Abstract
Broad-host-range IncP-1 plasmids generally encode two replication initiation proteins, TrfA1 and TrfA2. TrfA2 is produced from an internal translational start site within trfA1. While TrfA1 was previously shown to be essential for replication in Pseudomonas aeruginosa, its role in other bacteria within its broad host range has not been established. To address the role of TrfA1 and TrfA2 in other hosts, efficiency of transformation, plasmid copy number (PCN), and plasmid stability were first compared between a mini-IncP-1β plasmid and its trfA1 frameshift variant in four phylogenetically distant hosts: Escherichia coli, Pseudomonas putida, Sphingobium japonicum, and Cupriavidus necator. TrfA2 was sufficient for replication in these hosts, but the presence of TrfA1 enhanced transformation efficiency and PCN. However, TrfA1 did not contribute to, and even negatively affected, long-term plasmid persistence. When trfA genes were cloned under a constitutive promoter in the chromosomes of the four hosts, strains expressing either both TrfA1 and TrfA2 or TrfA1 alone, again, generally elicited a higher PCN of an IncP1-β replicon than strains expressing TrfA2 alone. When a single species of TrfA was produced at different concentrations in E. coli cells, TrfA1 maintained a 3- to 4-fold higher PCN than TrfA2 at the same TrfA concentrations, indicating that replication mediated by TrfA1 is more efficient than that by TrfA2. These results suggest that the broad-host-range properties of IncP-1 plasmids are essentially conferred by TrfA2 and the intact replication origin alone but that TrfA1 is nonetheless important to efficiently establish plasmid replication upon transfer into a broad range of hosts.
INTRODUCTION
Plasmids belonging to incompatibility group P-1 (IncP-1 group) are generally considered to have a broad host range owing to their ability to replicate in at least two classes within the phylum Proteobacteria (e.g., Alphaproteobacteria and Gammaproteobacteria). The majority of IncP-1 plasmids have been identified as agents that mediate transfer of beneficial traits between these phylogenetically distinct bacteria, such as antibiotic resistance, resistance to heavy metals, or catabolic activity of xenobiotic compounds (2, 51, 60). Since the worldwide spread of antibiotic resistance is a growing concern in human health care (40), it is important that we improve our understanding of the mechanisms by which IncP-1 plasmids are maintained in a broad range of hosts.
Plasmid RK2, identified in Pseudomonas aeruginosa, has been studied as a model IncP-1 plasmid to gain understanding of basic plasmid functions (i.e., replication, stable inheritance, and conjugative transfer) and of the promiscuous nature of the IncP-1 group (32). It is known that the replication initiation protein (trans-acting factor A, TrfA) and the replication origin (oriV) are responsible for RK2 replication in a broad range of hosts (20, 52). RK2 encodes two replication initiation proteins (Rep proteins), TrfA-44 and TrfA-33 (54, 56). TrfA-33 is the truncated version of TrfA-44 and is produced from a second translational start site in the trfA-44 reading frame. According to homology modeling, TrfA-33 shows structural similarity to Rep proteins from narrow-host-range plasmids such as the F plasmid and pPS10 (49). The N terminus of TrfA-44 is absent in the Rep proteins of those narrow-host-range plasmids (Fig. 1).
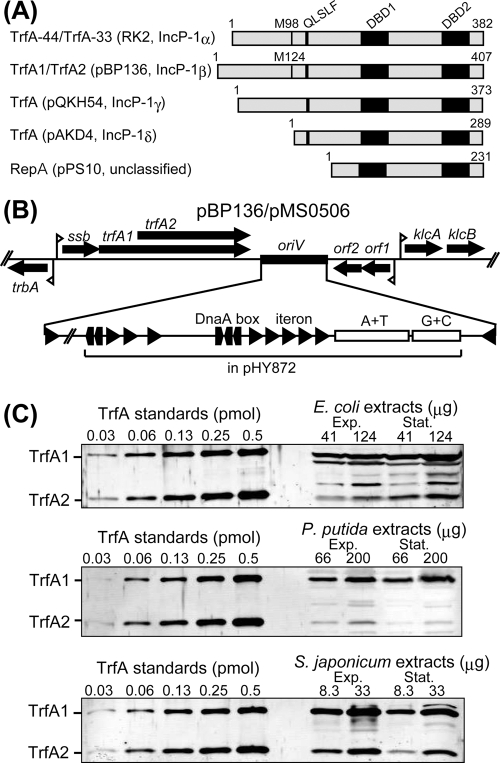
Fig 1.
(A) Schematic representation of the diversity in TrfA structure. M98 and M124 indicate the start methionines of TrfA-33 in RK2 and TrfA2 in pBP136, respectively. QLSLF indicates the conserved amino acid sequence motif responsible for interaction with DNA polymerase III (33). DBD1 and DBD2 indicate DNA-binding (winged-helix) domains deduced from secondary structure comparisons (22, 49). (B) The trfA gene locus of pBP136 and its mini-pBP136 derivative pMS0506. A+T and G+C indicate the A/T- and G/C-rich regions, respectively. Triangles and pentagons indicate iterons (TrfA-binding motif) and DnaA-boxes (DnaA-binding motif), respectively. Flags indicate promoters. The region cloned in pHY872 is indicated. (C) Western blots showing TrfA production from pMS0506 in three hosts: E. coli, P. putida, and S. japonicum. The quantities of cell extracts were normalized to total protein content using a Bradford assay (11). Exp, exponential phase; Stat, stationary phase.
It has been shown that TrfA-44 is essential for replication of plasmid RK2 in P. aeruginosa (16, 55). In this host TrfA-33 functions poorly in positioning the host's DnaB helicase at the plasmid oriV with or without the presence of the host's DnaA, while TrfA-44 is able to position DnaB at the oriV of plasmid RK2 (oriVRK2) without the assistance of DnaA (27). In fact, all DnaA boxes in oriVRK2 are dispensable for RK2 replication in P. aeruginosa (15). For RK2 replication in Pseudomonas putida, one of the four DnaA-boxes in oriVRK2 is indispensable, but in vitro TrfA-44 is able to position DnaB at oriVRK2 without DnaA of P. putida, just as observed for P. aeruginosa (15, 27). These observations suggested that TrfA-44 is able to modulate the plasmid's replication to be less dependent on the host's DnaA. Therefore, TrfA-44 has been postulated to play a role in extending the host range of IncP-1 plasmids (27, 63). However, P. aeruginosa is the only host reported so far in which RK2 required the longer TrfA-44 for replication. An experimentally generated mini-RK2 encoding only TrfA-33 was shown to replicate in several hosts currently classified as Alphaproteobacteria or Gammaproteobacteria (16). Furthermore, two naturally occurring plasmids of the IncP-1δ subgroup that encode only the TrfA-33 homolog have been found in natural hosts belonging to Betaproteobacteria (see below) (53, 68).These findings imply that the shorter protein alone might confer broad-host-range replication to the plasmid. Thus, the role of TrfA-44 and its homologs in broad-host-range replication of IncP-1 plasmids in hosts other than P. aeruginosa still needs to be elucidated.
IncP-1 plasmids have been classified into six subgroups (α, β, γ, δ, ε, and ζ) according to the phylogeny of the trfA gene or other plasmid backbone gene sequences (6, 25, 28, 46, 68). Plasmid sequence analyses have suggested that IncP-1 plasmids belonging to the α, β, ε, and ζ subgroups encode a long and shorter Rep proteins, just like the IncP-1α plasmid RK2. In contrast, plasmids from γ and δ subgroups encode only a single species of TrfA (Fig. 1A) (6, 25, 53, 68). The TrfA-44 and TrfA-33 homologs encoded by IncP-1 plasmids other than RK2 have been referred to as TrfA1 and TrfA2, respectively (61). From here on we will refer to TrfA1 and TrfA2 for all non-RK2 IncP-1 Rep protein homologs. The TrfA protein of IncP-1γ plasmid pQKH54 is equivalent to TrfA1, whereas the TrfA proteins of IncP-1δ plasmids pEST1044 and pAKD4 are essentially TrfA2 (25, 53, 68). In contrast to rapidly increasing information about the genetic diversity of IncP-1 plasmids, the significance of encoding TrfA1 alone, TrfA2 alone, or both proteins together is poorly understood. It was previously shown that the copy number of RK2 in Escherichia coli decreased when translation of TrfA-33 was eliminated by point mutation at the translational start site (17). On the other hand, elimination of TrfA-44 translation decreased the copy number of RK2 in Agrobacterium tumefaciens but not in E. coli or Azotobacter vinelandii (17). While these observations seemed to suggest that the presence of the shorter TrfA is important for a high plasmid copy number (PCN) in E. coli, it is unclear if the longer TrfA participates in the plasmid's replication in hosts other than P. aeruginosa or A. tumefaciens.
