Abstract
Streptococcus pyogenes (group A streptococcus [GAS]) is a human-specific pathogen that causes a variety of diseases ranging from superficial infections to life-threatening diseases. SpeB, a potent extracellular cysteine proteinase, plays an important role in the pathogenesis of GAS infections. Previous studies show that SpeB expression and activity are controlled at the transcriptional and posttranslational levels, though it had been unclear whether speB was also regulated at the posttranscriptional level. In this study, we examined the growth phase-dependent speB mRNA level and decay using quantitative reverse transcription-PCR (qRT-PCR) and Northern blot analyses. We observed that speB mRNA accumulated rapidly during exponential growth, which occurred concomitantly with an increase in speB mRNA stability. A closer observation revealed that the increased speB mRNA stability was mainly due to progressive acidification. Inactivation of RNase Y, a recently identified endoribonuclease, revealed a role in processing and degradation of speB mRNA. We conclude that the increased speB mRNA stability contributes to the rapid accumulation of speB transcript during growth.
INTRODUCTION
Streptococcus pyogenes (group A streptococcus [GAS]) is a Gram-positive pathogen that causes a variety of human diseases. GAS infections range from mild superficial infections, such as pharyngitis and impetigo, to life-threatening systemic diseases, such as toxic shock syndrome and necrotizing fasciitis (15). GAS also plays a significant role in the development of poststreptococcal infection sequelae, including acute rheumatic fever, acute glomerulonephritis, and reactive arthritis (15). The pathogenesis of GAS infection involves a complex host-pathogen interaction in which the streptococcal proteinase SpeB (streptococcal pyrogenic exotoxin B) plays a crucial role (47).
SpeB is a secreted cysteine proteinase with a broad spectrum of activities. SpeB cleaves human extracellular matrix proteins, such as fibrin, fibronectin, vitronectin, and matrix proteoglycans (16, 21, 41), and degrades human immunoglobulins (13, 14) and inflammatory mediators, such as complement factor C3b (46) and cathelicidin LL-37 (19). In addition, SpeB cleaves S. pyogenes surface proteins, releasing C5a peptidase and M protein (8). These observations indicate that SpeB can facilitate bacterial dissemination and survival and induce inflammation and tissue damage in the host. Clinical observations and animal experiments have clearly demonstrated the importance of SpeB in the pathogenesis of GAS infection. Accordingly, it has been observed that SpeB was abundantly present in necrotic human tissue (19) and that a decreased SpeB proteinase activity led to reduced tissue damage in a primate model for necrotizing fasciitis (36).
Since SpeB is an important virulence factor in GAS infection, it is not surprising that SpeB production is tightly regulated. Under laboratory conditions, the SpeB proteinase is usually not detected during early and mid-exponential growth phases, but it becomes highly abundant when the culture reaches late exponential and stationary phases (11, 38). SpeB production is strongly affected by culture pH and nutrient availability; for example, the optimal pH for SpeB synthesis ranges from pH 6.0 to pH 6.5 (11, 12, 28, 35), and supplementation of glucose or peptides in the growth medium usually inhibits production of the proteinase (11, 12, 38). Molecular biological studies show that SpeB is controlled at both the transcriptional and posttranslational levels. The transcription of SpeB is repressed by CovR/S (17) and Srv (39) and is activated by Rgg, which is also referred to as RopB (30), CcpA (22), and Mga (40). Among these regulators, RopB is essential for speB transcription by binding to the promoter region and facilitating transcription initiation (4, 10, 29, 33). It has yet to be determined whether speB is regulated at the posttranscriptional level.
The steady-state level of mRNA is determined by both transcript synthesis and degradation. Traditionally, the regulation of mRNA decay has been considered insignificant in prokaryotes, though this opinion has slowly changed, as a growing body of literature suggested that the regulation of mRNA turnover is widely distributed in many bacterial species (5). Barnett et al. (7) reported that the expression of certain “growth phase-dependent” genes, such as sagA and sda (encoding streptolysin S and streptodornase, respectively), were primarily regulated at the mRNA decay level, indicating an important role of posttranscriptional regulation on S. pyogenes virulence. These transcripts were more abundant in stationary phase than in exponential phase, mainly because their stability increased dramatically in the stationary phase (7). It was later found that ribonucleases J1 and J2 were involved in the decay process of these genes (9). Additionally, the mRNAs of prominent genes (mga, covR, and ska) have been shown to exhibit widely differing half-lives (45).
The aim of this study was to determine how S. pyogenes regulates SpeB at the posttranscriptional level leading to the rapid accumulation of speB transcripts during growth. By combining Northern blot analysis and quantitative reverse transcription-PCR (qRT-PCR), we observed that speB mRNA stability increased gradually during exponential growth and that the mRNA degradation process was pH dependent. RNase Y (encoded by cvfA), a recently identified endoribonuclease of S. pyogenes (20), is involved in speB mRNA processing and degradation, but other yet unidentified nucleases are also required. We conclude that the increased speB mRNA stability contributes to the rapid accumulation of speB transcript during growth.
MATERIALS AND METHODS
Bacterial strains and growth condition.
Bacterial strains used in this study are listed in Table 1. S. pyogenes NZ131 (serotype M49) was routinely grown in C medium (0.5% proteose peptone 3, 1.5% yeast extract, 10 mM K2HPO4, 0.4 mM MgSO4, 17 mM NaCl) (30) at 37°C without aeration. Erythromycin and spectinomycin, when required, were added at a final concentration of 2 μg/ml and 100 μg/ml, respectively.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Bacterial strains | ||
| S. pyogenes | ||
| NZ131 | Wild type | 32 |
| ΔspeBmutant | speB mutant strain | This study |
| ΔpnpAmutant | pnpA mutant strain | This study |
| ΔacpAmutant | acpA mutant strain | This study |
| ΔcvfAmutant | cvfA mutant strain | This study |
| ΔcvfA+pDL278::cvfA | ΔcvfA mutant strain carrying pDL278::cvfA | This study |
| E. coliDH5α | Cloning strain | New England Biolabs |
| Plasmids | ||
| pDL278 | Shuttle vector | 24 |
| pDL278::cvfA | pDL278 carrying a gene encoding cvfA | This study |
Total RNA extraction, cDNA synthesis, and real-time PCR analysis.
