Abstract
The VicRK two-component signaling system modulates biofilm formation, genetic competence, and stress tolerance in Streptococcus mutans. We show here that the VicRK modulates bacteriocin production and cell viability, in part by direct modulation of competence-stimulating peptide (CSP) production in S. mutans. Global transcriptome and real-time transcriptional analysis of the VicK-deficient mutant (SmuvicK) revealed significant modulation of several bacteriocin-related loci, including nlmAB, nlmC, and nlmD (P < 0.001), suggesting a role for the VicRK in producing mutacins IV, V, and VI. Bacteriocin overlay assays revealed an altered ability of the vic mutants to kill related species. Since a well-conserved VicR binding site (TGTWAH-N5-TGTWAH) was identified within the comC coding region, we confirmed VicR binding to this sequence using DNA footprinting. Overexpression of the vic operon caused growth-phase-dependent repression of comC, comDE, and comX. In the vic mutants, transcription of nlmC/cipB encoding mutacin V, previously linked to CSP-dependent cell lysis, as well as expression of its putative immunity factor encoded by immB, were significantly affected relative to the wild type (P < 0.05). In contrast to previous reports that proposed a hyper-resistant phenotype for the VicK mutant in cell viability, the release of extracellular genomic DNA was significantly enhanced in SmuvicK (P < 0.05), likely as a result of increased autolysis compared with the parent. The drastic influence of VicRK on cell viability was also demonstrated using vic mutant biofilms. Taken together, we have identified a novel regulatory link between the VicRK and ComDE systems to modulate bacteriocin production and cell viability of S. mutans.
INTRODUCTION
Bacteriocins are ribosomally synthesized peptides that have antimicrobial activity against other strains of the same or related species. Although bacteriocin producers are resistant to their own bacteriocins via the action of immunity proteins, production of these bacteriolytic or bacteriostatic molecules can be costly due to constitutively expressed immunity, plasmid carriage (e.g., Escherichia coli), and possible lethality of production (41, 42, 43). Therefore, the synthesis of these molecules has to be stringently regulated to optimize survival and the persistence of a bacterial population. In most cases for bacteriolytic molecules, the mode of activity can occur by their adsorption to exposed cell envelope or membrane receptors, which in turn, activate endogenous autolytic enzymes (e.g., murein hydrolases) causing membrane destabilization or pore formation leading to cell death (43, 50).
Streptococcus mutans, one of the major etiological agents of dental caries, is an opportunistic pathogen that resides in the dental biofilm. Its pathogenicity is contingent on virulence attributes that include its ability to produce acid by metabolizing dietary carbohydrates (acidogenicity), its remarkable aciduricity, and its ability to form a biofilm. Furthermore, S. mutans produces several bacteriocins (mutacins), which may provide an ecological-edge when competing against close relatives for the same nutrient sources or niche (45). Based on a survey of 143 strains of S. mutans used to assess bacteriocin production in vitro, 98 strains were reported to produce at least one bacteriocin (44). One of the major pathways of S. mutans for regulating bacteriocin production is via the ComDE two-component signal transduction system (TCSTS) that responds to accumulation of the competence stimulating peptide (CSP) whose precursor peptide is encoded by comC (12, 21, 28, 37, 55, 58). The ComDE is 1 of 13 TCSTSs in S. mutans (3, 24) that help respond to environmental stimuli by transmitting signals from a membrane-bound histidine kinase (HK) to an intracellular response regulator (RR) protein via transphosphorylation, which in turn, regulates the transcription of its target genes (49). The ComDE pathway was shown to modulate expression of nlmC (SMU.1914) that encodes mutacin V, and its cognate immunity protein encoded by immB (SMU.925) transcribed at a distant locus in the S. mutans chromosome (19, 37, 55). Although nlmC has been also referred to as bsmA and cipB, we use nlmC here, based on its function as a nonlantibiotic mutacin (13). Together, the com-controlled NlmC/ImmB (also known as CipB/CipI) pathway was shown to effect cellular autolysis and extracellular genomic DNA (eDNA) release (37), which contributes to the structural and functional integrity of the S. mutans' biofilm (19, 36, 37). Involvement of ComDE in regulating the transcription and/or production of mutacins I, IV, V, and VI has also been demonstrated by several labs (12, 13, 15, 19, 21, 28, 37, 55, 57, 58). The antimicrobial spectrum of mutacin IV is specifically against members of the mitis group of oral streptococci, while those of mutacins I, II, and III are broader (38–40). More recently, the HdrRM and BrsRM systems in S. mutans were examined for their role in bacteriocin production (30, 34, 35, 57). DNA microarrays revealed a tight overlap in genes controlled by the BrsR regulator with that of HdrR (57). Although both regulated transformability and bacteriocin production, a greater impact on bacteriocinogenesis was observed by BrsR overexpression (57).
We demonstrate here a role for the VicRK signal transduction system in mutacin production and cell death, at least in part, by direct modulation of CSP production in S. mutans. The vicK and vicR genes of S. mutans encode an HK and RR, respectively, and regulate genetic transformation, biofilm formation, and stress tolerance (8, 10, 46, 47, 48, 52). An unusual feature of VicRK, which is highly conserved in low G+C Gram-positive bacteria, is that it is essential for viability in most bacteria (9). In S. mutans, only the VicR serves an essential function (47). Using global transcriptome analysis of a VicK-null mutant, we demonstrate that inactivation of the VicK sensor kinase resulted in increased transcription of nlmAB, nlmC, and nlmD genes encoding mutacins IV, V, and VI, respectively. In contrast to previous reports, we further show that the loss of VicK resulted in enhanced cell death and that these cells underwent increased autolysis, as evaluated by a substantial increase in eDNA. Using DNA footprinting and transcriptional analyses, we demonstrate that VicR bound the comC coding region, which, in turn, affected the transcription of comC, comDE, and comX. Taken collectively, our findings provide substantial insight into the molecular basis of mutacin production and cell death of S. mutans and further demonstrate how the VicRK system can modulate the CSP-activated cell signaling system of S. mutans.
MATERIALS AND METHODS
Strains and growth conditions.
The wild-type S. mutans UA159 strain (provided by J. Ferretti, University of Oklahoma), a VicK-deficient mutant derivative (SmuvicK), and a vicRKX overexpressing mutant (Smuvic+) (47) were utilized in the present study. Todd-Hewitt yeast extract broth (THYE; Becton Dickinson, Sparks, MD) was used to routinely culture all S. mutans strains, whereas THYE medium solidified with 1.5% (wt/vol) agar (Bioshop, Burlington, Ontario, Canada) was used for propagation on plates. Erythromycin at 10 μg/ml was used in agar plates to propagate the mutant strains. Unless otherwise specified, no antibiotics were added to mutant cultures. All S. mutans strains were grown at 37°C and 5% CO2 (vol/vol) air mixture.