To further examine the role of TrfA1 and TrfA2 in the host range of IncP-1 plasmids, here we have characterized the replication activity in vivo of a mini-IncP-1β replicon derived from plasmid pBP136 (29) in four phylogenetically distinct hosts. TrfA1 and TrfA2 of pBP136 (TrfA1pBP136 and TrfA2pBP136, respectively) show 65% and 83% amino acid identity to TrfA-44 and TrfA-33 of RK2, respectively, and 94% and 95% identity to TrfA1 and TrfA2 of the archetype IncP-1β plasmid R751. We chose this minireplicon as our model system for two reasons: (i) all previous work on the role of long and short TrfA proteins in IncP-1 plasmid host range was done with the IncP-1α plasmid RK2 and not with any of the very widespread IncP-1β plasmids, and (ii) our previous experimental evolution study showed that trfA1 frameshift variants of the mini-IncP-1β replicon had a different host range and stability than the wild-type replicon (57). We have compared efficiency of transformation, plasmid copy number, and plasmid stability between the mini-pBP136 replicon and its trfA1 frameshift variant. In addition, plasmid copy number in the presence of TrfA1 alone, TrfA2 alone, or both proteins was determined. The results suggest the following: (i) that TrfA2 is sufficient for plasmid replication in a broad range of hosts; (ii) that TrfA1, but not TrfA2, plays a role in maintaining high plasmid copy number and increasing transformation efficiency; and (iii) that TrfA1 does not contribute to long-term plasmid persistence in hosts wherein TrfA2 is sufficient for plasmid replication.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
Bacterial strains and plasmids used in this study are listed in Table 1. pMS0506, a deletion derivative of the cryptic IncP-1β plasmid pBP136 isolated from Bordetella pertussis (29, 57), was used as a model IncP-1β plasmid. This minireplicon is no longer self-transmissible due to the removal of transfer genes. Transcription of the trfA gene in pMS0506 is controlled by cognate promoter and transcriptional regulators encoded by pMS0506 itself (KorB and KorA). The trfA1 variant of pMS0506, pEvo-Sh15, has a frameshift mutation in the 5′ end of trfA1 caused by a 13-bp duplication and was obtained previously after experimentally evolving pMS0506 to higher stability in Shewanella oneidensis MR-1. This plasmid no longer replicates in Pseudomonas aeruginosa (57). Western analysis has confirmed the production of TrfA2 and the lack of production of TrfA1 from pEvo-Sh15 (see Fig. S1 in the supplemental material).
Table 1.
Strains and plasmids used in this study
| Strain (class) or plasmid | Genotypes and relevant characteristic(s)a | Reference or source |
|---|---|---|
| E. coli strains (Gammaproteobacteria) | ||
| BW25113 | F−rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | 4 |
| BW29427 | thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::(erm-pir) Ermr | B. L. Warner, Purdue University |
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 50 |
| DY380 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara-leu)7697 gal490 pglΔ8 rpsL nupG [λ cI857indl Δ(cro-bioA):: tet] Tcr | 39 |
| EC100Dpir+ | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara-leu)7697 galU galK rpsL nupG pir (DHFR) | Epicentre (Madison, WI) |
| HY390 | BW25113 attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA1(M124L) | This study |
| HY407 | BW25113 attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA2 | This study |
| HY414 | BW25113 attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA1/trfA2b | This study |
| P. putida strains (Gammaproteobacteria) | ||
| KT2440 | Type strain, ATCC 47054 | 5 |
| HY566 | KT2440 attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA1/trfA2; Gmr | This study |
| HY569 | KT2440 attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA1(M124L); Gmr | This study |
| HY570 | KT2440 attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA2; Gmr | This study |
| S. japonicum strains (Alphaproteobacteria) | ||
| UT26S | Type strain, JCM17232 | 45 |
| HY848 | UT26S attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA1/trfA2; Gmr | This study |
| HY849 | UT26S attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA1(M124L); Gmr | This study |
| HY850 | UT26S attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA2; Gmr | This study |
| C. necator strains (Betaproteobacteria) | ||
| JMP228 | JMP134 derivative cured of plasmid pJP4; Rifr | 3 |
| HY876 | JMP228 attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA1/trfA2; Rifr Gmr | This study |
| HY877 | JMP228 attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA1(M124L); Rifr Gmr | This study |
| HY878 | JMP228 attTn7::mini-Tn7::aacC1-lacIq-Ptac-trfA2; Rifr Gmr | This study |
| Plasmids | ||
| pBAD24 | pMB1 replicon, protein expression vector; Apr | 24 |
| pBAD24C | pBAD24 derivative in which bla was replaced by cat; Cmr | This study |
| pEvo-Sh15 | pMS0506 derivative with a frameshift mutation in 5′ end of trfA1, unable to replicate in P. aeruginosa; Kmr | 57 |
| pHSG399 | pMB1 replicon, cloning vector; Cmr | 59 |
| pHY819 | pBAD24C carrying trfA1(M124L); Cmr | This study |
| pHY820 | pBAD24C carrying trfA2; Cmr | This study |
| pHY835 | pUCR6KT-mini-Tn7T derivative in which the HindIII site in the R6K origin was disrupted; Apr Gmr | This study |
| pHY835LAC | R6K replicon, mini-Tn7::aacC1-lacIq-Ptac; Apr Gmr | This study |
| pHY837 | pHSG399 carrying trfA1(M124L); Cmr | This study |
| pHY838 | pHSG399 carrying trfA2; Cmr | This study |
| pHY853 | pHSG399 carrying trfA1/trfA2; Cmr | This study |
| pHY872 | pJP5608 ΔPlac carrying the 0.5-kb fragment of oriVpBP136; Tcr | This study |
| pHY884 | pHY835LAC carrying trfA1/trfA2; Apr Gmr | This study |
| pHY885 | pHY835LAC carrying trfA1(M124L); Apr Gmr | This study |
| pHY886 | pHY835LAC carrying trfA2; Apr Gmr | This study |
| pHY922 | pSTV28 carrying the 1.5-kb PstI-EcoRI fragment containing kleE of pBP136; Cmr | This study |
| pHY924 | pHY872 carrying the 2.0-kb EcoRI fragment containing atpB of the BW25113 chromosome; Tcr | This study |
| pHY925 | pTXB1 carrying trfA2; Apr | This study |
| pHY926 | pTXB1 carrying trfA1(M124L); Apr | This study |
| pHY934 | pHY922 carrying the 2.0-kb EcoRI fragment containing atpB of the E. coli BW25113 chromosome; Cmr | This study |
| pHY950 | pHY922 carrying the 1.8-kb EcoRI fragment containing parB of the KT2440 chromosome; Cmr | This study |
| pHY951 | pHY872 carrying the 1.8-kb EcoRI fragment containing parB of the KT2440 chromosome; Tcr | This study |
| pHY974 | pHY872 carrying the 2-kb EcoRI fragment of SJA-00140 of the UT26S chromosome I; Tcr | This study |
| pHY976 | pHY922 carrying the 2-kb EcoRI fragment of SJA-00140 in the UT26S chromosome I; Cmr | This study |
| pHY982 | pHY872 carrying the 0.8-kb EcoRI fragment containing parB of the JMP228 chromosome I; Tcr | This study |
| pHY983 | pHY922 carrying the 0.8-kb EcoRI fragment containing parB of the JMP228 chromosome I; Cmr | This study |
| pJP5608 | R6K replicon, suicide vector; Tcr | 48 |
| pMS0506 | IncP-1β replicon, pBP136 Δ(trbB-traM)::(oriTRP4-kan); Kmr | 57 |
| pTXB1 | pMB1 replicon, protein expression vector; Apr | New England Biolabs (Ipswich, MA) |
| pUCR6KT-mini-Tn7T | R6K replicon, oriTRP4, mini-Tn7 vector; Apr | 14 |
| pUC18-mini-Tn7T-LAC | pUC18 replicon, mini-Tn7::aacC1-lacIq-Ptac; Apr Gmr | 14 |
| pUX-BF13 | R6K replicon, Tn7 transposase expression vector, tnsABCDE oriTRP4; Apr | 8 |
Abbreviations for are as follows: Tc, tetracycline; Ap, ampicillin; Gm, gentamicin; Cm, chloramphenicol; Km, kanamycin; Erm, erythromycin; Rif, rifampin. kan is the kanamycin resistance gene.
trfA1/trfA2, wild-type trfA1 encoding both TrfA1and TrfA2.
To address the role of the two Rep proteins in the ability of IncP-1 plasmids to establish themselves and be stably maintained in a wide range of bacteria, we used four model hosts. The two hosts, E. coli and P. putida (both belonging to the Gammaproteobacteria) were selected because the behavior of RK2 in these hosts has been relatively well studied. In addition, since IncP-1 plasmids have been found in strains within the Betaproteobacteria (for example, in the genera Achromobacter, Bordetella, Comamonas, Cupriavidus, Delftia, and Neisseria) and Alphaproteobacteria (genus Sphingomonas) (10, 26, 29, 37, 47, 67, 68), we also included representative hosts from those two classes: the betaproteobacterium Cupriavidus necator JMP228, a derivative of JMP134 (41), and the alphaproteobacterium Sphingobium japonicum UT26S (45).
Luria-Bertani (LB) medium (50) was used for cultivation of E. coli and P. putida, and 1/3-LB medium (3.3 g of Bacto tryptone, 1.7 g of yeast extract, and 5 g of NaCl/liter of H2O) was used for S. japonicum; 1/10-TSB (1/10 dilution of Bacto tryptic soy broth; BD Bioscience, Sparks, MD) was used for C. necator. Solid medium was made by addition of 1.5% agar to LB (referred to as LBA), 1/3-LB (1/3-LBA), or 1/10-TSB (1/10-TSBA). E. coli strains were incubated at 37°C for DNA cloning, while P. putida, S. japonicum, and C. necator strains were incubated at 30°C. Once the IncP-1 replicon was introduced, all strains including E. coli were incubated at 30°C. If necessary, the medium was supplemented with antibiotics at the following concentrations: for E. coli, 100 μg/ml ampicillin (Ap), 50 μg/ml kanamycin (Km), 10 μg/ml gentamicin (Gm), 25 μg/ml chloramphenicol (Cm), and 10 μg/ml tetracycline (Tc); for P. putida, 20 μg/ml Gm, 50 μg/ml Km, and 10 μg/ml Tc; for C. necator, 70 μg/ml Gm, 50 μg/ml Km, and 10 μg/ml Tc; for S. japonicum, 2.5 μg/ml Gm, 50 μg/ml Km, and 10 μg/ml Tc.
