TEXT
Many bacteria explore their immediate environment using flagellar locomotion, and on semisolid surfaces, this type of flagellum-driven motility is known as swarming (22). Dozens of genera exhibit swarming motility, and it can play an important role in biofilm formation (9, 28), virulence functions (1, 2, 31), and the colonization of new environments. Swarming generally relies upon the release of surfactants, compounds that reduce the critical surface tension of liquids. Bacterial biosurfactants are complex organic molecules, often lipids, glycolipids, or lipopeptides. Surfactant production is generally thought to reduce the drag of the bacterial cell on the surface during swarming and facilitate lateral movement. Given the functional linkage between flagellum-driven swarming locomotion and surfactant production, coregulation of these processes might be expected. Flagellar biogenesis is exquisitely controlled in a stepwise assembly process, providing abundant opportunities for additional regulatory inputs and outputs on motility and related functions. In addition to flagellar biogenesis (1, 12, 19, 37) and chemotaxis functions (8, 26), several other bacterial behaviors can be integrated with swarming motility; these behaviors include virulence functions (2) and the production of cell-surface polysaccharides (17).
In this issue of the Journal of Bacteriology, Burch et al. (7) report intriguing findings; they identified 3-(3-hydroxyalkanoyloxy) alkanoic acid (HAA), a lipid-type surfactant from the epiphytic plant pathogen Pseudomonas syringae pv. syringae B728a. P. syringae is the causative agent of plant brown spot disease and has served as an excellent model for plant leaf surface colonization (15). A transposon mutant screen led to the identification of P. syringae genes required for HAA synthesis. Several transposon mutants had incurred disruptions in flagellar biosynthetic genes, resulting in either diminished or elevated HAA production. Further exploration of these mutants revealed a highly nuanced coordination of flagellar assembly and function with surfactant production.
SURFACTANT PRODUCTION AND CONTROL
Among the pseudomonads, surfactants are best studied in the opportunistic human pathogen Pseudomonas aeruginosa. A family of related biosurfactants called rhamnolipids are important for swarming motility in P. aeruginosa; these biosurfactants include HAA, their rhamnose-free lipid precursor (7). Burch et al. (7) demonstrate that P. syringae produces HAA, in addition to the previously characterized lipopeptide surfactant syringafactin—which arguably plays a dominant role in P. syringae swarming (5). Swarming motility is thought to be important in the epiphytic fitness of this pathogen as well as its invasion of plant tissues (18, 30, 31).
Burch et al. (7) observed that P. syringae mutants disrupted for syringafactin biosynthesis exhibit limited surface motility, suggesting the production of additional biosurfactants. However, traditional methods used to detect surfactants failed to demonstrate the presence of a surfactant produced by the syringafactin-deficient (ΔsyfA) mutant. In earlier studies, however, the same authors developed a more sensitive assay for biosurfactants based on observing halos of dewetting surrounding colonies of surfactant-producing strains that have been misted with atomized oil droplets (6). This assay allowed them to detect production of a surfactant in the P. syringae ΔsyfA mutant that they subsequently characterized using mass spectrometry as HAA. Synthesis of HAA requires the rhlA acyltransferase gene, homologous to the rhlA gene required for rhamnolipid biosynthesis in P. aeruginosa.
FLAGELLAR ASSEMBLY AND COORDINATE REGULATION OF HAA SURFACTANT PRODUCTION
Burch et al. (7) report that mutations which disrupt flagellar biosynthesis can dramatically impact HAA synthesis. The bacterial flagellum is composed of three basic components, the basal body, hook, and flagellar filament (4). The biogenesis of the flagellar machinery is quite complex and costly, involving nearly 30 different proteins in many copies with ∼30,000 flagellin subunits required in filament formation (20). Many bacteria have therefore evolved precise control of flagellar assembly. Flagellar gene expression is regulated by a temporally fixed, structurally responsive mechanism (27). Several checkpoints exist to ensure that the flagellar biogenesis occurs in a specific sequence, so that early structures are always completed prior to subsequent additions to the nascent flagellum.
In contrast to the well-studied three-tier regulatory cascade controlling flagellar assembly in Escherichia coli and Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa has a four-tiered flagellar regulatory hierarchy (Fig. 1) (10). The NtrC-type transcription factor FleQ (designated a class I flagellar gene) is the major flagellar gene regulator. Class I genes also include the gene encoding the sigma factor FliA (σ28), which, when it is active, directs transcription of class IV genes. FleQ transcriptionally activates class II genes, which encode the two-component response regulators FleSR and include genes required for site selection and initiation of hook-basal body complex formation (33). Activated FleR controls expression of class III genes, which are required for completion of the hook-basal body complex. Class IV genes encode FliA-dependent products including flagellin and chemotaxis proteins. Upon the completion of the hook-basal body complex, the anti-σ factor FlgM is thought to be secreted through the hook-basal body complex, thereby relieving inhibition of FliA, resulting in expression of flagellin and other class IV genes (21) (Fig. 1). There are homologues of all these regulators in P. syringae, and it is likely that flagellar assembly occurs by a similar regulatory cascade.