Overnight cultures of S. pyogenes were diluted 1:40 in fresh C medium and grown at 37°C to the desired growth phase. Streptococcal cells were harvested by centrifugation (5,000 × g, 5 min, 4°C). Cell pellets were resuspended in TRIzol (Invitrogen) and stored at −80°C. To isolate RNA, cells were disrupted three times for 30 s each time using lysing matrix B (MP Biomedicals, Solon, OH) in a FastPrep FP210 homogenizer (Thermo Scientific) (speed setting of 6.5). Total RNA isolation was carried out according to the manufacturer's instructions (isolation of total RNA using TRIzol, Invitrogen). RNA samples were treated with Turbo DNase (Ambion) to remove traces of chromosomal DNA. RNeasy MiniElute cleanup kit (Qiagen) was used to purify RNA samples after DNase treatment. cDNA was synthesized from 1 μg of total RNA by using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed as described previously (2, 31). Briefly, the relative amounts of gene-specific cDNA were quantified by the comparative threshold cycle (CT) method using the Bio-Rad (Hercules, CA) MyiQ real-time PCR detection system with fluorescein-spiked SYBR green as the fluorophore. The primers were designed to have a melting temperature between 59.5°C and 60.5°C and to amplify 110- to 115-bp fragments. The amplification efficiency was between 90% and 110%, which was determined by analysis of the standard curve of real-time PCR with template dilution over 5 orders of magnitude. The CT value of each gene of interest (CT_goi) was normalized against the CT value of the 16S rRNA (CT_16S) (ΔCT = CT_goi − CT_16S). The 16S rRNA was used as the endogenous reference, because its abundance was consistent from early exponential phase to early stationary phase (data not shown). The relative abundance (RA) of each gene compared to that of 16S rRNA was calculated using the equation RA = 2−ΔCT.
Northern blot analysis.
Total RNA without DNase treatment was used for Northern blot analysis. RNA samples were frozen and thawed no more than once to minimize degradation. One to five micrograms of total RNA was separated on a 1% agarose–0.66 M formaldehyde gel in a buffer consisting of 40 mM 3-[N-morpholino]propanesulfonic acid (MOPS) (pH 7.0), 10 mM sodium acetate, and 0.2 mM EDTA. RNA was then transferred to a Hybond-N membrane (Amersham) in a Trans-Blot SD semidry transfer cell (Bio-Rad) and immobilized to the membrane by UV cross-linking (Stratagene). Gene-specific digoxigenin (DIG) probes were PCR generated by using a PCR DIG probe synthesis kit (Roche Diagnostic). Primers used for DIG probe synthesis are listed in Table 2. Hybridization of the DIG-labeled DNA probes to RNA on the membrane and visualization of the hybrid with CDP-Star were carried out according to the manufacturer's instructions (DIG application manual, Roche Diagnostic). The abundance of gene transcript, represented by the averaged pixel intensity of a band with defined size, was quantified with ImageJ software (1). The amount of 23S rRNA, which was visualized by ethidium bromide staining on an agarose gel prior to transfer, served as a loading control.
Table 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| Primers for qRT-PCR | |
| speB_1l | GTGGAGTCTCTGACGGCTTC |
| speB_1r | TGCCTACAACAGCACTTTGG |
| ropB_1l | TGCCTTGGTCAAGGTGTT |
| ropB_1r | GCACAGTCTCATAGTGACTCCA |
| 16S rRNA_1l | AAGCAACGCGAAGAACCTTA |
| 16S rRNA_1r | GTCTCGCTAGAGTGCCCAAC |
| Primers for construction of mutants | |
| Erm-L | CCGGGCCCAAAATTTGTTTGAT |
| Erm-R | AGTCGGCAGCGACTCATAGAAT |
| Erm-F(ptls) | GAAGGAGTGATTACATGAACAAAAA |
| cvfA_P1 | GGACGCTCAAAAGGTTCTCA |
| cvfA_P2(erm) | CAAATCAAACAAATTTTGGGCCCGGCCAATGAGGGCAGAAACAAT |
| cvfA_P3(erm) | ATAATTCTATGAGTCGCTGCCGACTGAGCGGTTGATTATGCCAAG |
| cvfA_P4 | AGCTGAAGGATCTGGGTGAA |
| speB_P1 | GGTCAATAGCCAGATGCGATA |
| speB_P2(erm) | CAAATCAAACAAATTTTGGGCCCGGTGATCGGCAAATACTGGGTTA |
| speB_P3(erm) | ATAATTCTATGAGTCGCTGCCGACTCGCACTAAACCCTTCAGCTC |
| speB_P4 | TCGAGACGAGTTTGGTGTTG |
| pnpA_P1 | CAGGTTTGGTCACAGGCTTT |
| pnpA_P2(erm_ptls) | TTTTTGTTCATGTAATCACTCCTTCGGTTTCCCTGCAAATGTTGT |
| pnpA_P3(erm) | ATAATTCTATGAGTCGCTGCCGACTCCCACCAAAACCAGAGAAAA |
| pnpA_P4 | TGAAGACTCCAGGAGCGATT |
| acpA_P1 | AGCCATGACGCTATTGATCC |
| acpA_P2(erm_ptls) | TTTTTGTTCATGTAATCACTCCTTCAGGAGACGATGCTCGTTAGC |
| acpA_P3(erm) | ATAATTCTATGAGTCGCTGCCGACTCCCTTGCCCAACTTAGTGAG |
| acpA_P4 | TCTCTGCTTCTTGGCCACTT |
| cvfA_L(BamHI) | AAAAAAGGATCCCATCATGGACGACTTGCTACA |
| cvfA_R(HindIII) | AAAAAAAAGCTTGAACATTTTCAAAGGCAAGTCA |
| Primers for DIG-labeled probe synthesis | |
| speB_L (probe) | CTATCAAAGCAGGTGCACGA |
| speB_R (probe) | TAATTTGAGCAGTTGCAGTAGCA |
| ropB_L (probe) | GGAAATTGGTGAAACCGTTG |
| ropB_R (probe) | AAACATATGATGGATCGTTTTGC |
mRNA decay assay.