General molecular techniques.
For DNA cloning, heat shock transformation of E. coli DH5α ultracompetent cells (50) and electroporation of E. coli EC100Dpir+ (Epicentre, Madison, WI) were routinely used. Plasmid DNA was isolated from E. coli using a GeneJET plasmid miniprep kit (Fermentas, Inc., Ontario, Canada). DNA fragment purifications from PCR mixtures and agarose gels were performed using a GeneJET PCR purification kit and a GeneJET gel extraction kit (Fermentas, Inc., Ontario, Canada), respectively. Phusion High-Fidelity DNA polymerase (New England BioLabs, Ipswich, MA) was used for conventional PCR. Total DNA extraction was performed using a GenElute bacterial genomic kit (Sigma, St. Louis, MO). Fast SYBR premix Taq (Applied Biosystems, Foster City, CA) was used for quantitative PCR (qPCR). Southern blotting experiments were performed using a DIG-High Prime DNA Labeling and Detection Kit (Roche Applied Science, Mannheim, Germany) (where DIG is a digoxigenin). Oligonucleotides used in this study were synthesized by Sigma (Sigma-Aldrich, St. Louis, MO) and are listed in Table S1 in the supplemental material.
For preparation of electrocompetent cells of E. coli, P. putida, and C. necator, 1 ml of overnight bacterial cultures was diluted in 100 ml of fresh medium, grown up to an optical density at 600 nm (OD600) of 0.8, washed with an electroporation solution (300 mM sucrose for E. coli and P. putida; 10% glycerol for C. necator), and resuspended in 2 ml of the solution. Cell suspensions (100 μl) containing 108 to 109 cells were used for electroporation. Electrocompetent cells of S. japonicum were prepared from bacterial colonies grown on 1/3-LBA for 48 h; the bacterial colonies were gently scraped from the agar surface, washed, and resuspended in distilled water. Electroporation was conducted using a 1-mm gap cuvette and a Gene Pulser Xcell (Bio-Rad, Hercules, CA) at 1.8 kV. After electroporation, cells were incubated in their appropriate liquid medium for 2 h and subsequently plated on plain agar medium for total cell counts and on agar medium containing Km for transformant counts.
Plasmid stability assay.
Plasmid stability assays were done essentially as described previously (57). Briefly, for E. coli and P. putida, each strain was first grown with Km overnight, and 4.9 μl of overnight culture was transferred to 5 ml of LB medium, while an aliquot of the overnight culture was serially diluted and plated on LBA to obtain single colonies. The following day, 52 single colonies were replicated on LBA and on LBA containing Km. The ratio of Km-resistant colonies to total colonies was considered the plasmid-containing fraction. This procedure was repeated every 24 h for 10 days. For C. necator, the strains were grown in 1/10-TSB. For S. japonicum, the strains were grown in 20 ml of 1/3-LB medium to prevent aggregation of bacterial cells, and the culture was transferred and plated every 2 days. Single colonies of S. japonicum were replicated on 1/3-LBA and 1/3-LBA containing Km.
Construction of strains carrying trfA in the chromosome.
The mini-Tn7 vector (14) was used to insert trfA genes into a specific site (attTn7) in the chromosomes. The mini-Tn7-aacC1-lacIq-Ptac region of pUC18T-mini-Tn7T-LAC (14) was cloned into the NdeI-HindIII region of pHY835, a derivative of pUCR6KT-mini-Tn7T carrying a mutation in the HindIII site in the R6K origin. The transcriptional terminator region in pColdIV (Takara, Shiga, Japan) was cloned into the HindIII-KpnI region of the pHY835 derivative. The resulting R6K replicon carrying mini-Tn7-aacC1-lacIq-Ptac was designated pHY835LAC. The trfA1 gene of pBP136 was first PCR amplified using primers that included a ribosome-binding binding site. The PCR products were cloned into the SacI-HindIII region of pHSG399 (59) to confirm the trfA sequences. To eliminate translation of TrfA2 from the trfA1 gene, the trfA2 start codon, a methionine (ATG) at 124, was replaced with the codon for leucine (CTG). This mutation (the M124L mutation) was introduced into trfA1 [trfA1(M124L)] in the pHSG399 derivatives by PCR, followed by phosphorylation and ligation. pHSG399 derivatives carrying either the wild-type trfA1 encoding both TrfA1 and TrfA2 (trfA1/trfA2), trfA1(M124L), or trfA2 were designated pHY853, pHY837, and pHY838, respectively. The SacI-HindIII region carrying trfA1/trfA2, trfA1(M124L), or trfA2 was cloned from pHY853, pHY837, or pHY838 into pHY835LAC, yielding pHY884, pHY885, and pHY886. To introduce the mini-Tn7::aacC1-lacIq-Ptac-trfA fragments to the recipient chromosomes, E. coli BW29427 harboring pHY884, pHY885, or pHY886, E. coli BW29427(pUX-BF13), and a recipient strain were mated on LBA or 1/3-LBA containing 200 μM diaminopimelic acid overnight. The recipient strains tagged by mini-Tn7 were selected on LBA or 1/3-LBA containing the appropriate concentration of Gm depending on the host. Mini-Tn7 insertion to attTn7 was confirmed by PCR amplification of the mini-Tn7-glmS junction. The presence of a single copy of trfA in the genomes of the constructed strains was confirmed by Southern hybridization analysis using the trfA2 region as a probe. PCR primers used to clone trfA genes or to confirm mini-Tn7 insertion are listed in Table S1 in the supplemental material. Location of the mini-Tn7-trfA insertion site in S. japonicum UT26S and C. necator JMP228 was determined by sequencing and was 26 bp downstream from the stop codon of the glmS gene in chromosome I for both strains.
Construction of TrfA expression plasmids.
Protein expression plasmid pBAD24C was constructed by replacing the ampicillin resistance gene on pBAD24 (24) with the pHSG399-derived chloramphenicol resistance gene in E. coli DY380 using λ-Red recombination according to a previously published protocol (39). The PCR-amplified trfA1(M124L) and trfA2 gene fragments were first cloned into pHSG399 to confirm the sequences and then moved into the EcoRI-HindIII region of pBAD24C. The resulting pBAD24C derivatives carrying either trfA1(M124L) or trfA2 were designated pHY820 or pHY819, respectively. For affinity protein purification, trfA1(M124L) and trfA2 were PCR amplified and cloned into the NdeI-XhoI region of pTXB1 (New England BioLabs, Ipswich, MA), yielding pHY926 and pHY927, respectively.
Determination of plasmid copy number.
To measure copy numbers of pMS0506 and pEvo-Sh15 by qPCR, a series of copy number reference plasmids was constructed carrying a pBP136 fragment and a chromosomal fragment (the oriC region) at a 1:1 ratio. The 1.7-kb PstI-HindIII fragment (the kleE region) of pBP136 was cloned from pMS0506 into pSTV28 (Takara, Shiga, Japan). The resulting plasmid was designated pHY922. The 0.8- to 2.0-kb EcoRI fragments near oriC in the chromosomes of E. coli, P. putida, C. necator, and S. japonicum were PCR amplified using primers shown in Table S1 in the supplemental material, and the PCR products were cloned into the EcoRI site of pHY922, yielding pHY934 (E. coli), pHY950 (P. putida), pHY982 (C. necator), and pHY974 (S. japonicum). The chromosomal fragment contained atpB for E. coli, parB for P. putida, parB of chromosome I for C. necator, or a partial fragment of the TonB-dependent receptor-like protein gene (SJA-0140) of chromosome I for S. japonicum. A copy number reporter plasmid, pHY872 (Fig. 1B), was constructed by PCR amplification of the pBP136 oriV (oriVpBP136) containing eight iteron copies, cloning of the PCR product into pJP5608 (48), and subsequent removal of the lac promoter in the pJP5608 backbone using PCR. To measure copy number of pHY872, copy number reference plasmids were constructed by cloning the chromosomal fragments described above into the EcoRI site of pHY872. The resulting copy number reference plasmids were designated pHY924 (for E. coli), pHY951 (P. putida), pHY983 (C. necator), and pHY976 (S. japonicum).
For the copy number analysis of pMS0506 and pEvo-Sh15, total DNA was purified from 1 ml of exponential-phase culture. For the copy number analysis of pHY872 in the strains carrying trfA genes in the chromosome, total DNA was purified from 0.5 ml of stationary-phase cultures grown with 10 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Relative copy numbers of IncP-1β replicons compared to a gene in the oriC region in the chromosome in total DNA were determined using qPCR. Prior to qPCR, purified total DNA was linearized by restriction digestion and recovered by ethanol precipitation. Digested total DNA (1.5 μl) at a concentration of 6.0 to 10.0 ng/μl was added to the 20-μl-scale qPCR mixtures. The EcoRI and PstI digests of pHY934, pHY950, pHY974, and pHY982 were used to generate standard curves in the copy number analysis for pMS0506 and pEvo-Sh15, while the EcoRI digests of pHY924, pHY951, pHY976, and pHY983 were used to generate standard curves in the copy number analysis of pHY872. Primers used in qPCR are listed in Table S1 in the supplemental material. Data analysis was performed using the SDS2 software supplied with the ABI 7900HT thermal cycler (Applied Biosystems, Foster City, CA). Quantities of the target sequence in the total DNA were determined on the standard curves with R2 of >0.994.
Proteins.