Microarray method.
S. mutans UA159 (control strain) and SmuvicK (experimental strain) were grown overnight in quadruplicate in sterile THYE. Subsequently, these cultures were diluted 20× in prewarmed THYE, incubated at 37°C with 5% CO2, and grown to mid-log phase (OD600 ∼ 0.4). The cells were harvested by centrifugation, snap-frozen in liquid nitrogen, and stored at −80°C until RNA isolation. When ready, frozen pellets were resuspended in TRIzol reagent (Invitrogen), and the FastPrep system (Bio 101 Savant) was used to isolate total RNA as specified by the manufacturer. Genomic DNA was eliminated from these samples by treatment with RQ1 DNase (Promega) according to the supplier's instructions. These RNA transcripts were then used for cDNA synthesis using a first-strand synthesis kit (MB1 Fermentas) as specified by the manufacturer. RNA derived from the UA159 wild-type strain was used as a reference for the control (UA159) and experimental (SmuvicK) samples. After labeling of both control and experimental cDNAs with cyanine 3 (Cy3) and reference cDNAs with cyanine 5 (Cy5), microarrays were conducted as specified in Senadheera et al. (46). A class comparison analysis was used to identify statistically significant genes, while statistical analysis was conducted using a two-sample t test (with random variance model) with the parametric P value cutoff set to P < 0.001. For robust statistical analysis of experimental data, we used cDNAs derived from four independent experiments for each strain. Selected genes that showed significant up- or downregulation from microarray analysis were validated utilizing quantitative real-time PCR. Gene identities were noted according to GenBank designations.
qRT-PCR.
Bacterial cultures were grown overnight in THYE medium, diluted 20-fold in fresh prewarmed THYE and grown to mid-log phase (optical density at 600 nm [OD600] of ∼0.4). Cells were collected using centrifugation, snap-frozen in liquid nitrogen, and stored at −80°C until needed. Isolation of total RNAs, DNase treatment, cDNA synthesis, quantitative real-time PCR (qRT-PCR), and analysis of expression was carried out as previously described (48). Synthesis of cDNAs was routinely carried out using total RNAs isolated from four independent cultures, whereas qRT-PCR was conducted in quadruplicate for each cDNA sample. Primers used for qRT-PCR are listed in Table S2 in the supplemental material. Since the expression of 16S rRNA was invariable under the test conditions (data not shown), it was used to normalize test gene expression. Statistical analysis was conducted using the Student t test: a P value of <0.05 or <0.001 was considered significant.
Deffered antagonism assays.
Bacteriocin overlay assays were performed using overnight cultures of S. mutans SmuvicK, Smuvic+, and UA159 strains as described previously (37). The indicator strains used included Streptococcus thermophilus, Streptococcus mitis, Streptococcus bovis (ATCC 9809), Streptococcus oralis SK23, Lactococcus lactis (ATCC 19435), Lactobacillus casei (ATCC 4961), Streptococcus gordonii (ATCC 10558), Streptococcus sobrinus (ATCC 27351), and Streptococcus ratti (ATCC 19645).
Overexpression and purification of VicR.
The coding sequence for VicR was cloned using primer oSG491 (GCGGGATCCAATGAAGAAAATTC) and oSG242 (CCCAAGCTTGCTAATAAAATTCGTAAAAATAAGGGAC) into pCRT7/NT-TOPO (Invitrogen) and overexpressed in OneShot BL21(DE3) pLysS (Invitrogen) cells to generate a fusion protein with an N-terminal His6 tag. To isolate purified VicR, 1 liter of cells was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an OD600 of ∼0.3. After 3 h, the cells were harvested by centrifugation and stored at −20°C. The cell pellet was thawed, resuspended in binding buffer (0.5 M NaCl, 50 mM NaH2PO4 [pH 8.0]), lysed by sonication, and treated with 0.5% Sarkosyl for 1 h to solubilize the fusion protein. After the Sarkosyl treatment, the cell lysate was harvested by centrifugation (30 min, 10,000 × g, 4°C) and loaded onto a ProBond nickel-chelating resin (Invitrogen) column. The column was washed with binding buffer containing 50 mM imidazole and eluted with binding buffer containing 500 mM imidazole. Protein fractions were diluted with glycerol to 25%, quantified, divided into aliquots, flash frozen, and stored at −80°C.
DNA footprinting analysis.
A 140-bp fragment of the nlmC promoter region (SMU.1914) was amplified by PCR using 32P-labeled primer oSG 578 (5′-GAATAATAAATCCAGCCTTG-3′) and an unlabeled primer, oSG579 (5′-GACTGATGAATTAGAGATTA-3′). Primer oSG578 was labeled using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Perkin-Elmer). The labeled substrate was gel purified using a QiaQuick gel extraction kit (Qiagen). Next, 22 nM the labeled substrate was incubated at room temperature with increasing concentrations of His-VicR for 30 min in 50 μl of binding buffer (25 mM Tris-HCl [pH 7.5], 12.5 mM KCl, 6.25 mM MgCl, 10% glycerol). The binding reactions were then subjected to DNase I digestion for 1 min by adding 0.25 U of DNase I (Promega) in the presence of 2.5 mM CaCl2 and 5 mM MgCl2. The reactions were stopped by adding 90 μl of stop buffer (200 mM NaCl, 30 mM EDTA, 1% sodium dodecyl sulfate, 100 μg of salmon sperm DNA/ml) and subsequently isolated by phenol-chloroform extraction and overnight ethanol precipitation. Samples were spun, dried, and dissolved in sequencing loading buffer (95% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue) and run on a 6% polyacrylamide denaturing sequencing gel. Sequencing reactions were prepared using 32P-labeled oSG578 and labeled nlmC fragment with the SequiTherm EXCEL II DNA Sequencing kit (Epicentre Biotechnologies). The gel was then dried and exposed to a phosphor screen (Fuji Film), scanned using a Typhoon phosphorimager (GE Healthcare), and analyzed using ImageQuant version 5.0 (Molecular Dynamics).
eDNA quantification.
