ± 0.03 mol of FAD. The b-type heme extracted from OnpA was confirmed by HPLC-MS (Fig. 3). The retention times of hemes extracted from OnpA and myoglobin were 14.17 and 14.13 min, respectively. Moreover, their mass spectra were found to be virtually identical to each other and also identical to results described elsewhere (4). The highest molecular ion peak at m/z 616.2 corresponded to the mass of b-type heme composed of only 12C, 14N, and 56Fe. The other peaks were consistent with the natural abundance of 13C, 15N, 54Fe, 56Fe, 57Fe, and 58Fe. Hemes extracted from both OnpA and myoglobin were also treated with pyridine under alkaline conditions, and their corresponding pyridine hemochromes had the same characteristic absorption spectra. This evidence strongly suggests that OnpA, like myoglobin, contains a b-type heme. Quantitative analysis based on absorption coefficient revealed that 1 mol of purified OnpA contained 0.2 ± 0.06 mol of b-type heme. As b-type heme is widely distributed among organisms, OnpA is able to exhibit its activity in several different genera, such as Alcaligenes (38), Pseudomonas (38, 40), and Escherichia (38).
Fig 3.
LC-MS analysis of the hemes extracted from equine myoglobin (A) and OnpA (B). (Insets) Mass spectra of the corresponding hemes.
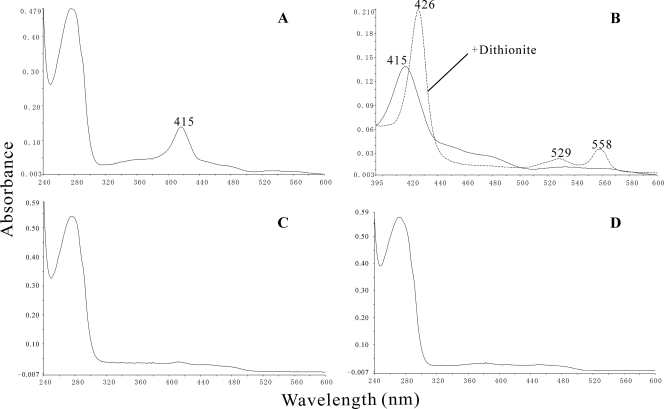
UV-visible absorbance spectra of OnpA and its mutants.
Intriguingly, in addition to a typical absorption peak of protein appeared at 280 nm, a distinct absorption maximum at 415 nm was also found in OnpA (Fig. 4A). This had previously been observed by Zeyer et al. for ortho-nitrophenol 2-monooxygenase from strain B2 but without further explanation (40). The molecular extinction coefficient of OnpA at 415 nm was calculated to be 25.7 ± 8.1 mM−1 cm−1, which was close to the value of 40 mM−1 cm−1 reported by Zeyer et al. (40). Based on the above results and other reports on Cyt b5 (5, 21, 44), we believe that the peak at 415 nm is due to the presence of a Cyt b5 domain. To confirm this hypothesis, sodium dithionite was added to the purified OnpA and a typical absorption spectrum of reduced cytochrome b was generated with peaks at 558, 529, and 426 nm, respectively (Fig. 4B), which are characteristic features of cytochrome b (5, 20, 44). The spectrum change strongly indicated the presence of a b-type heme in OnpA. However, this characteristic absorption spectrum change was not observed in OnpA with excess NADPH, differentiating OnpA from a flavin-containing reductase-Cyt b5 fusion, which was reduced by NADH via flavin (44). Notably, the spectra of both mutant proteins OnpA(H515D) and OnpA(H538D) lost the maximum absorption at 415 nm (Fig. 4C and D), confirming the importance of these residues as heme ligands in the Cyt b5 domain. Both mutants were also devoid of enzyme activity.
Fig 4.
UV-visible absorption spectra of OnpA (A), OnpA(H515D) (C), and OnpA(H538D) (D) and dithionite-reduced and air-oxidized OnpA (B) in 20 mM phosphate buffer, pH 7.5. The relevant absorption peaks are labeled.
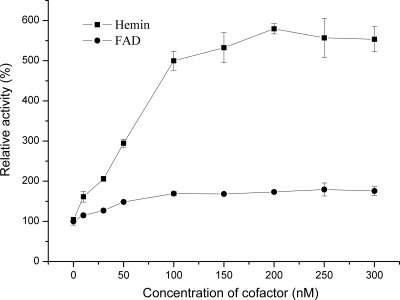
FAD and hemin enhance OnpA activity.