TrfA1 and TrfA2 were purified using an Impact kit (New England BioLabs, Ipswich, MA) and buffers used for purification of TrfA-33 and TrfA-44, with modifications (65). Protein expression was induced in E. coli Rosetta 2 (DE3) carrying pHY926 or pHY927 in 50 ml of LB medium. Cells were resuspended in 3 ml of a buffer (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, 0.05% Triton-X) and sheared by French press at 900 lb/in2 (repeated three times), followed by ultrasonic treatment for 2 min. After the cell extract was clarified by centrifugation at 20,000 × g for 25 min, phenylmethylsulfonyl fluoride (PMSF) was added to 1 mM. The supernatant was incubated with 1 ml of chitin beads for 1 h at 4°C to bind the expressed protein. The bead-TrfA complex was washed in a 2-ml column with 40 ml of wash buffer (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, 10% glycerol) and then incubated in 1 ml of wash buffer containing 50 mM dithiothreitol (DTT) and 1 mM PMSF at 30°C overnight to induce intein cleavage. The eluate containing tag-free TrfA was dialyzed overnight against 1 liter of stock buffer (50 mM sodium phosphate [pH 6.0], 100 mM NaCl, 10% glycerol). This protocol yielded >95% pure TrfA as assessed by SDS-PAGE gels. Protein concentration of purified TrfA was normalized to a bovine serum albumin (BSA) standard in SDS-PAGE gels stained with Coomassie brilliant blue (CBB).
To prepare whole-cell extracts, bacterial cells were suspended in an appropriate volume of buffer containing 10 mM sodium phosphate (pH 7.6) and 145 mM NaCl and then disrupted using a French press, followed by ultrasonic treatment as above. The disrupted cells were clarified by centrifugation at 20,000 × g for 25 min, and then PMSF was added to 1 mM. The supernatant containing 1 mM PMSF was used as whole-cell extract throughout this study. Protein concentrations of whole-cell extracts were determined by Bradford assay (11) using BSA as the standard.
Quantitative Western blotting.
Whole-cell extracts and purified TrfA2 (quantity standards) diluted in phosphate buffer containing 1 mg/ml BSA were denatured in sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 5% 2-mercaptoethanol, and 10% glycerol) and electrophoresed in an 11% SDS-polyacrylamide gel for 90 min at 120V to separate TrfA1 and TrfA2. Proteins were electrotransferred to polyvinylidene difluoride (PVDF) membrane (Immobilon-FL; Millipore, Billerica, MA) overnight (∼15 h) at 10V in Towbin buffer (66) using a TE 62 transfer unit (GE Healthcare, Piscataway, NJ). The membrane was hybridized with anti-TrfA2-serum diluted 1:2,000 in blocking buffer (1× phosphate-buffered saline [PBS], 5% dry milk). Polyclonal anti-TrfA2 antibodies were raised in rabbits immunized three times with denatured TrfA2pBP136 contained in an SDS-polyacrylamide gel slice (Harlan Laboratories, Inc., Indianapolis, IN). After being washed with blocking buffer, the membrane was incubated with IRDye 800CW goat anti-rabbit IgG (Rockland, Gilbertsville, PA), according to the manufacturer's protocols. Densitometry analysis was performed with an Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE). TrfA proteins were quantified on standard curves with an R2 of >0.99. Each experiment was performed at least three times.
RESULTS
Production of TrfA proteins encoded on IncP-1β plasmid pBP136.
Although plasmid sequence analysis suggests that pBP136 encodes both TrfA1 and TrfA2, no direct evidence had been obtained so far. Therefore, we first confirmed production of both proteins from the mini-pBP136 replicon, pMS0506, in our model hosts, E. coli, P. putida, and S. japonicum, using Western blotting and determined the ratio of TrfA1 to TrfA2. Production of both TrfA1 and TrfA2 was observed for the three hosts analyzed, but TrfA levels and the TrfA2/TrfA1 ratios differed among hosts (Fig. 1C). In exponential phase, TrfA1 and TrfA2 levels (fmol/μg of total protein) were, respectively, 2.9 ± 0.3 and <0.2 for E. coli, 2.1 ± 0.5 and 0.2 ± 0.1 for P. putida, and 40 ± 2 and 13 ± 1 for S. japonicum. In stationary phase, TrfA1 and TrfA2 levels were, respectively, 7.0 ± 0.9 and 0.7 ± 0.1 for E. coli, 1.3 ± 0.2 and <0.2 for P. putida, and 18 ± 1 and 7.0 ± 1.1 for S. japonicum. The TrfA2/TrfA1 ratios in exponential phase were <0.1:1 for E. coli, 0.1:1 for P. putida, and 0.3:1 for S. japonicum. The TrfA2/TrfA1 ratios in stationary phase were 0.1:1 for E. coli, <0.1:1 for P. putida, and 0.4:1 for S. japonicum. These results indicate that pBP136, indeed, encodes both TrfA1 and TrfA2 and that TrfA1 was consistently more abundant than TrfA2 regardless of growth phase and host. However, in previous studies using plasmid RK2 in P. putida, TrfA-44 was not produced at detectable levels (7). Thus, even within the IncP-1 group, there are differences in the levels of production of the larger TrfA protein.
TrfA2 is sufficient for broad host range but not for high transformation efficiency.
Previous studies have shown that elimination of TrfA1 expression abolished replication activity of IncP-1 plasmids in P. aeruginosa (16, 54, 57). This suggests that TrfA1 could be responsible for broad-host-range replication of IncP-1 plasmids. However, these studies used limited numbers of model hosts, and the requirement for TrfA1 in plasmid replication was not demonstrated outside P. aeruginosa. To address the role of TrfA1 in determining host range, we first determined whether the trfA1 frameshift variant of pMS0506, pEvo-Sh15 (57), could replicate in S. japonicum and C. necator when introduced into these hosts by electroporation. These two strains are phylogenetically distant from the two model hosts, E. coli and P. putida, in which the TrfA2 homolog TrfA-33 was previously shown to be sufficient for RK2 replication (16, 55). Efficiency of transformation was then compared between pMS0506 and pEvo-Sh15 for four hosts: E. coli, P. putida, S. japonicum, and C. necator (Table 2). All four hosts were successfully transformed with pEvo-Sh15, indicating that the presence of TrfA2 and the intact oriV was sufficient for the IncP-1β plasmid to replicate in a broad range of hosts. However, the transformation frequency of pEvo-Sh15 was decreased more than 10-fold in E. coli, 5-fold in P. putida and S. japonicum, and 3-fold in C. necator (Table 2). This suggests that the presence of TrfA1 increases the efficiency of establishment in a broad range of hosts even though TrfA2 is sufficient for broad-host-range replication.
Table 2.
Effect of the presence of TrfA1 on the transformation efficiency of mini-IncP-1β plasmids
| Plasmid (description) | No. of transformants/109 cells/μg of DNA (relative efficiency)a |
|||
|---|---|---|---|---|
| E. coli | P. putida | S. japonicum | C. necator | |
| pMS0506 (wt) | 6.7 × 105 ± 0.8 × 105 (1.0) | 1.4 × 104 ± 0.7 × 104 (1.0) | 4.6 × 104 ± 0.2 × 104 (1.0) | 1.4 × 102 ± 0.3 × 102 (1.0) |
| pEvo-Sh15 (trfA1) | 4.4 × 104 ± 0.2 × 104 (0.1)** | 3.4 × 103 ± 0.7 × 103 (0.2) | 9.8 × 103 ± 1.1 × 103 (0.2)** | 4.4 × 101 ± 1.1 × 101 (0.3)* |
The value indicates the mean and standard error of the mean obtained from four electroporation experiments. The concentration of plasmid DNA used per electroporation was normalized on a standard curve generated by densitometric analysis of linearized and serially diluted plasmid DNA in an agarose gel stained with ethidium bromide. Relative efficiency indicates transformation efficiency relative to that for pMS0506. Significance was determined in comparison to pMS0506 (t test): *, P < 0.05; **, P < 0.01.
Requirement of TrfA1 for a high plasmid copy number.
Several studies have shown a correlation between high transformation efficiency and high plasmid copy number (PCN) (19, 42, 69). To investigate if the observed change in transformation efficiency was associated with PCN, copy numbers of pMS0506 and pEvo-Sh15 were determined in the four hosts using qPCR, as described in Materials and Methods. The copy number of pMS0506 ranged from 3.0 to 5.4 copies per chromosomal oriC region in the four hosts, while that of pEvo-Sh15 was lower, 0.9 to 2.5 copies per oriC region (Table 3). Elimination of TrfA1 thus decreased the PCN in P. putida by a factor of 2 and by a factor of 3 in E. coli, S. japonicum, and C. necator. This suggests that TrfA1 plays a role in maintaining a high PCN in a broad range of hosts.
Table 3.
Effect of the presence of TrfA1 on the copy number of mini-IncP-1β plasmids
| Plasmid (description) | PCN (relative PCN)a |
|||
|---|---|---|---|---|
| E. coli | P. putida | S. japonicum | C. necator | |
| pMS0506 (wt) | 5.2 ± 0.2 (1.0) | 4.6 ± 0.3 (1.0) | 3.0 ± 0.3 (1.0) | 5.4 ± 0.4 (1.0) |
| pEvo-Sh15 (trfA1) | 1.7 ± 0.1 (0.3)** | 2.5 ± 0.2 (0.5)** | 0.9 ± 0.1 (0.3)* | 1.6 ± 0.4 (0.3)** |
Plasmid copy number (PCN) is represented by the ratio of kleE in the plasmids and a gene in the oriC region of the chromosomes (atpB for E. coli, parB for P. putida, SJA-00140 of chromosome I for S. japonicum, and parB of chromosome I for C. necator) at exponential phase. The values indicate the mean and standard error of the mean obtained from the analyses of four total DNA samples. Relative PCN is the ratio of PCN relative to that for pMS0506. Significance was determined by comparison to pMS0506 (t test): *, P < 0.01; **, P < 0.001. Total TrfA levels in three of the four hosts under this experimental condition were as follows (in fmol/μg of total protein for pMS0506 and pEvo-Sh15, respectively): 2.2 and <0.4 for E. coli; 2.3 and 0.5 for P. putida; 53 and 13 for S. japonicum.