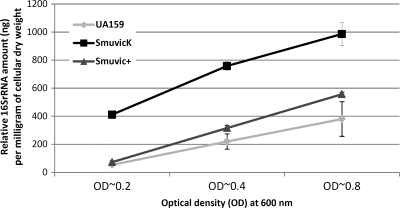
Overnight cultures were diluted 1:40 in prewarmed THYE and incubated until OD600s of 0.2, 0.4, and 0.8 were reached. At each density point, 10 ml of culture was extracted and subjected to centrifugation. The supernatant derived from centrifuged cultures was used for eDNA extraction using standard protocols with phenol, chloroform, and isoamyl alcohol solution. Cells pellets obtained from centrifuged cells were resuspended in 1 ml of phosphate-buffered saline (PBS), which were filtered in triplicate using preweighed 0.22-μm-pore-size membrane filters (Millipore), dried overnight at 37°C, and weighed to obtain the dry weight of cells. Each sample containing eDNA was amplified using 16SrRNA-specific primers using qRT-PCR. Cycle threshold (CT) values obtained from each amplification were plotted against a standard curve generated using a known amount of UA159 genomic DNA to quantify unknown amounts of eDNA present in each sample. These results were normalized using the dry weight of cell pellets.
Biofilm formation for confocal microscopy.
Overnight cultures were transferred to fresh Todd-Hewitt medium and incubated until an OD600 of ∼0.5 was reached. The ibidi Slide VI flow-cell system for Live Cell Analysis (Integrated BioDiagnostics, Munich, Germany) was used to form biofilms. Each flow cell chamber was inoculated with 150 μl of mid-exponential-phase cell suspensions, followed by incubation under static conditions for up to 48 h in an atmosphere of 5% CO2 at 37°C. Samples obtained at 1, 24, and 48 h were subjected to BacLight LIVE/DEAD staining.
Cell viability assays.
The LIVE/DEAD BacLight bacterial viability kit for microscopy (Molecular Probes) was used to assess membrane integrity of biofilm cells. The LIVE/DEAD mixture was prepared by mixing components A+B in the L-7007 kit, consisting of SYTO9 and propidium iodide, at a ratio of 1:100 with Todd-Hewitt inoculum medium. After removing the culture fluid from the flow chamber and a wash with PBS, 30 μl of the LIVE/DEAD mixture was added. The chambers were incubated at room temperature for 10 min. The fluorescence from stained cells was viewed using confocal scanning laser microscopy (CSLM).
CSLM and image analysis.
An Eclipse TE2000 inverted confocal scanning laser microscope (Nikon) was used for microscopy. By means of a motorized stage, 10 randomly selected image stack sections were imaged in each biofilm sample. Image stack sections were composed of 10 images, each taken with a variation of 2 μm along the z-position and covering an area of 0.05 mm2 per field of view. Confocal illumination for the green fluorescence signal was provided by an Ar laser (488-nm laser excitation). For detection of the red fluorescence, a G-HeNe laser was used (543-nm laser excitation).
CSLM images were analyzed in two and three dimensions by using the software bioImage_L (7), which allowed quantification of the total biofilm population and the independent subpopulations represented by green (viable) and red (damaged) fluorescence. The biovolume represented the overall volume of the biofilm, including an estimate of the biomass in the biofilm. Each experiment was repeated three times. The results are presented as mean calculations from a total of 30 biofilm sections, i.e., 10 sections/flow chamber-channels in three independent experimental rounds. Three independent biofilms were statistically compared by two-way analysis of variance (ANOVA). This statistical analysis detected any significant variation in the color-base identified subpopulations.
RESULTS AND DISCUSSION
Global transcriptome analysis of the VicK knockout strain.
We previously reported that loss of VicK affected biofilm formation, acid tolerance, genetic transformation, and oxidative stress tolerance in S. mutans (46–48). To elucidate the VicK regulon and understand the molecular pathways that control these phenotypes, we examined the global expression profiles of S. mutans UA159 wild-type parent and its VicK-deficient derivative (SmuvicK) using mid-log-phase cells grown in THYE. Microarrays revealed a total of 54 transcripts that were up- or downregulated by >2-fold in the VicK-null background (P < 0.001). Table S1 in the supplemental material shows numerical values and predicted functional categories of transcripts representing significantly affected genes. Of eight main functional categories that were affected by VicK mutagenesis, five were of major interest and included gene products with a putative or proven role in (i) bacteriocin production, (ii) cell wall metabolism, (iii) putative bacteriocin and bacteriophage immunity, (iv) fatty acid metabolism, and (v) cellular transport (see Table S1 in the supplemental material). Differential expression of at least 15 different genes obtained from microarrays was validated using transcriptional analysis (data not shown).
In the VicK regulon, six genes (SMU.1334, SMU.1335, SMU.1336, SMU.1341, SMU.1342, and SMU.1344) located within the largest genomic island in S. mutans, TnSmu2 (3), were downregulated 4-fold relative to the wild type. In strain UA159, genes in TnSmu2 encode nonribosomal peptide synthetases (NRPS), polyketide synthases (PKS), and accessory proteins involved in nonribosomal peptide (NRP) and polyketide (PK) biosynthesis (56). These were shown to be necessary for the biosynthesis of a hybrid NRP/PK pigment that was referred to as mutanobactin (56), whereas the secondary metabolite generated from this cluster was isolated and characterized as mutanobactin A (17). Whereas mutanobactin A was shown to suppress the morphological transition of Candida albicans from yeast to mycelium (17), mutational analysis of genes within the mutanobactin locus was shown to modulate biofilm formation and confer oxygen and hydrogen peroxide tolerance in S. mutans (56). In fact, we had previously reported the involvement of the VicRK in oxidative stress tolerance in S. mutans (48). Using qRT-PCR with primers specific for genes within the mutanobactin locus, we observed a significant decrease in transcription (P < 0.05) of these genes in the VicK mutant strain relative to the wild type (data not shown), which highlights a novel putative role for the VicRK in regulating the production of mutanobactin A in S. mutans.
In our microarray, SMU.2116 to SMU.2119 encoding an osmoprotectant transporter permease was significantly downregulated (P < 0.001), suggesting a putative role for the VicRK in osmotic stress tolerance. Interruption of vicR in Streptococcus pyogenes was shown to cause sensitivity to osmotic stress as well as growth defects, wherein five genes encoding an osmoprotectant transporter (OpuA) was shown to be upregulated in this mutant (26). It was shown that the VicR deficient mutant transported more osmoprotectants, betaine and proline, relative to the parent and therefore, demonstrated increased sensitivity to osmotic stress and increased susceptibility to osmotic pressure.
We had previously published microarray results of SmuvicK that was grown in tryptone yeast extract (TYE) medium exposed to acidic conditions (pH 5.5) (46). In that study, the VicK mutant exhibited a strong bias in regulating genes involved in fatty acid and phospholipid metabolism and in regulating cellular transport when induced by acid (46). The VicR/K homologs of Bacillus subtilis (known as YycFG) have a clear role in regulating fatty acid biosynthesis pathways and in determining fatty acid chain lengths in membrane lipids (31). While both genes under fatty acid metabolism and transport were also affected in THYE-grown mid-log-phase cells used in the present study, the actual number of affected transcripts was considerably lower compared to 121 transcripts obtained in our acid-induced microarray. These results suggest that nutrient availability and growth conditions have a significant impact on the VicK regulon in S. mutans.