Streptococcal cells were grown in C medium to the desired cell density. Rifampin was added to the culture to a final concentration of 1 mg/ml. Five-milliliter aliquots of the culture were withdrawn at different time points after the addition of rifampin, rapidly chilled to 0°C by mixing with 10 ml crushed ice, and harvested by centrifugation (5,000 × g, 5 min, 4°C). The transcript abundance of a particular gene at each time point was determined by qRT-PCR or Northern blot analyses. Excel software (Microsoft) was used to perform regression analysis and to calculate mRNA decay rates.
SpeB and RNase mutant construction.
Overlap extension PCR technique (44) was used to construct the S. pyogenes mutants shown in Table 1. Primer sequences for the construction of mutants are given in Table 2. For speB mutant (ΔspeB) construction, the upstream and downstream regions of the speB gene were PCR amplified with primer pairs speB_P1/speB_P2(erm) and speB_P3(erm)/speB_P4, respectively. An erythromycin resistance cassette (erm) was PCR amplified from a shuttle vector pHS17 (18) with primer pair Erm-L/Erm-R (L stands for left, and R stands for right). The speB_P2(erm) and speB_P3(erm) primers were oligonucleotide hybrids with 5′-end sequences complementary to the erm cassette and 3′-end sequences complementary to the flanking regions of the speB gene. PCR-generated fragments of the region upstream of speB, the erm cassette, and the region downstream of speB were mixed at 1:1:1 molar ratio and amplified with primer pair speB_P1/speB_P4. This amplification led to the “ligation” of the three fragments in the order of 5′-speB_upstream–erm–speB_downstream-3′. The resulting PCR product was used to transform S. pyogenes NZ131 to generate the speB mutant. The same strategy was applied for construction of other mutants. For pnpA and acpA mutant construction, a promoterless erm fragment [generated by a primer pair Erm-F(ptls)/Erm-R (ptls stands for promoterless)] was used to replace the pnpA or acpA gene so that expression of the gene downstream was not affected.
ΔcvfA mutant complementation.
A shuttle vector, pDL278 (24), was used to introduce the cvfA gene into the ΔcvfA mutant. The cvfA gene was PCR amplified with primer pairs cvfA_L(BamHI)/cvfA_R(HindIII). The PCR product was digested with restriction enzymes BamHI and HindIII (Promega) and ligated to the pDL278 vector that was similarly digested. The resulting plasmid, pDL278-cvfA, was introduced into S. pyogenes ΔcvfA mutant via electroporation to generate the ΔcvfA complemented strain (ΔcvfA mutant carrying plasmid pDL278::cvfA [ ΔcvfA+pDL278::cvfA]).
SpeB proteinase activity assay.
The overnight cultures of wild-type S. pyogenes and its derivatives were stab inoculated on a C-medium-based agar plate containing 1.5% skim milk. The plate was incubated at 37°C for 18 h in a candle jar. Caseinolytic activity results in a translucent zone around the stab site (10).
RESULTS
Increased speB mRNA abundance during exponential growth.
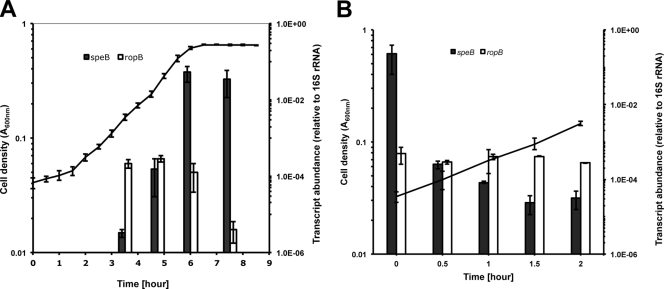
Previous studies demonstrated growth-dependent production of SpeB in S. pyogenes, with the highest proteinase activity detected in late exponential and stationary phases (11, 28). Furthermore, a novel peptide-mediated regulatory system has recently been shown to be involved in expression control of speB in late exponential and early stationary phase (43). In this study, we examined the level of speB transcript during bacterial growth with qRT-PCR. An overnight culture of S. pyogenes was diluted 1:100 in C medium and incubated at 37°C, and bacterial samples were withdrawn at different growth phases (Fig. 1A). The speB transcript level was extremely low in early exponential phase (A600 = 0.15). It increased by 50-fold from early to mid-exponential phase (A600 = 0.3) and increased by 340-fold from mid- to late exponential phase (A600 = 0.6). The overall speB transcript abundance increased by over 10,000-fold from early to late exponential phase. After the cells entered the stationary phase, the speB transcript abundance slightly decreased but was still considerably higher than in the early and mid-exponential phases. We also examined the transcription profile of the ropB gene, which encodes a positive transcriptional regulator of speB (33). The ropB gene was expressed at a constant level during exponential growth and diminished quickly upon entering stationary phase (Fig. 1A). These observations suggested that speB and ropB transcripts were regulated by distinct mechanisms.
Fig 1.
Changes in the levels of speB and ropB transcripts during growth. (A) S. pyogenes cells were continuously grown in C medium from early exponential phase to early stationary phase. (B) An aliquot of cell culture from the late exponential phase was diluted 1:20 in fresh C medium and incubated for 2 h. qRT-PCR analysis was used to determine the levels of speB and ropB transcripts at different time points. Data are presented as means ± standard deviations (error bars) from three (A) or two (B) independent experiments.
Our data showed a rapid increase of speB transcript abundance when cells grew from early to late exponential phase. Correspondingly, one would expect a rapid decrease of speB transcript abundance when cells are diluted to simulate a low-cell-density environment. To confirm this hypothesis, we grew S. pyogenes cells until late exponential phase and diluted the culture 1:20 in fresh prewarmed medium so that the cells reentered the early exponential phase. qRT-PCR analysis detected a rapid decrease of the speB transcript level immediately after dilution (Fig. 1B). The most significant change occurred within the first 30 min, when the total amount of speB transcripts decreased by almost 1,000-fold. After that, the speB transcript level continued to decrease and became 10,000-fold less than that of the inoculum after 1.5 h. In contrast, the ropB transcript remained at a constant level throughout the incubation. These data clearly show that S. pyogenes can accumulate or specifically destroy a large amount of speB transcripts over a short period of time and suggest that speB and ropB mRNA degradation is regulated by distinct mechanisms.