TrfA1 does not promote plasmid stability.
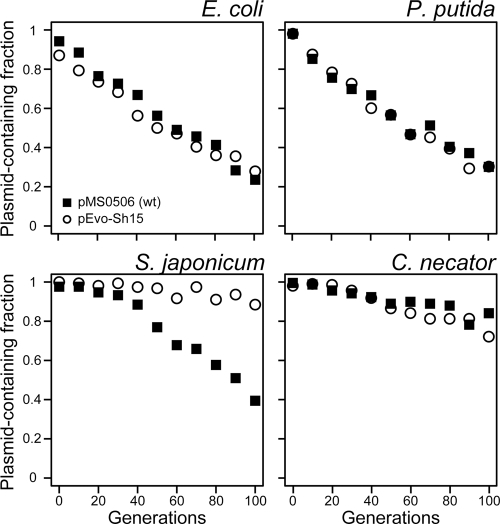
A decrease in plasmid copy number might negatively affect plasmid stability in a growing bacterial population by increasing the plasmid segregational loss rate. To assess this possibility, plasmid stability was analyzed for pMS0506 and pEvo-Sh15 in four hosts: E. coli, P. putida, S. japonicum, and C. necator. It should be noted that both plasmids used here carried intact partitioning genes. In E. coli, P. putida, and C. necator, the fraction of plasmid-bearing cells decreased at similar rates for both pMS0506 and pEvo-Sh15 (Fig. 2) even though there was a significant difference in PCN between the two plasmids in each host. Surprisingly, in S. japonicum plasmid pEvo-Sh15 carrying the trfA1 frameshift variant was drastically more stable than the wild-type pMS0506 (Fig. 2). These results indicate that when the plasmid carries the genes involved in plasmid inheritance, the presence of TrfA1 and the resulting high PCN do not contribute to, or even negatively affect, plasmid persistence.
Fig 2.
Persistence of pMS0506 and pEvo-Sh15 (the trfA1 frameshift variant) in four different bacterial hosts. Each data point represents the mean value obtained from four replicate assays for E. coli, S. japonicum, and C. necator, and three assays for P. putida.
Plasmid copy number in the presence of a single species of TrfA in the four hosts.
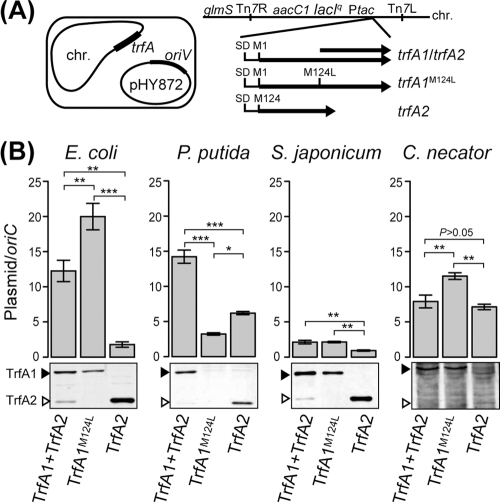
The significant decrease in both transformation frequency and PCN resulting from eliminating TrfA1 production in the cell prompted us to hypothesize that TrfA1 and TrfA2 are distinct in their abilities to generate plasmid copies. Alternatively, these decreases in transformation frequency and PCN may be simply due to decreased concentrations of total intracellular TrfA. In fact, total TrfA levels in pEvo-Sh15 were less than one-fourth of the wild-type level (see Fig. S1 and Table S2 in the supplemental material). To address this caveat, we inserted different variants of trfA in the chromosomes of the four hosts and measured the copy number of a synthetic plasmid, pHY872, which carried the pBP136 oriV but not trfA (Fig. 1B). This was done as follows. First, three trfA gene alleles were cloned under a tac promoter and a common ribosome-binding sequence: wild-type trfA (trfA1/trfA2), the trfA1 variant that does not produce TrfA2 [trfA1(M124L)], and trfA2 (Fig. 3A). These were inserted into the attTn7 site of the four host chromosomes, thus resulting in expression of the various TrfA proteins in the presence of IPTG. Subsequently, pHY872 was introduced into these 12 hosts, and its copy number was determined. It should be noted that, except in the presence of TrfA1(M124L) alone in P. putida, the total TrfA levels were higher than those produced by pMS0506 in E. coli, P. putida, and S. japonicum. The TrfA levels in C. necator could not be measured because the signal-to-noise ratio was too low in Western analysis. Below, we describe a pairwise comparison of the effect of the three TrfA variants on the PCN in each of the four hosts.
Fig 3.
Plasmid copy numbers in the presence of TrfA1 alone, TrfA2 alone, or both proteins. (A) Experimental design. TrfA proteins were expressed in the presence of 10 μM IPTG from the tac promoter inserted at the attTn7 site in the chromosomes. Plasmid copy number was determined as the ratio of the copy number of tetA in the introduced IncP-1β replicon pHY872 to the copy number of a gene in the chromosomal oriC region at stationary phase. (B) Plasmid copy numbers and TrfA levels in the four hosts. In the graphs, the value and the error bars indicate the mean and standard error of the mean obtained from at least three replicate experiments. Significance was determined by multiple comparisons (Tukey's test): *, P < 0.05; **, P < 0.01; ***, P < 0.001. The lower panels show the Western blots of the corresponding TrfA production under each condition. For each strain, the same amount of total protein was loaded in each lane of the SDS-PAGE gel: 6 μg for E. coli and P. putida, 3 μg for S. japonicum, and 25 μg for C. necator. Total TrfA levels in three hosts under this experimental condition were as follows (in fmol/μg of total protein, blots from left to right, respectively): 27, 12, and 68 for E. coli; 47, <10, and 30 for P. putida; 160, 80, and 290 for S. japonicum.
In three of the four hosts, the clones with wild-type trfA (producing both TrfA1 and TrfA2) showed higher PCNs than the clones with trfA2 alone. This was true even though similar or even higher levels of TrfA2 were found in the clones producing only TrfA2 than were found for TrfA1 produced from wild-type trfA (Fig. 3B, compare the third and first lanes of Western blots). This positive effect of the presence of TrfA1 on PCN was generally consistent with the result obtained for wild-type minireplicon pMS0506 and its derivative pEvo-Sh15 carrying the trfA1 frameshift variant (Table 2). Therefore, it is likely that the wild-type trfA gene of IncP-1β plasmids, which produces TrfA1 and TrfA2 at a certain host-specific ratio, typically generates equal or higher PCNs than the trfA2 gene, which produces only TrfA2.
When clones producing TrfA1(M124L) alone and TrfA2 alone were compared, a higher PCN was observed for TrfA1(M124L) in three of the four hosts (E. coli, S. japonicum, and C. necator) although the TrfA2 concentration was again higher than that of TrfA1(M124L) alone in E. coli and S. japonicum; in C. necator the TrfA2 level was lower than the TrfA1(M124L) level (Fig. 3B, compare third to second lanes in Western blots). In P. putida, the M124L mutation caused a decrease in the amount of TrfA1(M124L) to levels indistinguishable from the background, and the PCN was concurrently lowest in the presence of TrfA1(M124L) alone (Fig. 3B). In a second P. putida clone constructed in an independent experiment, TrfA1(M124L) was also produced at a similarly low level (data not shown), suggesting that poor production of TrfA1(M124L) was not due to a clone-specific DNA rearrangement or mutation in P. putida. The reasons for decreased TrfA1 production due to the M124L mutation are currently unclear. Together, these results suggest that when both TrfA1 and TrfA2 are synthesized at detectable levels in four phylogenetically distinct hosts, TrfA1 helps generate a higher PCN than TrfA2 can generate by itself.
Except for P. putida, clones carrying wild-type trfA produced equal or lower PCNs than clones carrying trfA1(M124L) even though the TrfA2 concentrations were much lower than those of TrfA1 (Fig. 3B, left lanes in Western blots). This suggests that a low level of expression of TrfA2 in the presence of a relatively high level of TrfA1 is sufficient to suppress the high PCN generated by TrfA1.
TrfA1 maintains higher plasmid copy number than TrfA2.