In other closely related streptococci, studies involving VicRK orthologs have clearly established a role in adapting to osmotic stress (26, 27), oxidative stress (11, 48), antibiotic stress (16, 27), and high temperature (27). Hence, it is possible that the VicR/K-mediated stress responses observed in various bacteria are caused by defective cell walls. In this microarray, a number of transcripts (e.g., Smu.20, Smu.22, and Smu.609) affected by VicK mutagenesis were linked to membrane biosynthesis and cell wall metabolism. Interestingly, these genes were also differentially regulated in our previous VicK microarray conducted under different growth conditions (46). Although the link between VicRK and cell viability has been reported previously (1), the genetic factors and events that mediate this event are not well understood.
Of genes affected by VicK mutagenesis, we focused our current work on the involvement of VicRK in regulating mutacin production and cell wall metabolism. When we compared our VicK-deficient arrays to the CSP-induced transcriptome of S. mutans (37) and to that of a comC-deficient mutant (unpublished data), a large overlap in mutacin-related genes (e.g., nlmA, nlmB, nlmC, nlmD, bsmB, and bsmL) was observed. Among other loci shared between the VicK and CSP mutant arrays, SMU.1913 (immA) and SMU.925 (immB) encoding putative immunity proteins and SMU.426 linked to copper transport were differentially expressed (see Fig. S1 in the supplemental material). Examination of the comC region for a VicR binding site revealed a conserved binding sequence for VicR within the comC gene, which was further subjected to binding analysis as described in the following sections.
Loss of VicK modulates the transcription of mutacin genes.
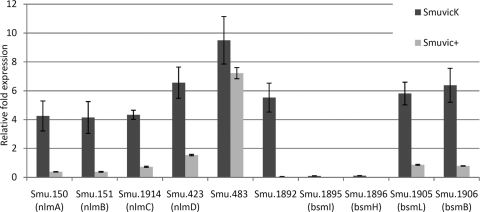
Mutagenesis of VicK significantly affected the expression of several genes with proven or putative roles in bacteriocin production (P < 0.001) (see Table S1 and Fig. S1 in the supplemental material). For example, nlmAB (SMU.150/SMU.151), nlmC, and nlmD (SMU.423) were upregulated over 5-fold in the VicK mutant microarray, suggesting a prominent role for VicK in mutacin IV, V, and VI production in S. mutans. Other putative bacteriocin-like peptides significantly affected by VicK mutagenesis included bsmBL (SMU.1905/SMU.1906), and bsmIH (SMU.1895/SMU.1896) encoding peptides harboring double-glycine motifs, which are typical of many bacteriocins. To validate our microarray results, we performed qRT-PCR using cDNAs derived from SmuvicK, Smuvic+, and their UA159 parent strain. Transcriptional analyses demonstrated that loss of VicK caused >4-fold upregulation of nlmAB, nlmC, nlmD, bsmBL, SMU.483, and SMU.1892 expression relative to the wild type (Fig. 1). The only exception to this was bsmIH transcription, which remained highly downregulated regardless of vicK deletion or vicRKX overexpression (Fig. 1). In contrast to SmuvicK, expression of nlmAB, nlmC, and SMU.1892 were downregulated in Smuvic+ relative to the wild-type UA159 strain. Taken collectively, these results highlight a clear role for the VicRK in regulating transcription of several putative and proven bacteriocin-encoding genes in S. mutans.
Fig 1.
Effect of vic mutagenesis on expression of putative or proven bacteriocin-related genes in S. mutans. The fold change in gene expression was calculated for cDNAs derived from SmuvicK and Smuvic+ strains relative to that in UA159, which was set at a user-defined value of 1.0. Mean expression values were calculated from three independent experiments, each subjected to triplicate amplifications ± the standard error.
Previously, van der Ploeg had shown that the VicK negatively affected nlmAB expression using LacZ transcriptional fusions harboring the mutated nlmAB promoter (55). A role for VicK in mutacin I production was also demonstrated by Tsang et al. using a random insertional mutagenesis approach in the virulent strain UA140 (53). In that study, disruption of the VicRK was shown to abolish mutacin I production. It was later discovered that while both VicK and a stress response regulator called HrcA were needed to produce mutacin I, VicK's influence on mutacin I synthesis was exerted by controlling transcription of a putative stress-responsive regulator designated irvA (inducible repressor of virulence A) for its role as a repressor for mutacin I synthesis in S. mutans (53, 54). In our microarray experiments, we observed that loss of VicK reduced the transcription of hrcA by >2.5-fold (see Table S1 in the supplemental material). Our examination of the comE and comX promoter regions did not reveal reasonably conserved VicR binding sites (data not shown). In addition to the previously described role for VicK in modulating mutacin I and mutacin IV production, this work has extended the role of VicRK in regulating the transcription of nlmC, nlmD, SMU.483, SMU.1892, bsmIH, and bsmBL in S. mutans.
The VicR regulates nlmC expression by binding to the comC coding sequence.
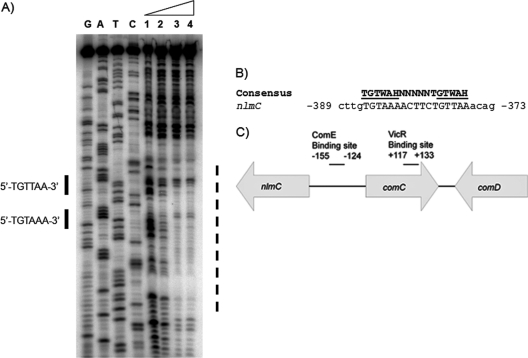
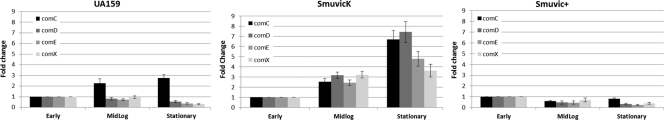
To investigate the underlying mechanism of VicRK's regulatory role on gene transcription, we examined the promoter regions of differentially regulated bacteriocins for direct repeat elements of the putative VicR binding consensus sequence (TGTWAH-5bp-TGTWAH, wherein W is A or T and H represent anything except G) (47). Using this search, we located a well conserved VicR-binding site, which intriguingly, was located within the comC coding region at −373 to −389 positions from the putative nlmC transcriptional start site (TSS) (Fig. 2). To examine whether VicR binds this sequence, we performed DNA footprinting analysis using purified His-VicR. Our results showed that His-VicR indeed bound to DNA containing these two direct repeats located within the comC coding region (Fig. 2). We reasoned that this site was perhaps the VicR binding site that inactivated comC, which in turn, modulated expression of genes encoding primary components of the S. mutans quorum sensing circuitry that included comD, comE, and the putative late competence transcription factor comX (24). Hence, we examined their expression in S. mutans UA159, SmuvicK and Smuvic+ strains grown to early-lag, mid-logarithmic-, and stationary-growth phases. Relative to their expression in the UA159 wild-type strain, the loss of VicK caused increased expression of the comC/D/E/X in the mid- and late-stationary-growth phases (Fig. 3). In contrast, overexpression of the vicRKX in strain Smuvic+ resulted in drastic repression of all of these genes at mid-log and late-stationary phases relative to the wild type (Fig. 3).