Increased speB mRNA stability during growth.
We initially hypothesized that the speB mRNA stability might increase during exponential growth, which might contribute to the rapid accumulation of speB mRNA from early to late exponential phase. To test this hypothesis, speB mRNA stability was measured in different growth phases with Northern blot analyses and qRT-PCR.
Northern blot analysis is a standard method for mRNA stability assays (3). It allows detection of differently sized transcripts from the same gene. Since each speB and ropB mRNA species has two transcripts of different sizes (33), Northern blot analysis makes it possible to measure the decay rate of each individual transcript. However, signal detection and quantification become difficult when the target mRNA level is very low. To circumvent the problem of low mRNA abundance, qRT-PCR detection was employed because of its high sensitivity, broad dynamic range, and direct quantitative measurement and because several studies have confirmed the validity of qRT-PCR in mRNA decay analysis (25, 34, 42). In this study, qRT-PCR primers were designed to target the protein-encoding regions of speB and ropB genes so that the results reflected the overall mRNA decay rate of each gene (Table 2). To verify qRT-PCR results for mRNA decay measurements, Northern blot analysis was performed to determine the decay rate of each transcript when target mRNA was sufficiently abundant for reliable detection.
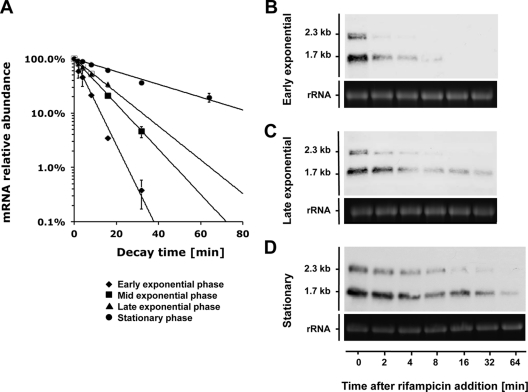
The speB mRNA decay rates appeared to follow first-order kinetics with corresponding exponential regression coefficients (R2) greater than 0.95 under all prevailing growth conditions. qRT-PCR analysis revealed an increasing trend of speB mRNA stability along the growth curve (Fig. 2A and Table 3). We could not directly measure the speB mRNA stability in the early exponential phase (A600 = 0.15), because the transcript abundance was extremely low in that condition (Fig. 1). An alternative strategy was used instead. Fifty milliliters of S. pyogenes culture was grown until early stationary phase when speB was highly expressed, and the cells were collected by centrifugation. The cell pellet was immediately suspended in 250 ml prewarmed C medium (1:5 dilution) to simulate the early exponential phase. qRT-PCR results showed that speB mRNA degraded rapidly under this condition (Fig. 2A). The stability of speB mRNA gradually increased as the cells grew into mid- and late exponential phases, and speB mRNA became very stable at early stationary phase (Fig. 2A). These findings were confirmed by Northern blot analysis (Fig. 2B to D). speB mRNA has two transcript sizes, 2.3 kb and 1.7 kb, with the short transcript (1.7 kb) being the dominant form. The short transcript was also more stable than the long transcript (2.3 kb) in a given growth phase. Nevertheless, both transcripts displayed an increasing trend of stability when cells grew from early exponential to stationary phase.
Fig 2.
Growth phase-dependent speB mRNA decay. Decay rates of speB mRNA in different growth phases were determined by qRT-PCR (A) and Northern blot analyses (B, C, and D). Note that to analyze speB mRNA decay in early exponential phase, we used stationary culture that was diluted in fresh medium to mimic the condition (see Results for details). qRT-PCR data are presented as means ± standard deviations (error bars) from two independent experiments. For Northern blot analysis, 23S rRNA was used as a loading control. One microgram of total RNA was loaded per lane.
Table 3.
Estimation of speB mRNA half-lives in different growth phases
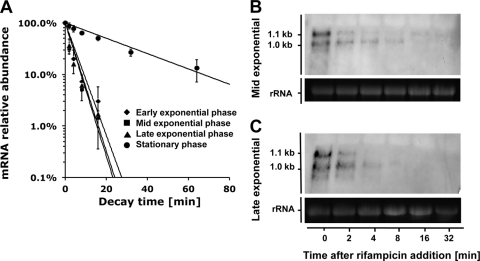
In contrast to speB, the stability of ropB mRNA remained unchanged during exponential growth and increased dramatically when cells entered the stationary phase (Fig. 3 and Table 4). ropB mRNA also has two sizes of transcripts, 1.1 kb and 1.0 kb. The two ropB transcripts were similar in abundance at the beginning of the decay assay (t = 0 min), though the long transcript (1.1 kb) diminished faster than the short transcript (1.0 kb), indicating that the short transcript was more stable.
Fig 3.
Growth phase-dependent ropB mRNA decay. Decay rates of ropB mRNA in different growth phases were determined by qRT-PCR (A) and Northern blot analyses (B and C). qRT-PCR data are presented as means ± standard deviations from two independent experiments. For Northern blot analysis, 23S rRNA was used as a loading control. Five micrograms of total RNA was loaded per lane.
Table 4.
Estimation of ropB mRNA half-lives in different growth phases
Effect of environmental pH on speB mRNA stability.
Two environmental factors, pH and nutrient availability, change continuously when bacteria actively grow in a batch culture. Because C medium has a weak buffering capacity, the culture pH decreased from pH 7.4 to pH 6.2 as S. pyogenes cells grew from early to late exponential phase and remained constant at pH 6.2 after the cells entered stationary phase (data not shown). C medium is used in this study because it supports a high level of expression of SpeB (30). The medium is rich in peptides and poor in carbohydrate (30). We assumed that the availability of an energy source was the limiting factor of bacterial growth, the depletion of which led to entry into the stationary phase. This assumption was supported by the fact that the bacterial culture in early stationary phase resumed growth immediately after the addition of exogenous glucose (data not shown). We hypothesized that either environmental pH or energy source availability or both affected speB mRNA stability.