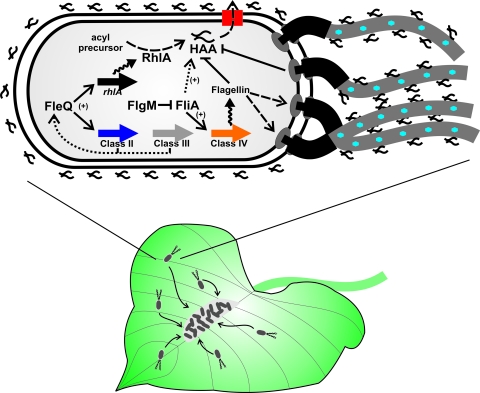
Fig 1.
P. syringae swarming with surfactant and flagella. A population of P. syringae cells exhibiting active swarming motility on a bean leaf surface are depicted (bacteria are not drawn to scale). Bacterial motility is indicated with curved arrows showing the cells converging on nutrient-rich sites on the plant surface where bacterial aggregates form. An enlarged bacterial cell is shown with a small tuft of polar flagella (extending off the page; P. syringae generally produces 2 to 4 polar flagella). HAA is synthesized by the activity of RhlA from acylated precursors. HAA is indicated as the linked squiggles reflective of its dual branched structure (not drawn to scale). HAA is exported from the cell by an uncharacterized export machinery (simplified as a red rectangle). As shown, HAA associates with the surface of the cell and the flagella, although this has not been proven. Blue hexagons on the flagella indicate putative glycosylation. Flagellar biosynthesis genes are diagrammatically simplified as thick, colored arrows (there are in fact multiple genes for each class). The FliA σ factor is inhibited by the FlgM anti-σ factor, which is exported through the hook-basal body complex (not shown). The black squiggly arrows indicate gene expression, the black dashed arrows indicate export or catalytic activity, the thin solid black arrows indicate putative positive regulatory interactions, and the thin solid black lines with bars indicate inhibition. Dashed or broken arrows are speculative but consistent with the findings reported.
Burch et al. (7) found that interruption of early or late flagellar genes could have opposing impacts on the production of HAA by P. syringae. For example, disruption of fleQ (class I) resulted in a complete deficiency of HAA production, and disruption of fliA (class I), fliF (class II; MS ring) and flgD (class III; scaffolding protein for hook assembly) all decreased surfactant production. In contrast, disruption in fliC (class IV; flagellin) or fgt1 and fgt2 (putatively required for flagellar glycosylation) resulted in surfactant overproduction. These findings suggest that HAA surfactant production is coordinately activated with early flagellar assembly but that late stages of flagellar maturation act conversely to limit surfactant production. Alternatively, fliC and fgt1 or fgt2 mutants may result in incomplete or easily sheared flagellar structures, respectively, that aberrantly result in elevated surfactant production. Either way, the increased surfactant in the class IV mutants indicates a mechanism by which HAA production is fine-tuned with flagellar assembly.
Expression analysis revealed that rhlA promoter activity is significantly reduced in a fleQ (class I) mutant, slightly reduced in a flgC (class III) mutant, but not obviously affected in a fliC (class IV) mutant (Fig. 1). Although unproven, it is plausible that FleQ directly regulates the transcription of rhlA, while class II and class III mutants, such as the flgC mutant, influence rhlA transcription and hence HAA production through a feedback response on fleQ (Fig. 1). Feedback regulation is common in flagellar biogenesis control (3). In Salmonella enterica, the FliT protein serves as an export chaperone for flagellar assembly but also inhibits the FlhDC complex, thereby limiting class II flagellar gene expression (36). In Proteus mirabilis, an flhA (class II; flagellum export) mutant decreased flhDC (class I) expression by 10-fold (16). Similar feedback control on fleQ could be readily tested in P. syringae. Elevated HAA levels in the fliC and fgt1 or fgt2 mutants are apparently due to posttranscriptional effects, although the mechanism by which this might occur remains undefined. It is interesting to note that mutations in both the fliC gene and the fgt1 or fgt2 gene increased expression of fliC itself, perhaps reflective of a common feedback mechanism for flagellar maturation triggered in these mutants, impacting surfactant and flagellin synthesis.