Both FAD and heme have been identified as cofactors in OnpA, whereas the ratio of both FAD and heme to protein was less than 1:1. Therefore, the cofactors were added individually to check for their effects on OnpA activity. FAD enhanced the ONP 2-monooxygenase activity by about 60% (Fig. 5), consistent with the observed content of 0.66 mol of FAD per mol in recombinant OnpA. This moderate enhancement by FAD was also observed for the ortho-nitrophenol 2-monooxygenase from strain B2 (40). However, 30% of OnpA activity was lost by addition of 10 μM FMN (data not shown).
Fig 5.
Enhancement of OnpA activity by hemin and FAD. OnpA activities were determined in a standard enzyme assay (40 nM OnpA, 0.05 mM ONP, 0.2 mM NADPH, and 4 mM Mg2+ in 500 μl [final volume] of phosphate buffer [20 mM, pH 7.5]). The OnpA activity without the addition of FAD or hemin is defined as 100%. The OnpA activity was increased by a further 10% in the presence of both hemin and FAD at a saturation concentration, in comparison with that obtained with hemin alone at a saturation concentration. Values are means ± standard deviations.
Recently, Kimura et al. reported that the neurotropic activity of neudesin is dependent on the binding of heme to its Cyt b5 domain (20). According to their method, hemin (a derivative of heme) was added to the enzymatic reaction of OnpA. Addition of hemin (dissolved in dimethyl sulfoxide) dramatically increased OnpA activity in a concentration-dependent manner (Fig. 5). When the concentration was over 0.1 μM, the activity was enhanced about 4.5 times. No apparent further improvement was found by adding more hemin, implying that the OnpA was saturated by hemin at this concentration. Given the earlier evidence that only 1/5 of the purified recombinant OnpA contained heme, it is entirely consistent that OnpA saturated with hemin had nearly 5 times the activity of the original OnpA without additional hemin. The activity was not increased by substituting protoporphyrin IX for hemin, strongly indicating that OnpA activity was dependent on heme binding to its Cyt b5 domain.
DISCUSSION
An ortho-nitrophenol 2-monooxygenase was previously purified from Pseudomonas putida B2 and characterized (40, 41). Although its sequence is still unknown, this enzyme shares many characteristics with OnpA, including size, substrate range, and UV-visible spectra (38, 40, 41; also this study). A peak at 415 nm in the absorption spectrum of the nitrophenol monooxygenase from strain B2 was also observed (40), but without amino acid sequence information, the presence of a heme-binding Cyt b5 domain was not recognized. Our results, including those obtained by sequence analysis, identification of b-type heme by LC-MS, site-directed mutagenesis, and enzymatic assay, show that a cytochrome b5 domain contributes the absorbance at 415 nm and is intimately involved in OnpA activity. Besides being heme dependent, OnpA was also identified as being FAD dependent. Addition of FAD gave a 10% increase in activity of the ortho-nitrophenol 2-monooxygenase from strain B2 (41), whereas with OnpA, activity was stimulated by about 60%, perhaps due to the overexpression of OnpA in E. coli. Probably, like OnpA, the P. putida B2 enzyme also contains two domains, an FAD-dependent monooxygenase domain and a heme-binding Cyt b5, both of which are essential to OnpA activity. Despite the ubiquity of Cyt b5 in eukaryotes (22, 25, 28, 31, 35), it is very rarely found in bacteria (31). A soluble form of Cyt b5 from Ectothiorhodospira vacuolata has been purified and structurally characterized, but its function is still unknown (21). In addition, an annotated but not verified desaturase with a cytochrome b5 fusion is present in the genome of Mycobacterium tuberculosis (10). Although many Cyt b5 fusion proteins have been found and studied in eukaryotic cells (22, 25, 28, 44), no functional Cyt b5 fusion protein has previously been identified in prokaryotes. This study therefore not only links Cyt b5 to flavin-dependent monooxygenases but also extends the biological range and functional role of Cyt b5.
Flavin-dependent monooxygenases are reported to be involved in many bioprocesses (1, 14, 34). In particular, flavin-dependent monooxygenases in liver microsomes catalyze the NADPH-dependent N- or S-oxygenation of heteroatom-containing compounds in coordination with Cyt P450, diversifying the metabolism of drugs and other compounds (6, 7, 34). Cyt P450 is an important enzyme associated with biotransformation and biodegradation of drugs in liver (3, 7), and its activity is affected by Cyt b5 through different mechanisms (11, 28, 42). However, there is no report on the interaction of flavin-dependent monooxygenase and Cyt b5. Our study proves that a Cyt b5 imposes a significant effect on the activity of a flavin-dependent monooxygenase. This raises an interesting question as to whether the activities of other flavin-dependent monooxygenases are affected by Cyt b5, particularly those involved in drug metabolism in the liver.