Fig 2.
(A) DNase I footprint analysis with increasing concentrations of VicR on the upstream region of nlmC. Lanes 1 to 4: 0, 0.25, 0.5, and 1 μM His-VicR. The putative binding site is indicated by solid lines, while the protected region is indicated by a broken line. (B) Putative VicR binding consensus sequence for nlmC regulation. Numbers preceding and following the nucleotide sequence represent the distance from the transcriptional start site for nlmC. W is A or T, H represents anything except G, and mismatches are shown in lowercase. (C) Depiction of the location of ComE and VicR binding sites for comC and nlmC (15). The putative VicR binding site for nlmC is found within the comC coding region, whereas the ComE binding site for comC is located within the comC/nlmC intergenic region as described previously (15, 19). Numbers indicate the nucleotide distance of each binding site from the first nucleotide of the comC start codon.
Fig 3.
Growth-phase-dependent expression of comC, comD, comE, and comX genes in cDNAs derived from SmuvicK, Smuvic+, and UA159 strains. S. mutans strains were grown to early-lag (OD of ∼0.2)-, mid-log (OD of ∼0.4)-, and late-stationary (OD of ∼0.8)-growth phases in THYE. Relative expression at the early-lag phase was set at a user-defined value of 1.0. The results are averages of triplicate samples from three independent experiments ± the standard error.
Previously, it was demonstrated that ComE can bind the intergenic region of comC and immA/nlmC to regulate their expression, as well as that of immB with a role in bacteriocin immunity (19, 21, 37, 55). Usually immunity proteins are expressed in concert with bacteriocins to counteract self-toxicity. Our expression analysis revealed that immA, another putative immunity gene, and immB were drastically repressed in Smuvic+ cells (see Fig. S2 in the supplemental material, P < 0.05). On the other hand, immA and immB were derepressed in VicK deficient cells relative to the wild type, suggesting a regulatory role for VicRK on the transcription of bacteriocin immunity genes in S. mutans.
Modulation of mutacin production and immunity against these toxic molecules by the com and vic systems is one of several phenotypes commonly regulated via both systems. It is also not surprising that we have previously observed >100-fold reduction in the natural transformation frequency of the vic overexpressing strain relative to the wild type (47), which is likely caused by VicR-mediated repression of CSP synthesis. Other phenotypes that overlap between the ComDE and VicRK signaling systems include biofilm formation, cell lysis, and stress tolerance in S. mutans (19–21, 23–25, 46–48, 55). Considering the regulatory effects of VicRK on CSP production, it is surprising that the com genes did not show up in our VicK-deficient microarray. It is likely that our microarray was not sensitive enough to detect differentially expressed transcripts relative to real-time PCR, which was used for quantification of these genes in the vic mutants.
The VicK sensor kinase harbors a PAS domain, which is a sensor of oxygen and cellular redox potential (51). We have previously reported that vicRKX transcription is drastically increased under intracellular oxidative stress (48). If vic expression and VicR synthesis are affected by oxidative stress as a consequence of a growing population, its regulatory effect on CSP synthesis and potentially ComE activity can have a substantial effect on genetic competence development versus bacteriocin production during growth. By analyzing the global transcriptome of S. mutans from the early-lag to late-stationary phases, we observed that expression of many competence-related genes is optimal at the early-lag phase, whereas bacteriocin-related genes were optimally transcribed at the late-stationary phase when the cell density and oxidative stress are high (data not shown). In addition to regulating mutacin expression by binding to comC, VicR may be able to regulate them directly by binding to their promoters or via other regulatory elements. We are currently pursuing a more comprehensive study of VicR binding analysis to understand the direct and/or indirect regulatory role of VicR in modulating the activity of competence and mutacin related genes that appeared as differentially regulated in our microarray validated by real-time data. Using mutagenesis and DNase footprinting, we know that both 6-bp repeats of the S. mutans VicR binding site are necessary for high-affinity VicR binding (E. A. Ayala and S. D. Goodman, unpublished data). Although an extensive binding analysis of VicR is beyond the scope of the present study, we believe that this information is imperative to understand how the VicRK regulates properties conducive to S. mutans virulence.
VicRK affects bacteriocinogenicity and bacteriocin immunity in S. mutans.
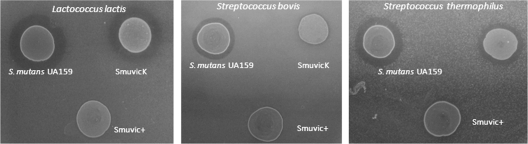
Our finding that His-VicR can bind the comC gene to disrupt its transcription and concomitantly downregulate comDE expression may contribute negatively to mutacin production. Previously, van der Ploeg (55) showed that S. mutans mutants deficient in comC, comDE, and comA (gene encoding the ABC transporter for CSP export) did not produce bacteriocins. To test whether bacteriocin-related expression correlated with the ability of SmuvicK and Smuvic+ strains to kill other species, we performed deferred antagonistic overlay assays using seven streptococcal and two lactococcal strains (see Materials and Methods). In these assays, drop inocula of overnight strains were deposited on THYE agar plates harboring indicator lawns and incubated overnight to assay inhibitory zones. Compared with UA159, results revealed altered lytic zones only when grown in the presence of S. bovis, S. thermophilus, and L. lactis indicator strains. For Smuvic+, the lytic zone was minimal and hardly detectable compared with the parent strain (Fig. 4), suggesting that VicRK perhaps inhibits mutacin production. As for SmuvicK, although the lytic zones were larger in diameter compared with that in Smuvic+, their diameters were smaller relative to that of the wild type (Fig. 4). The latter result was not anticipated, since a majority of mutacin-encoding genes were drastically upregulated in the absence of VicK. It is also possible that the effects of vic mutagenesis on immunity and cell viability may have contributed to what we observed as antagonistic effects against the indicator strains. Despite this, since mutagenesis of the vic locus impacted the ability of S. mutans to kill other streptococcal and lactococcal strains, our differed antagonistic experiments support a role for VicRK in the bacteriocinogenicity of S. mutans.