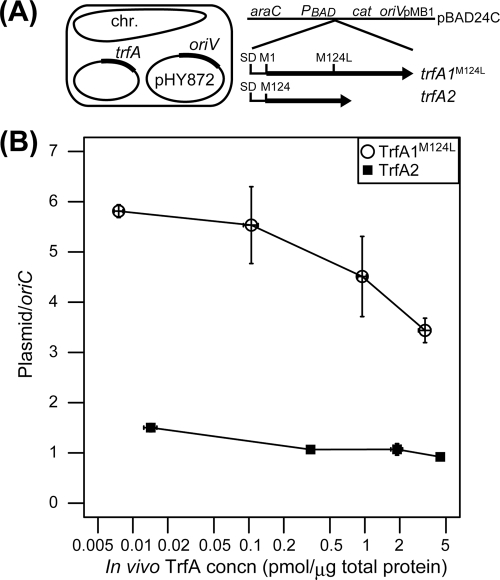
To clarify the differences in replication control between TrfA1 and TrfA2, the copy number of pHY872 was measured at different concentrations of TrfA1 or TrfA2 in E. coli. TrfA proteins were expressed from pBAD24C in the presence of different concentrations of arabinose (Fig. 4A). The TrfA concentration and the PCN (plasmid-to-oriC ratio) were determined by quantitative Western blotting and qPCR, respectively. By increasing the concentration of arabinose used for induction of protein expression, the TrfA1(M124L) concentration increased from 7.4 fmol/μg of total protein, which was approximately equivalent to the TrfA1 level in E. coli carrying pMS0506 (7.0 fmol/μg of total protein), to a level more than 400-fold higher. The TrfA2 concentration increased from twice the TrfA1 level in E. coli carrying pMS0506 to more than 600-fold the TrfA1 level. With increasing TrfA expression, the PCN decreased from 5.5 to 3.5 copies/oriC for TrfA1(M124L) and from 1.5 to 0.8 copies/oriC for TrfA2, indicating that overproduction of TrfA negatively affected replication initiation (Fig. 4B). Under these experimental conditions, TrfA1(M124L) maintained a 3- to 4-fold higher PCN than TrfA2 at any concentration of TrfA analyzed. This suggests that plasmid replication mediated by TrfA1 is more efficient than that by TrfA2.
Fig 4.
Plasmid copy numbers at different TrfA concentrations in E. coli. (A) Experimental design. Expression of TrfA1(M124L) and TrfA2 was induced from the pBAD24C constructs, pHY819 and pHY820, respectively, by addition of arabinose. Plasmid copy number was determined as the ratio of the copy number of tetA in the introduced IncP-1β replicon pHY872 to the copy number of a gene in the chromosomal oriC region at stationary phase. (B) A plot showing relationship between PCN and TrfA concentrations. Arabinose was added at the following concentrations (%, wt/vol): 0.2 × 10−5, 2 × 10−4, and 2 × 10−3 (data points from left to right). Each data point with error bars represents the mean and standard error of the mean obtained from three total DNA samples and more than three quantitative Western blot experiments. Note that the TrfA1(M124L) level without arabinose was equivalent to the TrfA1 level in E. coli harboring pMS0506.
DISCUSSION
Recent isolation and genome sequence analysis of a large number of IncP-1 plasmids have revealed a great diversity in replication initiation proteins encoded by these plasmids. Although the majority of sequenced plasmids encode both TrfA1 and TrfA2 homologs, the IncP-1β plasmid pADP-1 and IncP-1δ plasmids pEST1044 and pAKD4 encode TrfA2 alone (44, 53, 68), while the IncP-1γ plasmids encode TrfA1 alone (25; also H. Yano, L. M. Rogers, D. Sen, and E. M. Top, unpublished data) (Fig. 1A). Our study was aimed at investigating the significance of this diversity in the replication initiation protein gene, trfA, of IncP-1 plasmids. To address the role of the two Rep proteins in the ability of IncP-1 plasmids to establish themselves and be stably maintained in a wide range of bacteria, we used four model hosts belonging to three different proteobacterial subgroups, the IncP-1β minireplicon pMS0506 and its trfA1 frameshift variant pEvo-Sh15, as well as systems wherein the TrfA proteins were provided in trans to oriV.
Several reports have indicated the requirement of the TrfA1 N terminus for delivery and activation of DnaB helicase at oriV during replication initiation in P. aeruginosa (16, 55, 57). Discovery of this function of the TrfA1 N terminus led to the hypothesis that TrfA1 may be responsible for the wide host range of IncP-1 plasmids (27, 63, 70). However, there are some caveats to the assumption that the long TrfA1 protein is needed to ensure a broad host range for IncP-1 plasmids. As shown in this study, the trfA1 frameshift variant of mini-pBP136, pEvo-Sh15, was also able to replicate in bacteria from three proteobacterial subgroups (in not only the four model hosts but also A. tumefaciens [Alphaproteobacteria], Burkholderia multivorans [Betaproteobacteria], and P. fluorescens [Gammaproteobacteria]) (data not shown). Moreover, the stability of pEvo-Sh15 was equivalent to or higher than that of the wild-type plasmid in spite of decreased plasmid copy number (Fig. 2). Similarly, a trfA-44 null mutant of RK2 was previously shown to replicate in hosts currently classified as Alphaproteobacteria and Gammaproteobacteria (16). Thus, TrfA1 seems to be required for IncP-1 plasmid replication in only a limited number of bacterial species. The TrfA2 domain and the intact replication origin contain all the elements necessary for an IncP-1 plasmid to have a broad host range. Consistent with this idea is the similarity in secondary structure of the Rep protein of the broad-host-range IncW plasmid R388 (18) to TrfA2 rather than TrfA1 (data not shown).
To address the role of TrfA1, we analyzed two of the most fundamental plasmid traits, infection rate (transformation efficiency) and plasmid persistence, using pMS0506 (wild type) and pEvo-Sh15 (trfA1). First, differences in transformation efficiency were observed for most hosts analyzed (Table 2). This comparative analysis is analogous to an earlier study that used mini-RK2 and its trfA-44 frameshift variant (55). In that study, however, no differences in infection rates (transformation and conjugative transfer) between the two plasmids were observed in either E. coli or P. putida (55). Therefore, the role of the long TrfA protein in these and other hosts may be different for IncP-1α and IncP-1β plasmids. Here, we showed that TrfA1 seems to improve the plasmid's initial establishment efficiency even though TrfA2 is sufficient for broad-host-range replication.
Generally, high transformation efficiency correlates with high PCN (19, 42, 69) although this does not seem to be true for all iteron-containing plasmids (1). Plasmid pMS0506, indeed, showed a higher copy number than pEvo-Sh15 in all hosts tested (Table 3). This suggested that the coexpression of TrfA1 and TrfA2 ensures more efficient replication than the expression of TrfA2 alone. This result was confirmed in experiments using strains carrying trfA genes under a common promoter in the chromosome. Except for C. necator, hosts expressing both TrfA1 and TrfA2 produced a higher PCN than hosts expressing TrfA2 alone even though the TrfA2 level under the TrfA2-alone condition was as high as the TrfA1 level under the TrfA1-plus-TrfA2 condition. These findings suggest that the naturally occurring IncP-1 plasmids pADP-1 (44), pEST1044 (68), and pAKD4 (53), which do not encode the longer TrfA1, may exhibit lower PCNs than other IncP-1 plasmids encoding both Rep proteins. Under some environmental conditions, a low plasmid copy number may be advantageous for large plasmids encoding catabolic pathways like pADP-1 and pEST4011 because of a possibly high cost of maintaining more plasmid copies. Future studies should determine if the loss of TrfA1 production in these natural plasmids resulted in lower copy numbers than for other IncP-1 plasmids and, if so, if that in turn decreased the plasmid cost.
To eliminate TrfA2 production, we introduced the M124L mutation in the trfA1 gene, which was equivalent to the M98L mutation in trfA-44 of RK2 (17). It was previously demonstrated that the M98L mutation did not affect replication efficiency in vitro (17, 31). In our study, the equivalent M124L mutation reduced TrfA1 levels in most hosts analyzed (Fig. 3B). This was most striking in P. putida even though the introduced codon (CTG) is frequently used in P. putida. It has been reported that even a neutral mutation in the trfA-33 region of RK2 reduced both TrfA levels and replication efficiency in a temperature-dependent and host-specific manner, possibly by affecting local mRNA structure and translational efficiency (30). It is currently unclear how introduction of CTG to create the M124L mutation in trfA1 affected the mRNA structure. Another possible explanation for reduced TrfA1 levels is the potential requirement for TrfA2 in the cell in mediating stability or synthesis of TrfA1. However, contrary to our expectations, supplying TrfA2 using an additional expression plasmid vector in the P. putida strain carrying trfA1(M124L) in the chromosome did not significantly improve the TrfA1(M124L) level (data not shown). Interestingly, Fang and Helinski (17) observed a decrease in copy number of RK2 in E. coli upon introduction of the M98L mutation, and their results were also consistent with a reduction in the total TrfA levels. At this point we can assume that either elimination of TrfA2 expression from the same molecule or potential changes in the mRNA or protein structure of TrfA1 negatively affected the TrfA1 level, thus resulting in a lower PCN in P. putida. Recently, TrfA stability in vivo was shown to increase in a Lon protease-deficient host (38). The TrfA2-TrfA1 interaction may increase the stability of TrfA1 by protecting it from such proteases.
Even though the TrfA1(M124L) level was lower than the TrfA2 level, a higher PCN was observed in three of the four hosts when TrfA1(M124L) alone was expressed (Fig. 3). This suggests that replication control by TrfA1 is generally more relaxed than that by TrfA2. We confirmed this for E. coli by comparing PCNs at a range of concentrations that were similar for TrfA1 and TrfA2 (Fig. 4B). A 30% (TrfA2) or 40% [TrfA1(M124L)] decrease in the PCN of the IncP-1β replicon was observed when the TrfA level was increased to more than 45-fold (TrfA2) or 440-fold [TrfA1(M124L)] of the normal TrfA1 level of pMS0506. Replication inhibition by overproduction of the Rep protein (above the normal level) could be common throughout iteron-containing plasmids since similar conclusions were previously obtained for P1 replication and R6K γ-origin replication (13, 21).