Cyt b5, in either an independent or a fused form, is known to be an intermediate electron transfer component (22, 28, 35). In sulfite oxidase, the electrons are transferred from molybdenum to cytochrome c via Cyt b5 (27); in fatty acid desaturase, the electrons are transferred from NADH cytochrome b5 reductase to Cyt b5 and then to the terminal desaturase (26); in nitrate reductase, the electron flow is from NADH to FAD to Cyt b5 to molybdopterin to nitrate (23); and in cytochrome b2, the electron transfer is from the flavin to cytochrome c via Cyt b5 (37). Cyt b5 is also involved in the Cyt P450 system, and its roles were summarized in a review (28). In contrast, the role of the Cyt b5 domain in OnpA is intriguing. A characteristic of Cyt b5 is that histidine is coordinated to the sixth position of the heme iron, preventing its direct interaction with oxygen (28). Thus, as in other flavin-dependent monooxygenases, the flavin group is likely to be the site of oxygen activation, and this is consistent with the sequence similarity of the flavin domain to classical flavin-dependent monooxygenases. Indeed, para-nitrophenol 4-monooxygenase from Pseudomonas sp. strain WBC-3, which effectively catalyzes the same reaction but with a different regiochemistry, is a simple flavoprotein with no Cyt b5 domain (43). This observation indicates a possible role for Cyt b5. In the majority of flavin-dependent monooxygenases interacting with aromatic substrates, hydroxylation results from a nucleophilic attack of the substrate on the C4a-flavin hydroperoxide (1). However, the strong electron-withdrawing properties of the nitro group reduce the nucleophilicity of nitrophenols. Thus, the attack on nitrophenols must involve a nucleophilic attack by the C4a-flavin peroxide, which would then resolve to form the corresponding quinone. This is clearly the case with para-nitrophenol 4-monooxygenase, as para-benzoquinone has been identified as a product and a benzoquinone reductase gene forms part of the operon in strain WBC-3 (43). A quinone reductase gene also forms part of the onp operon in strain NyZ215, but in the assay of OnpA, no quinone was detected, only catechol (38). This is because ortho-benzoquinone is considerably less stable than para-benzoquinone, readily reducing to catechol in vitro (40). Although it is possible that Cyt b5 acts as an electron transfer route to reduce the unstable ortho-quinone in the active site of OnpA, we think that this is unlikely, given that consecutive 1e− reductions would deliberately produce the highly reactive semiquinone as an intermediate. Instead, we propose that the role of the Cyt b5 domain is to protect OnpA from accidental formation of the semiquinone in the active site. Although the lack of OnpA activity in the absence of heme binding might indicate that OnpA is being inactivated in the absence of this protective mechanism, it could be that heme binding triggers a conformational change which allows activity to occur; i.e., it has an additional regulatory role. Nevertheless, it is clear that more information, such as structural data, is needed to elucidate the actual roles of the Cyt b5 domain in OnpA. We also believe that more functions of cytochrome b5 will be discovered in the future, enabling us to further understand the interaction between monooxygenases and cytochrome b5.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (grant 30730002) and the National Basic Research Program of China (973 Program) (grant 2012CB721003). Y.X. is a recipient of funds from the Knowledge Innovation Program of the Chinese Academy of Sciences for young scientists.