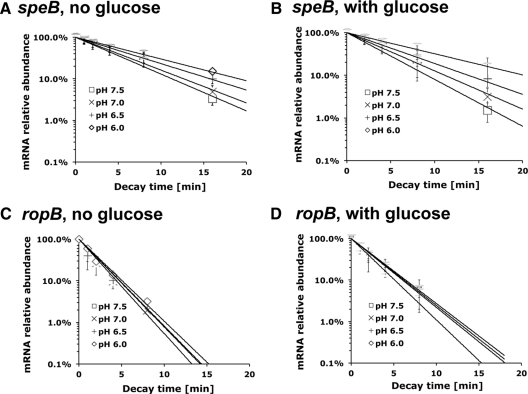
To test this hypothesis, S. pyogenes cells were grown until late exponential phase, when speB mRNA was highly abundant. The culture was then divided into four aliquots. Each aliquot was adjusted with 2 M Bis-Tris buffer (the final concentration of Bis-Tris buffer is 0.1 M) to pH 7.5, pH 7.0, pH 6.5, or pH 6.0 and was supplemented with 1% glucose or not supplemented with glucose. Rifampin was added simultaneously with Bis-Tris buffer and glucose to inhibit new mRNA synthesis. qRT-PCR results showed that the adjusted culture pH had an instant effect on speB mRNA stability (Fig. 4A and B and Table 5). The speB mRNA degraded rapidly at neutral pH and became more stable when the culture medium was gradually acidified. This pattern was observed in cultures supplemented with glucose or cultures not supplemented with glucose. At a given environmental pH, the supplementation of glucose did not obviously change speB mRNA stability. We conclude that environmental pH, rather than energy source availability, played a major role in regulating speB mRNA stability. This finding can at least partially explain the increased SpeB activity measured under low pH as shown before (28). We also measured ropB mRNA stability in different environmental conditions. qRT-PCR results showed that ropB mRNA degraded at a constant rate under all pH conditions. The presence of glucose led to a slight increase of ropB mRNA stability, though the change was too small to be considered biologically significant (Fig. 4C and D and Table 5). These findings indicated that in contrast to speB mRNA variations in stability, ropB mRNA stability was not affected by environmental pH or energy source availability.
Fig 4.
speB and ropB mRNA decay under different environmental conditions. S. pyogenes NZ131 was grown to late exponential phase (A600 = 0.55) and divided into four aliquots. The culture pH was artificially adjusted from pH 7.5 to pH 6.0 with Bis-Tris buffer (final concentration of 0.1 M). Glucose (1% [wt/vol]) was added to the culture as indicated. The mRNA decay assay was conducted immediately after the medium was modified. qRT-PCR was used to determine the decay rates of speB mRNA (A and B) and ropB mRNA (C and D). Data are presented as the averages ± standard deviations from two independent experiments.
Table 5.
Estimation of speB and ropB mRNA half-lives under different pH conditions based on qRT-PCR analysis in Fig. 4
| pH | Estimated half-life (min) of: |
|||
|---|---|---|---|---|
|
speBmRNA |
ropBmRNA |
|||
| Without glucose | With glucose | Without glucose | With glucose | |
| 7.5 | 3.3 ± 0.2 | 2.8 ± 0.5 | 1.5 ± 0.1 | 2.0 ± 0.5 |
| 7.0 | 3.7 ± 0.5 | 3.6 ± 0.8 | 1.5 ± 0.0 | 1.8 ± 0.0 |
| 6.5 | 5.1 ± 0.5 | 5.2 ± 0.1 | 1.6 ± 0.1 | 1.7 ± 0.3 |
| 6.0 | 6.1 ± 0.2 | 8.4 ± 2.0 | 1.6 ± 0.1 | 2.0 ± 0.1 |
Search for RNase(s) involved in speB mRNA degradation.
The degradation of RNA molecules is a tightly controlled process involving different types of RNases. The genome of S. pyogenes NZ131 contains genes that encode at least 11 RNases (32), 4 exoribonucleases, and 9 endoribonucleases (RNases J1 and J2 have both exo- and endoribonuclease activities). In this study, RNases polynucleotide phosphorylase (PNPase) (encoded by pnpA), RNase III (encoded by acpA), and RNase Y (encoded by cvfA) were chosen and investigated for their possible roles in speB mRNA degradation. RNases J1 and J2 are essential and cannot be inactivated (9).
PNPase is an exoribonuclease that catalyzes the 3′-5′ phosphorolytic degradation of RNA (6). PNPase in S. pyogenes is involved in the decay of at least two gene transcripts, sagA and sda (7). RNase III is an endoribonuclease that specifically cleaves double-stranded RNAs (37). Its function in S. pyogenes remains to be determined. RNase Y from Bacillus subtilis is an endoribonuclease that is the functional equivalent of RNase E from Escherichia coli (26, 27). Its ortholog in S. pyogenes (CvfA) is involved in the expression of multiple virulence factors, including SpeB (20).
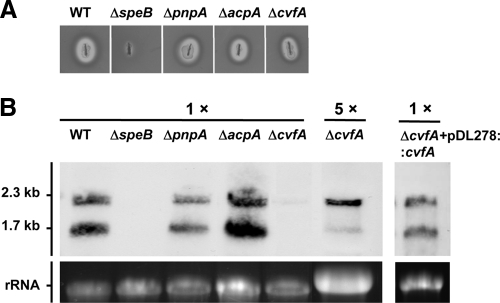
We constructed isogenic null mutants of PNPase (ΔpnpA), RNase III (ΔacpA), and RNase Y (ΔcvfA) by allelic exchange. All RNase mutants were viable, suggesting that these RNases were dispensable for the organism. All three mutants formed a translucent zone on a skim milk plate (Fig. 5A), indicating that functionally active SpeB proteinase was produced and secreted in these strains. This observation is at variance with a previous study that the proteinase activity was undetected in the cvfA mutant (20). We attribute this variance to the different serotypes of the strains used in the two studies (strain NZ131 [serotype M49] versus strain HSC5 [serotype M5]). Northern blot analysis was then performed to determine the level of speB mRNA in these strains at early stationary phase. No hybridization signal was detected in the ΔspeB mutant, confirming that the speB probe was highly specific (Fig. 5B). The wild type and PNPase (ΔpnpA) and RNase III (ΔacpA) mutants produced similar amounts of speB mRNA, with the short transcript being more abundant than the long transcript (Fig. 5B). These two RNase mutants were not further studied. In contrast, the RNase Y mutant (ΔcvfA) produced considerably less speB mRNA than the other strains. The hybridization signal was detected only when the sample was concentrated (Fig. 5B). One striking feature was that the relative levels of two speB transcripts were inverted in the ΔcvfA mutant, with the long transcript being much more abundant than the short transcript. To rule out a possible polar effect or unexpected mutation that may occur during mutant construction, the cvfA gene was reintroduced into the ΔcvfA mutant on a shuttle vector, pDL278. Northern blot analysis showed that the speB transcript pattern was restored to the wild-type level after the cvfA gene complementation (Fig. 5B), which confirmed that the altered speB transcript pattern was due to the cvfA gene deletion.