It was unexpected that the PCNs were higher in strains wherein TrfA, especially TrfA1, was expressed from the chromosome (Fig. 3) than when it was expressed from the plasmid (Fig. 4). We note that the levels of expressed TrfA1 and TrfA2 shown in Fig. 3 (12 to 68 fmol/μg of total protein in E. coli) were in the range shown in Fig. 4, and the same copy number reference plasmid was used to generate a standard curve in qPCR in both experiments. This suggests that an additional PCN-limiting factor other than the TrfA level influenced the results in the two different experimental systems. The PCN-limiting factor involved in these experiments is currently unresolved.
Replication initiation proteins of iteron-containing plasmids mediate both activation and inhibition of replication initiation to keep the PCN constant (12). Several mechanisms could be involved in the observed differences in PCN generated by TrfA1 and TrfA2; we describe the three major ones below. The first mechanism is related to the process that generates the active form of TrfA. Both in vivo and in vitro, TrfA is present mostly as a dimer that cannot bind to iterons efficiently (64, 65). The active monomer form of TrfA, which binds to an iteron with high affinity and mediates oriV melting, can be generated by the DnaK/DnaJ/GrpE chaperone during protein synthesis or by conversion from inactive dimers by the protein chaperone ClpX or the ClpB/DnaK/DnaJ/GrpE bichaperone system (34, 36). These protein chaperones may act on inactive molecules of TrfA1 and TrfA2 with different efficiencies. Alternatively, the active fraction of TrfA1 may be higher than that of TrfA2 even without protein chaperones.
A second possible mechanism is the DnaB helicase loading and activation after oriV melting. TrfA-44 interacts with DnaB more efficiently than TrfA-33 at oriV, depending on the source of the DnaB (27, 70). In the case of pPS10, the increased interaction of RepA mutants with DnaA resulted in an increased PCN in E. coli (42, 43). In E. coli, DnaA is required in the process of DnaB loading to oriV during RK2 replication (35). Thus, it is possible that the difference in the interactions of TrfA1 versus TrfA2 with host proteins such as DnaA and DnaB affected efficiency in the DnaB loading and activation and resulted in the difference in PCN.
A third mechanism that may underlie the difference in PCNs between TrfA1 and TrfA2 is the inhibition of oriV melting by blocking the replication origin by TrfA dimers that bridge iteron clusters on separate molecules (so-called handcuffing) (31) or iteron clusters on the same molecule (71). Although the polypeptides involved in the TrfA dimer formation seem to be localized in the C-terminal region of TrfA2 (9, 64), it is unclear how TrfA monomers and dimers interact in the replication inhibition process. The presence of the N-terminal extension in TrfA1 may affect the efficiency of inhibition through monomer-monomer or monomer-dimer interactions. Future work will address these possibilities.
In addition to efficient plasmid establishment and replication in new hosts, plasmid stability is an important factor that affects the fate of a plasmid in natural bacterial communities. In three of the four hosts, no difference was observed in levels of plasmid stability between pMS0506 and pEvo-Sh15, yet in S. japonicum, pEvo-Sh15 was more stable than pMS0506. Thus, elimination of TrfA1 was beneficial for plasmid persistence in an S. japonicum population, in spite of the reduced PCN. It is likely that as long as the plasmids carry functional partitioning genes, an increase in PCN greater than 1 to 2 copies/oriC does not necessarily improve plasmid persistence. Greater plasmid stability of the pEvo-Sh15 than pMS0506 was also previously observed by us in the gammaproteobacterium Shewanella oneidensis MR-1, and in that host the copy numbers of both plasmids were indistinguishable (57). Factors other than segregational plasmid loss may limit plasmid persistence, such as fitness cost of the plasmid and low transferability (58). Since our model plasmid is conjugation deficient, we hypothesize that the low stability of pMS0506 compared to pEvo-Sh15 is due to a high fitness cost of wild-type TrfA1 in some hosts. One possible molecular mechanism is the titration of DnaB and DnaA by the TrfA1 N terminus. A second is related to the unstructured (disordered) nature of the N terminus that has been suggested by bioinformatic analysis (57, 70). Proteins carrying intrinsically unstructured regions are generally sensitive to proteolysis (23, 62), which seems to be consistent with our observation of the low TrfA1 level in the absence of TrfA2 in P. putida (Fig. 3). Expression of unstructured proteins is tightly controlled in eukaryotic systems, which implies that it imposes a high cost to the cells (23). It is therefore possible that a high rate of degradation of TrfA1 or the nature of structural disorder itself negatively affects host fitness, thereby resulting in occasional poor plasmid stability of plasmids encoding TrfA1. These hypotheses obviously need to be tested in future studies. In sum, while the molecular mechanism has not yet been elucidated, the negative effect of TrfA1 on long-term persistence of an IncP-1β plasmid has now been demonstrated in members of two different proteobacterial subgroups (Alphaproteobacteria and Gammaproteobacteria). These findings suggest that IncP-1 plasmid persistence does not always benefit from encoding the long TrfA1 protein.
In conclusion, we presented evidence that the IncP-1β plasmid-encoded TrfA1 protein participates in plasmid replication in a broad range of hosts but is not essential for replication in these hosts. However, in natural, highly diverse bacterial communities, the relaxed replication control mediated by TrfA1 could play an important role in the efficient establishment of theta replication in a broad range of hosts to which the plasmids transfer. Thus, in spite of the poor stability of plasmids encoding TrfA1 in some hosts, the promiscuous nature of IncP-1 plasmids may present strong enough selective pressure for plasmids to maintain the rather unique genetic system of a trfA locus that produces two replication initiation proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank Herbert P. Schweizer at Colorado State University for providing the mini-Tn7 constructs, Donald L Court at National Cancer Institute for E. coli strain DY380, and National BioResource Project E. coli (NIG, Japan) for strain BW25113. We also thank Patricia Hartzell at University of Idaho for use of laboratory equipment. We are grateful to Laura Frost and Igor Konieczny for helpful discussions and to three anonymous reviewers for useful suggestions.
This work was supported by NIH grants P20 RR16448 from the COBRE program of the National Center for Research Resources and R01 AI084918 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print 6 January 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abhyankar MM, Reddy JM, Sharma R, Bullesbach E, Bastia D. 2004. Biochemical investigations of control of replication initiation of plasmid R6K. J. Biol. Chem. 279:6711–6719 [DOI] [PubMed] [Google Scholar]
- 2. Adamczyk M, Jagura-Burdzy G. 2003. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim. Pol. 50:425–453 [PubMed] [Google Scholar]
- 3. Amy PS, Schulke JW, Frazier LM, Seidler RJ. 1985. Characterization of aquatic bacteria and cloning of genes specifying partial degradation of 2,4-dichlorophenoxyacetic acid. Appl. Environ. Microbiol. 49:1237–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bagdasarian M, et al. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237–247 [DOI] [PubMed] [Google Scholar]
- 6. Bahl MI, Hansen LH, Goesmann A, Sørensen SJ. 2007. The multiple antibiotic resistance IncP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established α, β and δ sub-groups. Plasmid 58:31–43 [DOI] [PubMed] [Google Scholar]
- 7. Banack T, Kim PD, Firshein W. 2000. TrfA-dependent inner membrane-associated plasmid RK2 DNA synthesis and association of TrfA with membranes of different gram-negative hosts. J. Bacteriol. 182:4380–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bao Y, Lies DP, Fu H, Roberts GP. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167–168 [DOI] [PubMed] [Google Scholar]
- 9. Blasina A, Kittell BL, Toukdarian AE, Helinski DR. 1996. Copy-up mutants of the plasmid RK2 replication initiation protein are defective in coupling RK2 replication origins. Proc. Natl. Acad. Sci. U. S. A. 93:3559–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boon N, Goris J, De Vos P, Verstraete W, Top EM. 2001. Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl. Environ. Microbiol. 67:1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 12. Chattoraj DK. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37:467–476 [DOI] [PubMed] [Google Scholar]
- 13. Chattoraj DK, Snyder KM, Abeles AL. 1985. P1 plasmid replication: multiple functions of RepA protein at the origin. Proc. Natl. Acad. Sci. U. S. A. 82:2588–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi KH, et al. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448 [DOI] [PubMed] [Google Scholar]
- 15. Doran KS, Helinski DR, Konieczny I. 1999. Host-dependent requirement for specific DnaA boxes for plasmid RK2 replication. Mol. Microbiol. 33:490–498 [DOI] [PubMed] [Google Scholar]
- 16. Durland RH, Helinski DR. 1987. The sequence encoding the 43-kilodalton trfA protein is required for efficient replication or maintenance of minimal RK2 replicons in Pseudomonas aeruginosa. Plasmid 18:164–169 [DOI] [PubMed] [Google Scholar]
- 17. Fang FC, Helinski DR. 1991. Broad-host-range properties of plasmid RK2: importance of overlapping genes encoding the plasmid replication initiation protein TrfA. J. Bacteriol. 173:5861–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fernandez-López R, et al. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30:942–966 [DOI] [PubMed] [Google Scholar]