The studies reported by Burch et al. (7) suggest that in wild-type P. syringae, assembly of the hook-basal body complex stimulates HAA surfactant production, and once the hook-basal body complex is completed, increased levels of class IV gene products limit HAA production. This model would be more convincing if the fliA mutant phenocopied the fliC null mutant in terms of HAA production, since fliA turns on flagellin synthesis. However, the fliA mutant exhibits a modest reduction in HAA synthesis, in contrast to the fliC mutant, suggesting a more complex mechanism than simply lack of flagellin. Indeed, flagella are recognized to function directly in regulation and can provide a sensory mechanism for surface detection. For example, in Vibrio parahaemolyticus, a polar flagellum is thought to act as a mechanosensor for switching on production of lateral flagella required for swarming (3, 35). Similarly, in Caulobacter crescentus and Agrobacterium tumefaciens, surface contact stimulates production of a polar surface adhesin, and this may involve interactions of flagella with surfaces (24). It is conceivable that in P. syringae structural and functional perturbation of flagella may similarly modulate surfactant production.
The observations reported by Burch et al. (7) lead to an appealing proposal for the role of the HAA surfactant in P. syringae swarming motility, in which HAA serves primarily as a lubricant of the flagella and perhaps to some extent for the cell, whereas syringafactin plays the major role in reducing surface tension between the cell and the surface. However, there are other possible explanations accounting for HAA's role in swarming motility. For example, HAA could be the primary surfactant under environmental conditions not achieved in the laboratory—with the synthesis of HAA under strict environmental control.
This work raises many questions that warrant further investigation and particularly highlights the limitations on our understanding of surfactant function and its production during swarming. Both syringafactin and HAA contribute to swarming motility, but they are clearly regulated very differently. FleQ and flagellar biogenesis regulate HAA but apparently do not impact syringafactin, perhaps reflecting the different spectrum of functions for HAA and syringafactin. How is production of these two surfactants coordinated so that each can fulfill its proper activity? In P. aeruginosa, rhamnolipids are regulated by acylhomoserine lactone (AHL) quorum sensing (29), and in P. syringae, AHLs have a profound impact on colonization and the distribution of bacterial populations across the leaf surface, as well as in virulence (32). For P. syringae, quorum sensing inhibits motility and thereby promotes aggregate formation on leaves (Fig. 1). It, however, remains to be elucidated whether the HAA surfactant control observed here is also influenced by quorum sensing. Syringafactin and a second surfactant activity, likely to be HAA, are both influenced by a regulator called PmpR, and this may provide a mechanism for coordinating their activity (5). In addition, iron and the presence of other bacteria in the phyllosphere community can have important effects on P. syringae swarming motility and hence surfactants (13, 14, 25). In P. aeruginosa, specific nutrients promote more efficient biosurfactant production, modulating swarming motility, and HAA synthesis may be similarly environmentally responsive (11, 23, 34). Burch et al. also provide results that suggest that HAA synthesis, and thus perhaps swarming motility, is influenced by the AlgT stress response pathway (7).
Even with these unanswered questions, the findings reported by Burch et al. (7) reveal an intricate level of integration between flagellar function and surfactant synthesis. The flagellum is a remarkably complex molecular machine in its assembly and activity. As with other mechanical systems that function through the dynamic interactions of many individual components, appropriate lubrication is a crucial feature for optimal performance. HAA may function as a lubricating surfactant, enhancing flagellar performance, and also perhaps contributing in additional ways to flagellum-driven propulsion across surfaces. Coregulation of HAA with flagellar assembly, both positively and negatively, may ensure that surfactant production is balanced or calibrated so that sufficient levels are synthesized to facilitate flagellar function but prevent surfactant overproduction, which would waste resources and could compromise motility.
ACKNOWLEDGMENTS
Research in the Fuqua lab is supported through the National Institutes of Health (GM092660 and GM080546).
We thank Daniel Kearns for commenting on the manuscript.