Fig 4.
Deferred antagonism assays for S. mutans SmuvicK, Smuvic+, and UA159 strains. Aliquots (20 μl) of each strain were spotted as drop inocula (11 mm) and overlaid with soft-agar containing indicator strains. The zone of growth inhibition was noted after incubation for 24 h at 37°C. The results shown are representative of at least three independent experiments. Inhibitory diameters (in mm) when spotted with S. mutans UA159, SmuvicK, and Smuvic+, respectively: L. lactis (18.5, 17, and 11.5), S. bovis (15.5, 11.5, and 11), and S. thermophilus (16, 15, and 12).
Due to the unusual expression of bsmIH, whose transcription was drastically repressed in both vic mutants, we further investigated each strain's ability to kill closely related indicator strains in bacteriocin overlay assays. Hence, we constructed mutants deficient in individual bsmI or bsmH genes, as well as a double mutant lacking both genes in S. mutans UA159, which were then used in bacteriocin overlay assays. The results revealed that none of these mutants had an altered ability to kill indicator strains relative to UA159, under our test conditions (data not shown), perhaps suggesting a different role for these in the physiology of S. mutans.
In a previous study, Ahn et al. linked VicK to cellular autolysis by influencing the maturation of AtlA, a major autolysin in S. mutans (1). We further observed that expression of nlmC and its cognate immunity gene immB, which were previously linked with cellular autolysis (22, 37), was drastically affected by vic mutagenesis in the present study (see Fig. S2 in the supplemental material). In addition to their involvement in bacteriocin production, it was likely that reduced killing of related strains by the vic mutants was also correlated with effects of vic expression on cell viability.
Loss of VicK results in decreased cell viability.
Emerging reports from VicKR homologs in other bacteria suggest that one of its primary functions is in the maintenance of cell wall metabolism (5, 9). Ahn et al. previously showed that mutagenesis of VicK affected maturation of AtlA (2). Using planktonic culture-based autolysis assays, these authors showed that a VicK-deficient mutant was hyper-resistant to autolysis (2). Using our SmuvicK mutant and its UA159 parent, we successfully validated these results via autolysis assays (data not shown). However, when we conducted these assays, we suspected that the VicK mutant's tendency to coaggregate and collect at the bottom of the microtiter-plate wells (47) impacted our readings to some extent, which we thought might hinder our conclusions derived from OD measurements. Hence, we performed an alternate experiment to investigate VicK's effects on cell lysis by extracting eDNAs from supernatants of cultures grown to different growth phases, which were then subjected to qRT-PCR amplification using 16S rRNA-specific primers. Instead of using OD values, we normalized the expression results using the dry weights of cell pellets whose supernatants were used for eDNA extraction, at each time interval. We argued that the amount of eDNA released may positively correlate with bacteria undergoing cellular autolysis. Our results showed that relative to the wild type, the amount of eDNA released by SmuvicK at all growth phases was significantly increased (P < 0.001) (Fig. 5). In contrast to SmuvicK, overexpression of vicRKX drastically diminished eDNA release relative to the VicK mutant, although this amount was still slightly higher than UA159 levels at mid-log and late stationary growth phases (Fig. 5). In contrast to previous studies that suggested a hyper-resistant phenotype for the VicK-deficient mutant, our results suggest that the loss of VicK remarkably increased cell lysis, as judged by the substantial enhancement in eDNA release.
Fig 5.
DNA release assays. Quantification of eDNA present in supernatants derived from S. mutans UA159, SmuvicK, and Smuvic+ strains using supernatant extracted from early-, mid-log-, and late-growth-phase THYE-grown cultures. Total eDNA was quantified with qRT-PCR using 16S rRNA primers, and the values are expressed as nanograms of eDNA per milligram of dry cell weight. The results indicate mean expression values from three independent experiments, each amplified in triplicate samples ± the standard error.
Involvement of VicRK in cell viability using vic mutant biofilms.
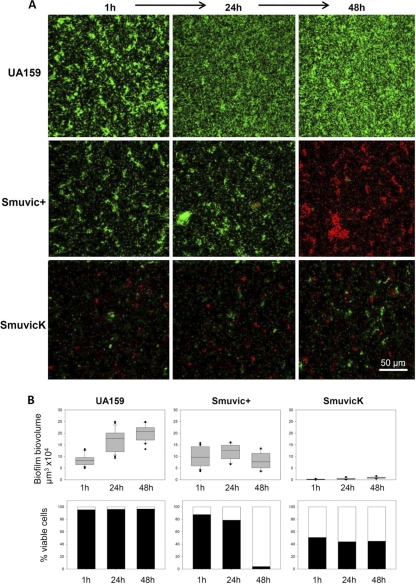
Despite accumulating evidence for VicRK in regulating cell wall metabolism and perhaps cellular autolysis, its role on cell viability in a biofilm mode of growth has not been studied. Since our eDNA quantification results suggested that loss of VicK enhanced cellular lysis relative to its wild-type parent, we further examined the effects of the VicRK on cell viability using confocal microscopy. Briefly, SmuvicK, Smuvic+, and UA159 biofilms grown under static conditions for 1, 24, and 48 h were subjected to LIVE/DEAD staining and cell viability counts. Compared with vic mutants whose biovolume remained considerably low, the biofilm biovolume of the control UA159 strain increased 2-fold from 1 to 48 h (Fig. 6). The ability of S. mutans to adhere to the surface was severely compromised upon deletion of VicK. For instance, only a few microcolonies were seen adhered to the surface for SmuvicK (Fig. 6), although this observation was not surprising since we had previously documented the inability of SmuvicK to adhere properly and form a robust biofilm (47). Particularly, the “washing step” with PBS, incorporated into the protocol prior to staining may have resulted in the removal of the bulk of loosely attached cells belonging to the VicK mutant biofilm. In the UA159 biofilm, more than 95% of the population remained viable at all three time points tested. In contrast, the percent viability of SmuvicK cells was significantly reduced by >40% at these time points (P < 0.05). Although viability of Smuvic+ was reduced at 1 h and 24 h relative to the wild type, a significant level of reduction in live cells was only noted in 48-h biofilms (P < 0.05). Taken together, our quantification of viable cells in vic mutant biofilms strongly supports a role for VicRK in modulating cell viability in S. mutans.