Fig 5.
SpeB expression in RNase mutants and ΔcvfA complementation strain. (A) SpeB proteinase activities in RNase mutants were visualized on a 1.5% skim milk agar plate. The wild type (WT) and ΔspeB mutant were included as positive and negative controls, respectively. (B) The level of speB mRNA transcript in each strain at early stationary phase was determined by Northern blot analysis. The samples were used as found (1 ×) or concentrated fivefold (5 ×). One microgram of total RNA was loaded per lane for each strain except for the ΔcvfA mutant. For the ΔcvfA mutant, 5 μg of total RNA was used so that the hybridization signal was comparable to the other strains.
Role of RNase Y in speB mRNA degradation.
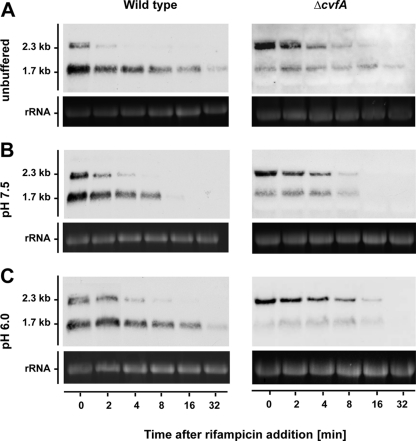
The speB mRNA abundance in ΔcvfA strain was further examined. qRT-PCR results showed that, unlike the wild-type strain, the level of speB mRNA in the ΔcvfA mutant was very low during exponential growth and increased rapidly only immediately before the cells entered stationary phase (data not shown). This expression pattern made it technically difficult to collect ΔcvfA samples for an mRNA decay assay, because the assay required samples expressing high levels of speB mRNA but were still exponentially growing. To solve this problem, 1% glucose was added to bacterial cultures in the early stationary phase (ES plus 1% glucose) so that the cells “reentered” the exponential phase. A similar speB mRNA decay rate under this condition (ES plus 1% glucose) and in late exponential phase (compare Fig. 6A and Fig. 2C) in the wild-type strain was observed. Cells treated this way (ES plus 1% glucose) should behave similarly to those in late exponential phase.
Fig 6.
speB mRNA decay in wild type and ΔcvfA strains under different pH conditions. Bacterial strains were grown until early stationary phase, and 1% glucose was added to the culture so that the cells resumed growth. The culture pH was either unbuffered (pH 6.2) (A) or buffered to pH 7.5 (B) or pH 6.0 (C). Rifampin was added immediately after the pH was modified. For Northern blot analysis, 23S rRNA was used as a loading control. One microgram (wild type) or 5 μg (ΔcvfA) of total RNA was loaded per lane.
Subsequently, speB mRNA decay rates in the wild type and ΔcvfA mutant were compared. Cells were grown until early exponential phase (30 min after reaching the maximal A600) and supplemented with 1% glucose. The culture was either unbuffered (pH 6.2), or buffered to pH 7.5 or 6.0 with Bis-Tris buffer (final concentration of 0.1 M), and mRNA decay was determined immediately after modification. The ΔcvfA mutant showed a fivefold-reduced abundance of speB mRNA compared to the wild type. To compensate for the low abundance of speB mRNA, the total RNA amount was therefore increased 5-fold and used in the Northern blot assay. Again, we observed an altered speB transcript pattern in the ΔcvfA strain, with the long transcript being the most dominant form under all three conditions (Fig. 6). We also observed the following. (i) In a given strain (wild type or ΔcvfA) under a given environmental condition (unbuffered, pH 7.5, or pH 6.0), the speB short transcript was always more stable than the long transcript was. (ii) In a given strain, both short and long transcripts were more stable at low pH (unbuffered, pH 6.0) than at high pH (pH 7.5). (iii) Under a given environmental condition, RNase Y contributes to the stability of the speB short transcript, but not long transcript (Fig. 6 and Table 6). On the basis of these observations, we conclude that RNase Y is not responsible for pH-dependent regulation of speB mRNA degradation, although it apparently plays a general role in determining the stability of the speB short transcript.
Table 6.
Estimation of speB transcript half-lives based on Northern blot analysis in Fig. 6
| pH | Estimated speB mRNA half-life (min) |
|||
|---|---|---|---|---|
| WT |
ΔcvfA mutant |
|||
| Short transcript | Long transcript | Short transcript | Long transcript | |
| 6.2 (unbuffered) | 16.9 | 3.5 | 27.0 | 3.7 |
| 7.5 | 3.3 | 1.7 | 5.4 | 1.6 |
| 6.0 | 10.0 | 2.8 | 14.5 | 3.4 |
DISCUSSION
The degradation of mRNA appears to play an important regulatory role in the actual availability of several virulence factors in GAS (9). Furthermore, it has been suggested that growth phase-dependent regulation of GAS gene expression is predominantly regulated at the level of mRNA stability (9, 20). In the present study, we used the important GAS virulence gene speB to further dissect posttranscriptional regulation in S. pyogenes. Using Northern blot analysis and qRT-PCR to detect the level of speB mRNA, we measured speB mRNA abundance and decay under several environmental conditions. The main results of mRNA abundance and decay measurements suggest that the apparent increase in speB mRNA abundance is the result of increased speB mRNA stability in the late logarithmic and early stationary phase. The smooth accumulation processes of speB mRNA abundance suggest that speB gene expression is controlled by the same regulatory mechanism from early to late exponential phase.