Footnotes
Published ahead of print 20 January 2012
REFERENCES
- 1. Ballou DP, Entsch B, Cole LJ. 2005. Dynamics involved in catalysis by single-component and two-component flavin-dependent aromatic hydroxylases. Biochem. Biophys. Res. Commun. 338:590–598 [DOI] [PubMed] [Google Scholar]
- 2. Barber MJ, Desai SK, Marohnic CC, Hernandez HH, Pollock VV. 2002. Synthesis and bacterial expression of a gene encoding the heme domain of assimilatory nitrate reductase. Arch. Biochem. Biophys. 402:38–50 [DOI] [PubMed] [Google Scholar]
- 3. Bernhardt R. 2006. Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 124:128–145 [DOI] [PubMed] [Google Scholar]
- 4. Buchenau B, Kahnt J, Heinemann IU, Jahn D, Thauer RK. 2006. Heme biosynthesis in Methanosarcina barkeri via a pathway involving two methylation reactions. J. Bacteriol. 188:8666–8668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burchell A. 1985. A simple method for purification of rat hepatic microsomal cytochrome b5. Biochem. J. 226:339–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cashman JR. 2004. The implications of polymorphisms in mammalian flavin-containing monooxygenases in drug discovery and development. Drug Discov. Today 9:574–581 [DOI] [PubMed] [Google Scholar]
- 7. Cashman JR. 2005. Some distinctions between flavin-containing and cytochrome P450 monooxygenases. Biochem. Biophys. Res. Commun. 338:599–604 [DOI] [PubMed] [Google Scholar]
- 8. Cho HP, Nakamura M, Clarke SD. 1999. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J. Biol. Chem. 274:37335–37339 [DOI] [PubMed] [Google Scholar]
- 9. Cho HP, Nakamura MT, Clarke SD. 1999. Cloning, expression, and nutritional regulation of the mammalian delta-6 desaturase. J. Biol. Chem. 274:471–477 [DOI] [PubMed] [Google Scholar]
- 10. Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 11. Durr UH, Waskell L, Ramamoorthy A. 2007. The cytochromes P450 and b5 and their reductases—promising targets for structural studies by advanced solid-state NMR spectroscopy. Biochim. Biophys. Acta 1768:3235–3259 [DOI] [PubMed] [Google Scholar]
- 12. Durr UH, Yamamoto K, Im SC, Waskell L, Ramamoorthy A. 2007. Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b5. J. Am. Chem. Soc. 129:6670–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eppink MH, Boeren SA, Vervoort J, van Berkel WJ. 1997. Purification and properties of 4-hydroxybenzoate 1-hydroxylase (decarboxylating), a novel flavin adenine dinucleotide-dependent monooxygenase from Candida parapsilosis CBS604. J. Bacteriol. 179:6680–6687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eppink MH, Schreuder HA, Van Berkel WJ. 1997. Identification of a novel conserved sequence motif in flavoprotein hydroxylases with a putative dual function in FAD/NAD(P)H binding. Protein Sci. 6:2454–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garrett RM, Bellissimo DB, Rajagopalan KV. 1995. Molecular cloning of human liver sulfite oxidase. Biochim. Biophys. Acta 1262:147–149 [DOI] [PubMed] [Google Scholar]
- 16. Graham SE, Peterson JA. 1999. How similar are P450s and what can their differences teach us? Arch. Biochem. Biophys. 369:24–29 [DOI] [PubMed] [Google Scholar]
- 17. Hongsthong A, et al. 2006. Revealing the complementation of ferredoxin by cytochrome b 5 in the Spirulina-Δ6-desaturation reaction by N-terminal fusion and co-expression of the fungal-cytochrome b 5 domain and Spirulina-Δ6-acyl-lipid desaturase. Appl. Microbiol. Biotechnol. 72:1192–1201 [DOI] [PubMed] [Google Scholar]
- 18. Hyde GE, Crawford NM, Campbell WH. 1991. The sequence of squash NADH:nitrate reductase and its relationship to the sequences of other flavoprotein oxidoreductases. A family of flavoprotein pyridine nucleotide cytochrome reductases. J. Biol. Chem. 266:23542–23547 [PubMed] [Google Scholar]
- 19. Kay CJ, Lippay EW. 1992. Mutation of the heme-binding crevice of flavocytochrome b2 from Saccharomyces cerevisiae: altered heme potential and absence of redox cooperativity between heme and FMN centers. Biochemistry 31:11376–11382 [DOI] [PubMed] [Google Scholar]
- 20. Kimura I, et al. 2008. Neurotrophic activity of neudesin, a novel extracellular heme-binding protein, is dependent on the binding of heme to its cytochrome b5-like heme/steroid-binding domain. J. Biol. Chem. 283:4323–4331 [DOI] [PubMed] [Google Scholar]
- 21. Kostanjevecki V, et al. 1999. Structure and characterization of Ectothiorhodospira vacuolata cytochrome b558, a prokaryotic homologue of cytochrome b5. J. Biol. Chem. 274:35614–35620 [DOI] [PubMed] [Google Scholar]