- 19. Fernandez-Tresguerres ME, Martin M, Garcia de Viedma D, Giraldo R, Diaz-Orejas R. 1995. Host growth temperature and a conservative amino acid substitution in the replication protein of pPS10 influence plasmid host range. J. Bacteriol. 177:4377–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Filutowicz M, McEachern MJ, Helinski DR. 1986. Positive and negative roles of an initiator protein at an origin of replication. Proc. Natl. Acad. Sci. U. S. A. 83:9645–9649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Giraldo R, Fernandez-Tresguerres ME. 2004. Twenty years of the pPS10 replicon: insights on the molecular mechanism for the activation of DNA replication in iteron-containing bacterial plasmids. Plasmid 52:69–83 [DOI] [PubMed] [Google Scholar]
- 23. Gsponer J, Futschik ME, Teichmann SA, Babu MM. 2008. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 322:1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haines AS, et al. 2006. Plasmids from freshwater environments capable of IncQ retrotransfer are diverse and include pQKH54, a new IncP-1 subgroup archetype. Microbiology 152:2689–2701 [DOI] [PubMed] [Google Scholar]
- 26. Harada KM, Aso Y, Hashimoto W, Mikami B, Murata K. 2006. Sequence and analysis of the 46.6-kb plasmid pA1 from Sphingomonas sp. A1 that corresponds to the typical IncP1-β plasmid backbone without any accessory gene. Plasmid 56:11–23 [DOI] [PubMed] [Google Scholar]
- 27. Jiang Y, Pacek M, Helinski DR, Konieczny I, Toukdarian A. 2003. A multifunctional plasmid-encoded replication initiation protein both recruits and positions an active helicase at the replication origin. Proc. Natl. Acad. Sci. U. S. A. 100:8692–8697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jobanputra RS, Datta N. 1974. Trimethoprim R factors in enterobacteria from clinical specimens. J. Med. Microbiol. 7:169–177 [DOI] [PubMed] [Google Scholar]
- 29. Kamachi K, et al. 2006. Plasmid pBP136 from Bordetella pertussis represents an ancestral form of IncP1-β plasmids without accessory mobile elements. Microbiology 152:3477–3484 [DOI] [PubMed] [Google Scholar]
- 30. Karunakaran P, Blatny JM, Ertesvag H, Valla S. 1998. Species-dependent phenotypes of replication-temperature-sensitive trfA mutants of plasmid RK2: a codon-neutral base substitution stimulates temperature sensitivity by leading to reduced levels of trfA expression. J. Bacteriol. 180:3793–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kittell BL, Helinski DR. 1991. Iteron inhibition of plasmid RK2 replication in vitro: evidence for intermolecular coupling of replication origins as a mechanism for RK2 replication control. Proc. Natl. Acad. Sci. U. S. A. 88:1389–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kolatka K, Kubik S, Rajewska M, Konieczny I. 2010. Replication and partitioning of the broad-host-range plasmid RK2. Plasmid 64:119–134 [DOI] [PubMed] [Google Scholar]
- 33. Kongsuwan K, Josh P, Picault MJ, Wijffels G, Dalrymple B. 2006. The plasmid RK2 replication initiator protein (TrfA) binds to the sliding clamp β subunit of DNA polymerase III: implication for the toxicity of a peptide derived from the amino-terminal portion of 33-kilodalton TrfA. J. Bacteriol. 188:5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Konieczny I, Helinski DR. 1997. The replication initiation protein of the broad-host-range plasmid RK2 is activated by the ClpX chaperone. Proc. Natl. Acad. Sci. U. S. A. 94:14378–14382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Konieczny I, Helinski DR. 1997. Helicase delivery and activation by DnaA and TrfA proteins during the initiation of replication of the broad host range plasmid RK2. J. Biol. Chem. 272:33312–33318 [DOI] [PubMed] [Google Scholar]
- 36. Konieczny I, Liberek K. 2002. Cooperative action of Escherichia coli ClpB protein and DnaK chaperone in the activation of a replication initiation protein. J. Biol. Chem. 277:18483–18488 [DOI] [PubMed] [Google Scholar]
- 37. Król JE, et al. 2012. Role of IncP1-β plasmids pWDL7::rfp and pNB8c in chloroaniline catabolism as determined by genomic and functional analyses. Appl. Environ. Microbiol. 78:828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kubik S, Wegrzyn K, Pierechod M, Konieczny I. 2011. Opposing effects of DNA on proteolysis of a replication initiator. Nucleic Acids Res. [Epub ahead of print.] doi:10.1093/nar/gkr813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee EC, et al. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56–65 [DOI] [PubMed] [Google Scholar]
- 40. Levy SB, Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122–S129 [DOI] [PubMed] [Google Scholar]
- 41. Lykidis A, et al. 2010. The complete multipartite genome sequence of Cupriavidus necator JMP134, a versatile pollutant degrader. PLoS One 5:e9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maestro B, Sanz JM, Diaz-Orejas R, Fernandez-Tresguerres E. 2003. Modulation of pPS10 host range by plasmid-encoded RepA initiator protein. J. Bacteriol. 185:1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maestro B, et al. 2002. Modulation of pPS10 host range by DnaA. Mol. Microbiol. 46:223–234 [DOI] [PubMed] [Google Scholar]
- 44. Martinez B, Tomkins J, Wackett LP, Wing R, Sadowsky MJ. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagata Y, et al. 2011. Genomic organization and genomic structural rearrangements of Sphingobium japonicum UT26, an archetypal gamma-hexachlorocyclohexane-degrading bacterium. Enzyme Microb. Technol. 49:499–508 [DOI] [PubMed] [Google Scholar]
- 46. Norberg P, Bergstrom M, Jethava V, Dubhashi D, Hermansson M. 2011. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat. Commun. 2:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pachulec E, van der Does C. 2010. Conjugative plasmids of Neisseria gonorrhoeae. PLoS One 5:e9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Penfold RJ, Pemberton JM. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145–146 [DOI] [PubMed] [Google Scholar]
- 49. Pierechod M, et al. 2009. Conformation of a plasmid replication initiator protein affects its proteolysis by ClpXP system. Protein Sci. 18:637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 51. Schlüter A, Szczepanowski R, Pühler A, Top EM. 2007. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 31:449–477 [DOI] [PubMed] [Google Scholar]
- 52. Schmidhauser TJ, Helinski DR. 1985. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacteria. J. Bacteriol. 164:446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sen D, et al. 2010. Comparative genomics of pAKD4, the prototype IncP-1δ plasmid with a complete backbone. Plasmid 63:98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shingler V, Thomas CM. 1984. Analysis of the trfA region of broad host-range plasmid RK2 by transposon mutagenesis and identification of polypeptide products. J. Mol. Biol. 175:229–249 [DOI] [PubMed] [Google Scholar]
- 55. Shingler V, Thomas CM. 1989. Analysis of nonpolar insertion mutations in the trfA gene of IncP plasmid RK2 which affect its broad-host-range property. Biochim. Biophys. Acta 1007:301–308 [DOI] [PubMed] [Google Scholar]
- 56. Smith CA, Thomas CM. 1984. Nucleotide sequence of the trfA gene of broad host-range plasmid RK2. J. Mol. Biol. 175:251–262 [DOI] [PubMed] [Google Scholar]
- 57. Sota M, et al. 2010. Shifts in the host range of a promiscuous plasmid through parallel evolution of its replication initiation protein. ISME J. 4:1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stewart FM, Levin BR. 1977. The population biology of bacterial plasmids: a priori conditions for the existence of conjugationally transmitted factors. Genetics 87:209–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. 1987. High-copy-number and low-copy-number plasmid vectors for lacZα-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63–74 [DOI] [PubMed] [Google Scholar]
- 60. Thomas CM, Smith CA. 1987. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu. Rev. Microbiol. 41:77–101 [DOI] [PubMed] [Google Scholar]
- 61. Thorsted PB, Shah DS, Macartney D, Kostelidou K, Thomas CM. 1996. Conservation of the genetic switch between replication and transfer genes of IncP plasmids but divergence of the replication functions which are major host-range determinants. Plasmid 36:95–111 [DOI] [PubMed] [Google Scholar]
- 62. Tompa P, Prilusky J, Silman I, Sussman JL. 2008. Structural disorder serves as a weak signal for intracellular protein degradation. Proteins 71:903–909 [DOI] [PubMed] [Google Scholar]
- 63. Toukdarian A. 2004. Plasmid strategies for broad-host-range replication in Gram-negative bacteria, p 259–270 Phillips G, Funnell B. (ed), Plasmid biology. ASM Press, Washington, DC [Google Scholar]
- 64. Toukdarian AE, Helinski DR. 1998. TrfA dimers play a role in copy-number control of RK2 replication. Gene 223:205–211 [DOI] [PubMed] [Google Scholar]
- 65. Toukdarian AE, Helinski DR, Perri S. 1996. The plasmid RK2 initiation protein binds to the origin of replication as a monomer. J. Biol. Chem. 271:7072–7078 [DOI] [PubMed] [Google Scholar]
- 66. Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Trefault N, et al. 2004. Genetic organization of the catabolic plasmid pJP4 from Ralstonia eutropha JMP134 (pJP4) reveals mechanisms of adaptation to chloroaromatic pollutants and evolution of specialized chloroaromatic degradation pathways. Environ. Microbiol. 6:655–668 [DOI] [PubMed] [Google Scholar]
- 68. Vedler E, Vahter M, Heinaru A. 2004. The completely sequenced plasmid pEST4011 contains a novel IncP-1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J. Bacteriol. 186:7161–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xia G, Manen D, Yu Y, Caro L. 1993. In vivo and in vitro studies of a copy number mutation of the RepA replication protein of plasmid pSC101. J. Bacteriol. 175:4165–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhong Z, Helinski D, Toukdarian A. 2003. A specific region in the N terminus of a replication initiation protein of plasmid RK2 is required for recruitment of Pseudomonas aeruginosa DnaB helicase to the plasmid origin. J. Biol. Chem. 278:45305–45310 [DOI] [PubMed] [Google Scholar]
- 71. Zzaman S, Bastia D. 2005. Oligomeric initiator protein-mediated DNA looping negatively regulates plasmid replication in vitro by preventing origin melting. Mol. Cell 20:833–843 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.