Fig 6.
Effects of vic mutagenesis on biofilm cell viability. (A) Representative sections were obtained from CLSM images of biofilms formed by vic mutants and their UA159 parent strain. Biofilms were grown as static cultures for 1, 24, and 48 h, and membrane integrity was assessed using LIVE/DEAD BacLight viability kit (Molecular Probes), wherein viable bacteria fluoresce green and those with damaged membranes fluoresce red. Bar, 50 μm. (B) Biofilm biovolume and percentage cell viability over time. Each experiment was repeated in triplicate, and the results shown are presented as mean calculations from a total of 30 biofilm sections, i.e., 10 sections/flow chamber-channels ± the standard error. Three independent biofilms were statistically compared by two-way ANOVA.
The VicK mutant's impaired cell shape, its size, and its enhanced sensitivity to environmental stresses (8, 10, 47, 48) suggest an important role for VicRK in sustaining cell wall integrity. One reason for diminished cell viability of the SmuvicK mutant is likely due to VicK's central role in maintaining cell wall integrity by modulating cellular murein hydrolase activity. Murein hydrolases serve important biological functions during cell growth and division, which include cell wall growth, peptidoglycan recycling, and turnover (6, 14). These enzymes contribute to bacterial susceptibility to antibiotics and may have a role in bacterial pathogenicity (18). Murein hydrolases which cause destruction of the cell wall leading to cell lysis are known as autolysins. In Streptococcus pneumoniae, VicRK has been shown to mediate activation of the cell wall hydrolase-encoding genes pcsB (gbpB), lytB, and lytN (32, 33). Our group and others have previously reported that S. mutans VicRK modulates gbpB transcription (10, 47). In S. mutans and also in several Gram-positive organisms, gbpB is flanked by two genes, mreC and mreD, whose products have a role in determining cell shape (29). In our VicK mutant microarray, the expression of mreC was downregulated (see Table S1 in the supplemental material), perhaps contributing to the defects in cell shape and size previously observed.
In addition to gbpB, transcription of smaA (SMU.609) encoding a putative 40-kDa cell wall protein precursor affecting murein hydrolase activity (6), was upregulated nearly 12-fold by VicK mutagenesis (see Table S1 in the supplemental material), whose increased expression was validated using qRT-PCR (data not shown). In S. mutans, SmaA has been linked to the activity of the major surface autolysin, AtlA, required for biofilm maturation and biogenesis of a normal cell surface (1). In addition, in the VicK mutant microarray, expression of SMU.984, a putative autolysin gene, whose product was shown to harbor a cysteine, histidine-dependent amidohydrolase/peptidase (CHAP) domain, was upregulated by >6-fold relative to the wild type (4). The CHAP domain is often associated with other domains that function in peptidoglycan hydrolysis (4).
In a review discussing the cellular roles and essentiality of VicRK (designated WalRK) across Gram-positive bacteria, Dubrac et al. proposed that VicRK senses accumulation of the peptidoglycan biosynthetic precursor (9), the d-Ala-d-Ala moiety of lipid II, thus signaling active synthesis of the cell wall and the requirement for autolysin production to incorporate lipid II-linked cell wall precursors to the nascent cell wall. These researchers used this model to explain the VicRK link to cell viability, which they proposed was required to sense the “healthy” status of actively dividing cells and perform the crucial task of adjusting cell wall metabolism to cell growth needs. Although this may be the case, more research is warranted to understand the link between VicRK and cell viability in S. mutans.
Conclusions.
In the present study, we report the involvement of VicRK in regulating mutacin production and cell death in S. mutans by modulating the transcription of several bacteriocins and cell wall hydrolases. A significant finding is our demonstration of the binding of VicR to the comC gene, thereby negatively affecting transcription of comC, comDE, comX, and nlmC. VicR regulation of comC transcription also perhaps explains the overlap between VicRK-modulated and CSP-dependent phenotypes that include mutacin production, genetic transformation, cell death, biofilm formation, and acid tolerance in S. mutans. Our finding that VicK modulates not only the expression of mutacins, including nlmC encoding mutacin V acting as an intracellular death peptide, but also its immunity gene encoded by immB, perhaps provides an important link between bacteriocin regulation and cell death. A comprehensive analysis of VicR binding is under way to delineate the direct and indirect regulatory role of VicR in modulating genes that contribute to these phenotypes. Manipulating the production of indigenous bacteriostatic or bacteriolytic molecules as a means of interfering with colonization and virulence of pathogenic strains is gaining interest, since the widespread use of antibiotic treatment as a strategy for microbial interference in bacterial infections has increased the emergence of antibiotic-resistant strains at an alarming rate, and it is difficult to develop effective, novel antibiotics to combat infections. Taken collectively, this study enhances our understanding of the intricate pathways and cross-regulatory mechanisms underlying cellular signal transduction networks in modulating virulence properties in bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Krastel and R. Mair for technical assistance in this study and J. Azavedo for review of the manuscript.
D.G.C. and S.D.G. are recipients of NIH grant R01DE013230-03. D.G.C. is a recipient of CIHR-MT15431 and a Canada Research Chair position.