- 22. Lederer F. 1994. The cytochrome b5-fold: an adaptable module. Biochimie 76:674–692 [DOI] [PubMed] [Google Scholar]
- 23. Lu G, Lindqvist Y, Schneider G, Dwivedi U, Campbell W. 1995. Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain. J. Mol. Biol. 248:931–948 [DOI] [PubMed] [Google Scholar]
- 24. Lubben M, Morand K. 1994. Novel prenylated hemes as cofactors of cytochrome oxidases. Archaea have modified hemes A and O. J. Biol. Chem. 269:21473–21479 [PubMed] [Google Scholar]
- 25. Napier JA, Sayanova O, Stobart AK, Shewry PR. 1997. A new class of cytochrome b5 fusion proteins. Biochem. J. 328(Pt. 2):717–718 [PMC free article] [PubMed] [Google Scholar]
- 26. Rioux V, Pedrono F, Legrand P. 2011. Regulation of mammalian desaturases by myristic acid: N-terminal myristoylation and other modulations. Biochim. Biophys. Acta 1811:1–8 [DOI] [PubMed] [Google Scholar]
- 27. Rudolph MJ, Johnson JL, Rajagopalan KV, Kisker C. 2003. The 1.2 Å structure of the human sulfite oxidase cytochrome b5 domain. Acta Crystallogr. D Biol. Crystallogr. 59:1183–1191 [DOI] [PubMed] [Google Scholar]
- 28. Schenkman JB, Jansson I. 2003. The many roles of cytochrome b5. Pharmacol. Ther. 97:139–152 [DOI] [PubMed] [Google Scholar]
- 29. Seibold B, et al. 1996. 4-Hydroxybenzoate hydroxylase from Pseudomonas sp. CBS3. Purification, characterization, gene cloning, sequence analysis and assignment of structural features determining the coenzyme specificity. Eur. J. Biochem. 239:469–478 [DOI] [PubMed] [Google Scholar]
- 30. Somerville CC, Nishino SF, Spain JC. 1995. Purification and characterization of nitrobenzene nitroreductase from Pseudomonas pseudoalcaligenes JS45. J. Bacteriol. 177:3837–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sperling P, Ternes P, Zank TK, Heinz E. 2003. The evolution of desaturases. Prostaglandins Leukot. Essent. Fatty Acids 68:73–95 [DOI] [PubMed] [Google Scholar]
- 32. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Torres Pazmino DE, Winkler M, Glieder A, Fraaije MW. 2010. Monooxygenases as biocatalysts: classification, mechanistic aspects and biotechnological applications. J. Biotechnol. 146:9–24 [DOI] [PubMed] [Google Scholar]
- 34. van Berkel WJ, Kamerbeek NM, Fraaije MW. 2006. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 124:670–689 [DOI] [PubMed] [Google Scholar]
- 35. Vergeres G, Waskell L. 1995. Cytochrome b5, its functions, structure and membrane topology. Biochimie 77:604–620 [DOI] [PubMed] [Google Scholar]
- 36. White-Stevens RH, Kamin H. 1972. Studies of a flavoprotein, salicylate hydroxylase. I. Preparation, properties, and the uncoupling of oxygen reduction from hydroxylation. J. Biol. Chem. 247:2358–2370 [PubMed] [Google Scholar]
- 37. Xia ZX, et al. 1987. Three-dimensional structure of flavocytochrome b2 from baker's yeast at 3.0-A resolution. Proc. Natl. Acad. Sci. U. S. A. 84:2629–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiao Y, Zhang JJ, Liu H, Zhou NY. 2007. Molecular characterization of a novel ortho-nitrophenol catabolic gene cluster in Alcaligenes sp. strain NyZ215. J. Bacteriol. 189:6587–6593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamamoto S, Katagiri M, Maeno H, Hayaishi O. 1965. Salicylate hydroxylase, a monooxygenase requiring flavin adenine dinucleotide. I. Purification and general properties. J. Biol. Chem. 240:3408–3413 [PubMed] [Google Scholar]
- 40. Zeyer J, Kocher HP. 1988. Purification and characterization of a bacterial nitrophenol oxygenase which converts ortho-nitrophenol to catechol and nitrite. J. Bacteriol. 170:1789–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zeyer J, Kocher HP, Timmis KN. 1986. Influence of para-substituents on the oxidative metabolism of o-nitrophenols by Pseudomonas putida B2. Appl. Environ. Microbiol. 52:334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang H, Myshkin E, Waskell L. 2005. Role of cytochrome b5 in catalysis by cytochrome P450 2B4. Biochem. Biophys. Res. Commun. 338:499–506 [DOI] [PubMed] [Google Scholar]
- 43. Zhang JJ, Liu H, Xiao Y, Zhang XE, Zhou NY. 2009. Identification and characterization of catabolic para-nitrophenol 4-monooxygenase and para-benzoquinone reductase from Pseudomonas sp. strain WBC-3. J. Bacteriol. 191:2703–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu H, Qiu H, Yoon HW, Huang S, Bunn HF. 1999. Identification of a cytochrome b-type NAD(P)H oxidoreductase ubiquitously expressed in human cells. Proc. Natl. Acad. Sci. U. S. A. 96:14742–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]