The rapid accelerating degradation rate of speB mRNA implies the existence of a strong mRNA degradation mechanism in S. pyogenes. A search for putative RNases in the genome of S. pyogenes NZ131 revealed 11 candidates (32). Our previous microarray data showed that most RNase-encoding genes were actively transcribed in both exponential and early stationary phases (data not shown) (23). On the basis of these findings, we propose that the mRNA degradation mechanism is highly active throughout exponential growth. The accumulation of speB mRNA is because more transcripts are synthesized than can be serviced by the degradosome. A highly active degradation mechanism would predict that disruption of speB mRNA synthesis would result in a quick degradation of the accumulated mRNA. This was observed in our dilution experiment and during the challenge experiment with different pHs. We observed an immediate increase in speB mRNA degradation during the shift to pH 7.5, demonstrating the presence of an active degradosome and excluding new synthesis since rifampin was administered simultaneously. If speB mRNA was simply stabilized by a stopped degradation after entry into stationary phase, it is unlikely that the message would be so rapidly degraded upon the dilution of stationary-phase cells. Our data also suggest a pH-dependent regulation of the speB-specific degradosome.
What could be the evolutionary rationale for not simply stopping mRNA degradation entirely to accumulate speB transcripts during exponential growth? A highly active degradation process at any given time means that the cell can eliminate the speB transcript and therefore SpeB production by preventing its translation. Because SpeB is a potent extracellular proteinase, control of speB mRNA abundance by its synthesis and degradation rates may enable S. pyogenes to respond to environmental changes in a more time-efficient manner. One example shown in this study is the dilution experiment, in which the speB mRNA abundance decreases by 1,000-fold within 30 min after the cells from late exponential phase are diluted in fresh medium (Fig. 1B). This drastic change can occur only when (i) the synthesis of new speB mRNA is completely stopped and (ii) the existing mRNA molecules are rapidly degraded.
One major environmental signal influencing speB mRNA stability and SpeB abundance is the culture pH (Fig. 4) (28). The speB transcript is more stable at a low culture pH than neutral pH. This explains why the speB mRNA stability increased during exponential growth, when the culture medium was gradually acidified by metabolic products (lactic acid, etc.). In contrast to speB, ropB mRNA stability is not affected by environmental pH. This observation further supports the notion that the pH-dependent regulation of mRNA stability is not a universal phenomenon but seems to be gene specific, since ropB mRNA abundance remains constant during exponential growth with a short half-life. The constitutive expression of the ropB gene suggests a balanced synthesis and degradation of ropB mRNA. This observation is at variance with a previous study (33), in which the ropB gene was expressed in the late stage of exponential growth. We consider that the variance may be due to the different serotypes of the strains used in the two studies (serotypes M14 and M49). However, in the same study (33), the authors also demonstrated that ectopic expression of ropB (i.e., ropB was expressed in the early stage of exponential growth) had no influence on the pattern of speB gene expression. This result is consistent with the results of our study, since we did not detect any correlation between speB mRNA abundance and ropB expression.
RNase Y (also known as CvfA) is a recently identified endoribonuclease that controls SpeB abundance in S. pyogenes (20). RNase Y seems to be part of the RNA degradosome in Gram-positive bacteria, which also includes the metabolic enzyme enolase and phosphofructokinase. We found in the present study that the speB mRNA abundance was obviously reduced in the RNase Y mutant, confirming previous findings (20). We also report for the first time that the mutation of RNase Y led to an altered speB transcript pattern. The long transcript, but not the short transcript, was the dominant form of speB mRNA species. The exact mechanism of how S. pyogenes produces two sizes of speB transcripts is still unclear. Our findings favor the hypothesis that the short transcript might be a processed product from the primary transcript through endonucleolytic cleavage and that RNase Y might be a major enzyme responsible for the cleavage. Since the speB short transcript is significantly more stable than the long transcript, the conversion of speB mRNA from long to short transcript may increase the overall stability of speB mRNA, which in turn promotes the production of protein. We are currently testing this hypothesis.
ACKNOWLEDGMENTS
J.K. was supported by NIH/NIDCR grant 4R00DE018400.
We thank Jimmy Ballard and Justin Merritt (Department of Microbiology and Immunology, University of Oklahoma Health Sciences Center) for helpful discussions.