Footnotes
Published ahead of print 20 January 2012
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1. Allison C, Coleman N, Jones PL, Hughes C. 1992. Ability of Proteus mirabilis to invade human urothelial cells is coupled to motility and swarming differentiation. Infect. Immun. 60:4740–4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allison C, Lai HC, Hughes C. 1992. Coordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol. Microbiol. 6:1583–1591 [DOI] [PubMed] [Google Scholar]
- 3. Anderson JK, Smith TG, Hoover TR. 2010. Sense and sensibility: flagellum-mediated gene regulation. Trends Microbiol. 18:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Apel D, Surette MG. 2008. Bringing order to a complex molecular machine: the assembly of the bacterial flagella. Biochim. Biophys. Acta 1778:1851–1858 [DOI] [PubMed] [Google Scholar]
- 5. Berti AD, Greve NJ, Christensen QH, Thomas MG. 2007. Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 189:6312–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burch AY, Shimada BK, Browne PJ, Lindow SE. 2010. Novel high-throughput detection method to assess bacterial surfactant production. Appl. Environ. Microbiol. 76:5363–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burch AY, et al. 2012. Pseudomonas syringae coordinates production of a motility-enabling surfactant with flagellar assembly. J. Bacteriol. 194:1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burkart M, Toguchi A, Harshey RM. 1998. The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:2568–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caiazza NC, Shanks RMQ, O'Toole GA. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187:7351–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dasgupta N, et al. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809–824 [DOI] [PubMed] [Google Scholar]
- 11. Davey ME, Caiazza NC, O'Toole GA. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dufour A, Furness RB, Hughes C. 1998. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol. Microbiol. 29:741–751 [DOI] [PubMed] [Google Scholar]
- 13. Dulla GFJ, Krasileva KV, Lindow SE. 2010. Interference of quorum sensing in Pseudomonas syringae by bacterial epiphytes that limit iron availability. Environ. Microbiol. 12:1762–1774 [DOI] [PubMed] [Google Scholar]
- 14. Dulla GFJ, Lindow SE. 2009. Acyl-homoserine lactone-mediated cross talk among epiphytic bacteria modulates behavior of Pseudomonas syringae on leaves. ISME J. 3:825–834 [DOI] [PubMed] [Google Scholar]
- 15. Freeman BC, Chen C, Beattie GA. 2010. Identification of the trehalose biosynthetic loci of Pseudomonas syringae and their contribution to fitness in the phyllosphere. Environ. Microbiol. 12:1486–1497 [DOI] [PubMed] [Google Scholar]
- 16. Furness RB, Fraser GM, Hay NA, Hughes C. 1997. Negative feedback from a Proteus class II flagellum export defect to the flhDC master operon controlling cell division and flagellum assembly. J. Bacteriol. 179:5585–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gygi D, et al. 1995. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol. Microbiol. 17:1167–1175 [DOI] [PubMed] [Google Scholar]
- 18. Haefele DM, Lindow SE. 1987. Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae. Appl. Environ. Microbiol. 53:2528–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harshey RM, Matsuyama T. 1994. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. U. S. A. 91:8631–8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones CJ, Aizawa S. 1991. The bacterial flagellum and flagellar motor: structure, assembly and function. Adv. Microb. Physiol. 32:109–172 [DOI] [PubMed] [Google Scholar]
- 21. Karlinsey JE, et al. 2000. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 37:1220–1231 [DOI] [PubMed] [Google Scholar]
- 22. Kearns DB. 2010. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kohler T, Curty LK, Barja F, van Delden C, Pechere JC. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li G, et al. 2012. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol. Microbiol. 83:41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindow S. 2009. Plant factors and other bacterial residents modulate iron levels on leaves thereby influencing quorum sensing controlled epiphytic fitness and virulence in Pseudomonas syringae. Phytopathology 99:S154–S154 [Google Scholar]
- 26. Mariconda S, Wang QF, Harshey RM. 2006. A mechanical role for the chemotaxis system in swarming motility. Mol. Microbiol. 60:1590–1602 [DOI] [PubMed] [Google Scholar]
- 27. McCarter LL. 2006. Regulation of flagella. Curr. Opin. Microbiol. 9:180–186 [DOI] [PubMed] [Google Scholar]
- 28. Mireles JR, Toguchi A, Harshey RM. 2001. Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J. Bacteriol. 183:5848–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ochsner UA, Koch AK, Fiechter A, Reiser J. 1994. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176:2044–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Panopoulos NJ, Schroth MN. 1974. Role of flagellar motility in invasion of bean leaves by Pseudomonas phaseolicola. Phytopathology 64:1389–1397 [Google Scholar]
- 31. Quinones B, Dulla G, Lindow SE. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant Microbe Interact. 18:682–693 [DOI] [PubMed] [Google Scholar]
- 32. Quinones B, Pujol CJ, Lindow SE. 2004. Regulation of AHL production and its contribution to epiphytic fitness in Pseudomonas syringae. Mol. Plant Microbe Interact. 17:521–531 [DOI] [PubMed] [Google Scholar]
- 33. Ritchings BW, Almira EC, Lory S, Ramphal R. 1995. Cloning and phenotypic characterization of FleS and FleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 63:4868–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shrout JD, et al. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264–1277 [DOI] [PubMed] [Google Scholar]
- 35. Stewart BJ, McCarter LL. 2003. Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 185:4508–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamamoto S, Kutsukake K. 2006. FliT acts as an anti-FlhD(2)C(2) factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:6703–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Young GM, Smith MJ, Minnich SA, Miller VL. 1999. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181:2823–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]