Footnotes
Published ahead of print 6 January 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Ahn SJ, Burne RA. 2007. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J. Bacteriol. 189:6293–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahn SJ, Rice KC, Oleas J, Bayles KW, Burne RA. 2010. The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology 156:3136–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ajdic D, et al. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434–14439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bateman A, Rawlings ND. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234–237 [DOI] [PubMed] [Google Scholar]
- 5. Bisicchia P, et al. 2010. Peptidoglycan metabolism is controlled by the WalRK (YycFG) and PhoPR two-component systems in phosphate limited Bacillus subtilis cells. Mol. Microbiol. 75:972–989 [DOI] [PubMed] [Google Scholar]
- 6. Catt DM, Gregory RL. 2005. Streptococcus mutans murein hydrolase. J. Bacteriol. 187:7863–7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chavez de Paz LE. 2009. Image analysis software based on color segmentation for characterization of viability and physiological activity of biofilms. Appl. Environ. Microbiol. 75:1734–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deng DM, Liu MJ, ten Cate JM, Crielaard W. 2007. The VicRK system of Streptococcus mutans responds to oxidative stress. J. Dent. Res. 86:606–610 [DOI] [PubMed] [Google Scholar]
- 9. Dubrac S, Msadek T. 2008. Tearing down the wall: peptidoglycan metabolism and the WalK/WalR (YycG/YycF) essential two-component system. Adv. Exp. Med. Biol. 631:214–228 [DOI] [PubMed] [Google Scholar]
- 10. Duque C, et al. 2010. Downregulation of GbpB, a component of the VicRK regulon, affects biofilm formation and cell surface characteristics of Streptococcus mutans. Infect. Immun. 79:786–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Echenique JR, Trombe MC. 2001. Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J. Bacteriol. 183:4599–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hale JD, Heng NC, Jack RW, Tagg JR. 2005. Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans. J. Bacteriol. 187:5036–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hale JD, Ting YT, Jack RW, Tagg JR, Heng NC. 2005. Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection. Appl. Environ. Microbiol. 71:7613–7617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holtje JV. 1995. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch. Microbiol. 164:243–254 [DOI] [PubMed] [Google Scholar]
- 15. Hung DCI, et al. 2011. Characterization of DNA binding sites of ComE response regulator from Streptococcus mutans J. Bacteriol. 193:3642–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jansen A, et al. 2007. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int. J. Med. Microbiol. 297:205–215 [DOI] [PubMed] [Google Scholar]
- 17. Joyner PM, et al. 2010. Mutanobactin A from the human oral pathogen Streptococcus mutans is a cross-kingdom regulator of the yeast-mycelium transition. Org Biomol. Chem. 8:5486–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Korsak D, Liebscher S, Vollmer W. 2005. Susceptibility to antibiotics and beta-lactamase induction in murein hydrolase mutants of Escherichia coli. Antimicrob. Agents Chemother. 49:1404–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kreth J, et al. 2007. The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology 153:1799–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kreth J, Merritt J, Shi W, Qi F. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighboring species. Mol. Microbiol. 57:392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kreth J, Merritt J, Zhu L, Shi W, Qi F. 2006. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol. Lett. 265:11–17 [DOI] [PubMed] [Google Scholar]
- 22. Lemme A, Grobe L, Reck M, Tomasch J, Wagner-Dobler I. 2011. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J. Bacteriol. 193:1863–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li YH, Hanna MN, Svensater G, Ellen RP, Cvitkovitch DG. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li YH, et al. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu M, et al. 2006. Defects in ex vivo and in vivo growth and sensitivity to osmotic stress of group A Streptococcus caused by interruption of response regulator gene vicR. Microbiology 152:967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin PK, Li T, Sun D, Biek DP, Schmid MB. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matsumoto-Nakano M, Kuramitsu HK. 2006. Role of bacteriocin immunity proteins in the antimicrobial sensitivity of Streptococcus mutans. J. Bacteriol. 188:8095–8102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mattos-Graner RO, Porter KA, Smith DJ, Hosogi Y, Duncan MJ. 2006. Functional analysis of glucan binding protein B from Streptococcus mutans. J. Bacteriol. 188:3813–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Merritt J, Zheng L, Shi W, Qi F. 2007. Genetic characterization of the hdrRM operon: a novel high-cell-density-responsive regulator in Streptococcus mutans. Microbiology 153:2765–2773 [DOI] [PubMed] [Google Scholar]
- 31. Mohedano ML, et al. 2005. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 187:2357–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ng WL, Kazmierczak KM, Winkler ME. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53:1161–1175 [DOI] [PubMed] [Google Scholar]
- 33. Ng WL, et al. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 50:1647–1663 [DOI] [PubMed] [Google Scholar]
- 34. Okinaga T, Niu G, Xie Z, Qi F, Merritt J. 2010. The hdrRM operon of Streptococcus mutans encodes a novel regulatory system for coordinated competence development and bacteriocin production. J. Bacteriol. 192:1844–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Okinaga T, Xie Z, Niu G, Qi F, Merritt J. 2010. Examination of the hdrRM regulon yields insight into the competence system of Streptococcus mutans. Mol. Oral Microbiol. 25:165–177 [DOI] [PubMed] [Google Scholar]
- 36. Perry JA, Cvitkovitch DG, Levesque CM. 2009. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol. Lett. 299:261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Levesque CM. 2009. Peptide alarmone signaling triggers an auto-active bacteriocin necessary for genetic competence. Mol. Microbiol. 72:905–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qi F, Chen P, Caufield PW. 1999. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl. Environ. Microbiol. 65:652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qi F, Chen P, Caufield PW. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 67:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qi F, Chen P, Caufield PW. 1999. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl. Environ. Microbiol. 65:3880–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Riley MA. 1998. Molecular mechanisms of bacteriocin evolution. Annu. Rev. Genet. 32:255–278 [DOI] [PubMed] [Google Scholar]
- 42. Riley MA, Gordon DM. 1999. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 7:129–133 [DOI] [PubMed] [Google Scholar]
- 43. Riley MA, Wertz JE. 2002. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84:357–364 [DOI] [PubMed] [Google Scholar]
- 44. Rogers AH. 1976. Bacteriocin patterns of strains belonging to various serotypes of Streptococcus mutans. Arch. Oral Biol. 21:243–249 [DOI] [PubMed] [Google Scholar]
- 45. Rogers AH. 1976. Bacteriocinogeny and the properties of some bacteriocins of Streptococcus mutans. Arch. Oral Biol. 21:99–104 [DOI] [PubMed] [Google Scholar]
- 46. Senadheera D, et al. 2009. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J. Bacteriol. 191:6415–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Senadheera MD, et al. 2005. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 187:4064–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Senadheera MD, et al. 2007. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J. Bacteriol. 189:1451–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 50. Tagg JR, Dajani AS, Wannamaker LW. 1976. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40:722–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tremblay YD, Lo H, Li YH, Halperin SA, Lee SF. 2009. Expression of the Streptococcus mutans essential two-component regulatory system VicRK is pH and growth-phase dependent and controlled by the LiaFSR three-component regulatory system. Microbiology 155:2856–2865 [DOI] [PubMed] [Google Scholar]
- 53. Tsang P, Merritt J, Nguyen T, Shi W, Qi F. 2005. Identification of genes associated with mutacin I production in Streptococcus mutans using random insertional mutagenesis. Microbiology 151:3947–3955 [DOI] [PubMed] [Google Scholar]
- 54. Tsang P, Merritt J, Shi W, Qi F. 2006. IrvA-dependent and IrvA-independent pathways for mutacin gene regulation in Streptococcus mutans. FEMS Microbiol. Lett. 261:231–234 [DOI] [PubMed] [Google Scholar]
- 55. van der Ploeg JR. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187:3980–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu C, et al. 2010. Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance. Appl. Environ. Microbiol. 76:5815–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xie Z, Okinaga T, Niu G, Qi F, Merritt J. 2010. Identification of a novel bacteriocin regulatory system in Streptococcus mutans. Mol. Microbiol. 78:1431–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yonezawa H, Kuramitsu HK. 2005. Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrob. Agents Chemother. 49:541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.