Footnotes
Published ahead of print 20 January 2012
REFERENCES
- 1. Abramoff MD, Magelhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 2. Ajdic D, Pham VT. 2007. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J. Bacteriol. 189:5049–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alwine JC, Kemp DJ, Stark GR. 1977. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl. Acad. Sci. U. S. A. 74:5350–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anbalagan S, McShan WM, Dunman PM, Chaussee MS. 2011. Identification of Rgg binding sites in the Streptococcus pyogenes chromosome. J. Bacteriol. 193:4933–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson KL, Dunman PM. 2009. Messenger RNA turnover processes in Escherichia coli, Bacillus subtilis, and emerging studies in Staphylococcus aureus. Int. J. Microbiol. 2009:525491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andrade JM, Pobre V, Silva IJ, Domingues S, Arraiano CM. 2009. The role of 3′-5′ exoribonucleases in RNA degradation. Prog. Mol. Biol. Transl. Sci. 85:187–229 [DOI] [PubMed] [Google Scholar]
- 7. Barnett TC, Bugrysheva JV, Scott JR. 2007. Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus. J. Bacteriol. 189:1866–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berge A, Bjorck L. 1995. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J. Biol. Chem. 270:9862–9867 [DOI] [PubMed] [Google Scholar]
- 9. Bugrysheva JV, Scott JR. 2010. The ribonucleases J1 and J2 are essential for growth and have independent roles in mRNA decay in Streptococcus pyogenes. Mol. Microbiol. 75:731–743 [DOI] [PubMed] [Google Scholar]
- 10. Chaussee MS, Ajdic D, Ferretti JJ. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SpeB production. Infect. Immun. 67:1715–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chaussee MS, Phillips ER, Ferretti JJ. 1997. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect. Immun. 65:1956–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cohen JO. 1969. Effect of culture medium composition and pH on the production of M protein and proteinase by group A streptococci. J. Bacteriol. 99:737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collin M, Olsen A. 2001. Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect. Immun. 69:7187–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Collin M, Olsen A. 2001. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 20:3046–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cunningham MW. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elliott SD. 1945. A proteolytic enzyme produced by group A streptococci with special reference to its effect on the type-specific M antigen. J. Exp. Med. 81:573–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Graham MR, et al. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. U. S. A. 99:13855–13860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haake SK, Yoder SC, Attarian G, Podkaminer K. 2000. Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems. J. Bacteriol. 182:1176–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johansson L, et al. 2008. Cathelicidin LL-37 in severe Streptococcus pyogenes soft tissue infections in humans. Infect. Immun. 76:3399–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kang SO, Caparon MG, Cho KH. 2010. Virulence gene regulation by CvfA, a putative RNase: the CvfA-enolase complex in Streptococcus pyogenes links nutritional stress, growth-phase control, and virulence gene expression. Infect. Immun. 78:2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kapur V, et al. 1993. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb. Pathog. 15:327–346 [DOI] [PubMed] [Google Scholar]
- 22. Kietzman CC, Caparon MG. 2010. CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect. Immun. 78:241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kreth J, Chen Z, Ferretti J, Malke H. 2011. Counteractive balancing of transcriptome expression involving CodY and CovRS in Streptococcus pyogenes. J. Bacteriol. 193:4153–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130–145 [DOI] [PubMed] [Google Scholar]
- 25. Leclerc GJ, Leclerc GM, Barredo JC. 2002. Real-time RT-PCR analysis of mRNA decay: half-life of beta-actin mRNA in human leukemia CCRF-CEM and Nalm-6 cell lines. Cancer Cell Int. 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lehnik-Habrink M, et al. 2011. RNase Y in Bacillus subtilis: a natively disordered protein that is the functional equivalent to RNase E from Escherichia coli. J. Bacteriol. 193:5431–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lehnik-Habrink M, et al. 2011. RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol. Microbiol. 81:1459–1473 [DOI] [PubMed] [Google Scholar]
- 28. Loughman JA, Caparon M. 2006. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J. Bacteriol. 188:399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lyon WR, Caparon MG. 2003. Trigger factor-mediated prolyl isomerization influences maturation of the Streptococcus pyogenes cysteine protease. J. Bacteriol. 185:3661–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lyon WR, Gibson CM, Caparon MG. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Malke H, Steiner K, McShan WM, Ferretti JJ. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296:259–275 [DOI] [PubMed] [Google Scholar]
- 32. McShan WM, et al. 2008. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J. Bacteriol. 190:7773–7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neely MN, Lyon WR, Runft DL, Caparon M. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 185:5166–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Driscoll L, Daly C, Saleh M, Clynes M. 1993. The use of reverse transcriptase-polymerase chain reaction (RT-PCR) to investigate specific gene expression in multidrug-resistant cells. Cytotechnology 12:289–314 [DOI] [PubMed] [Google Scholar]
- 35. Ogburn CA, Harris TN, Harris S. 1958. Extracellular antigens in steady-state cultures of the hemolytic Streptococcus: production of proteinase at low pH. J. Bacteriol. 76:142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Olsen RJ, Musser JM. 2010. Molecular pathogenesis of necrotizing fasciitis. Annu. Rev. Pathol. 5:1–31 [DOI] [PubMed] [Google Scholar]
- 37. Panganiban AT, Whiteley HR. 1983. Purification and properties of a new Bacillus subtilis RNA processing enzyme. Cleavage of phage SP82 mRNA and Bacillus subtilis precursor rRNA. J. Biol. Chem. 258:12487–12493 [PubMed] [Google Scholar]
- 38. Podbielski A, Woischnik M, Kreikemeyer B, Bettenbrock K, Buttaro BA. 1999. Cysteine protease SpeB expression in group A streptococci is influenced by the nutritional environment but SpeB does not contribute to obtaining essential nutrients. Med. Microbiol. Immunol. 188:99–109 [DOI] [PubMed] [Google Scholar]
- 39. Reid SD, et al. 2006. Inactivation of the group A Streptococcus regulator srv results in chromosome wide reduction of transcript levels, and changes in extracellular levels of Sic and SpeB. FEMS Immunol. Med. Microbiol. 48:283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ribardo DA, McIver KS. 2006. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol. Microbiol. 62:491–508 [DOI] [PubMed] [Google Scholar]
- 41. Schmidtchen A, Frick IM, Bjorck L. 2001. Dermatan sulphate is released by proteinases of common pathogenic bacteria and inactivates antibacterial alpha-defensin. Mol. Microbiol. 39:708–713 [DOI] [PubMed] [Google Scholar]
- 42. Schmittgen TD, et al. 2000. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 285:194–204 [DOI] [PubMed] [Google Scholar]
- 43. Shelburne, et al. 2011. An amino-terminal signal peptide of Vfr protein negatively influences RopB-dependent SpeB expression and attenuates virulence in Streptococcus pyogenes. Mol. Microbiol. 82:1481–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Song JH, et al. 2005. Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol. Cells 19:365–374 [PubMed] [Google Scholar]
- 45. Steiner K, Malke H. 2002. Dual control of streptokinase and streptolysin S production by the covRS and fasCAX two-component regulators in Streptococcus dysgalactiae subsp. equisimilis. Infect. Immun. 70:3627–3636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Terao Y, et al. 2008. Group A streptococcal cysteine protease degrades C3 (C3b) and contributes to evasion of innate immunity. J. Biol. Chem. 283:6253–6260 [DOI] [PubMed] [Google Scholar]
- 47. von Pawel-Rammingen U, Bjorck L. 2003. IdeS and SpeB: immunoglobulin-degrading cysteine proteinases of Streptococcus pyogenes. Curr. Opin. Microbiol. 6:50–55 [DOI] [PubMed] [Google Scholar]