Abstract
This study examined differences in antibiotic-resistant soil bacteria and the presence and quantity of resistance genes in soils with a range of management histories. We analyzed four soils from agricultural systems that were amended with manure from animals treated with erythromycin and exposed to streptomycin and/or oxytetracycline, as well as non-manure-amended compost and forest soil. Low concentrations of certain antibiotic resistance genes were detected using multiplex quantitative real-time PCR (qPCR), with tet(B), aad(A), and str(A) each present in only one soil and tet(M) and tet(W) detected in all soils. The most frequently detected resistance genes were tet(B), tet(D), tet(O), tet(T), and tet(W) for tetracycline resistance, str(A), str(B), and aac for streptomycin resistance, and erm(C), erm(V), erm(X), msr(A), ole(B), and vga for erythromycin resistance. Transposon genes specific for Tn916, Tn1549, TnB1230, Tn4451, and Tn5397 were detected in soil bacterial isolates. The MIC ranges of isolated bacteria for tetracycline, streptomycin, and erythromycin were 8 to >256 μg/ml, 6 to >1,024 μg/ml, and 0.094 to >256 μg/ml, respectively. Based on 16S rRNA gene similarity, isolated bacteria showed high sequence identity to genera typical of soil communities. Bacteria with the highest MICs were detected in manure-amended soils or soils from agricultural systems with a history of antibiotic use. Non-manure-amended soils yielded larger proportions of antibiotic-resistant bacteria, but these had lower MICs, carried fewer antibiotic resistance genes, and did not display multidrug resistance (MDR).
INTRODUCTION
Antibiotic resistance is a major health concern which has developed over time from resistance to single classes of antibiotics to multidrug resistance and extreme drug resistance. Many of the problematic resistance genes have spread due to their relocation from the chromosomes of environmental bacteria to a mobile element and then to clinical pathogens (8, 35, 37, 39, 43). The soil is also a natural reservoir of antibiotic-producing bacteria containing both intrinsic resistance mechanisms and transferable resistance genes. Approximately 50% of Actinomycetes organisms isolated from soil are capable of synthesizing antibiotics, which provides a natural antibiotic residue in soil (36).
Agricultural practices have a major impact on the selection and promotion of antibiotic-resistant bacteria, as they provide a positive selective pressure for these bacteria (7, 13, 15, 34, 49, 54, 56, 59, 66). Antibiotic residues in agricultural soils resulting from direct applications or indirect exposure via manure/biosolid amendments can range from a few μg/kg up to g/kg (58). Numerous reports describe the positive selection pressure of antibiotic use in animal growth promotion and animal husbandry on the selection of antibiotic-resistant bacterial pathogens (55, 60, 64). Antibiotic-resistant pathogens have also been isolated from fruit orchards and vegetable farms, which use antibiotics as prophylactic treatments and also use soils that have been amended with manure. The use of antibiotics in agriculture, as growth promoters, and in veterinary medicine has been suggested to contribute to increased rates of resistance in human pathogens (21, 23, 50). Antibiotic resistance in the environment is also affected directly by the release of antibiotic-resistant bacteria and antibiotic residues via waste from humans receiving therapeutic antibiotics. Most antibiotics are degraded only partially in sewage treatment plants (32), and erythromycin and tetracycline appear to undergo no degradation during sewage treatment processes. Antibiotic degradation rates in the environment vary considerably and can be dependent upon environmental conditions: tetracycline degrades by 24% within 10 to 180 days, and erythromycin degrades by 25% within 30 days, but streptomycin is not degraded for up to 30 days after environmental release (58). Degradation rates in soil are particularly dependent upon antibiotic properties (molecular structure and chemical and physical properties), edaphic properties (pH, minerals, and organic matter), and/or other conditions (temperature, aerobicity, adsorption, and absorption). Residual antibiotics can affect the soil microbiome, resulting in differential inhibition of certain microorganisms and in perturbations in community composition (11, 12, 18, 44, 45, 48). The increasing use of antibiotics in medicine, veterinary medicine, and agricultural production systems has coincided with increasing development of high levels of antibiotic resistance and novel antibiotic resistances (1, 2, 3, 4, 32, 53). Despite mounting concern over the existence, rising detection, and levels of resistant bacteria, particularly multidrug-resistant (MDR) bacteria, in clinical and natural environments, few studies have focused on characterization of antibiotic resistance in natural environments (30, 44).
This study examined soils with different management histories in order to compare the influence of the use of antibiotics, manure, and intensive farming on the selection of tetracycline-, streptomycin-, or erythromycin-resistant bacteria and antibiotic resistance genes. Tetracycline, streptomycin, and erythromycin resistance genes and associated resistance elements were detected in cultured bacteria and quantified in DNAs isolated from arable farmland, vegetable garden, fruit orchard, composted, and forest soils.
MATERIALS AND METHODS
Soils and compost.
Investigations were performed on six soils and compost collected from different management systems, including (i) compost from a biodynamic garden obtained by composting plant residues (manure and/or antibiotics were not used), (ii) soil from a pine forest in the Masuria region of Poland (manure and/or antibiotics were not used), (iii) soil from a vegetable garden (where manure was applied), (iv) soil from an apple orchard (Warka, Poland), (v) soil from a mixed fruit orchard (Warka), and (vi) soil from arable farmland in the area of Lesznowola (Masovian Voivodeship, Poland). Soils 3 to 6 were used for intensive cultivation of vegetables and fruits, and soils 4 to 6 were from cultivation systems with a history of antibiotic use (streptomycin and oxytetracycline). The manure was obtained from animals which were treated with erythromycin. Soil samples (two 1-kg replicates for each source) were taken from the humus profile (depth of 10 to 15 cm) into sterile glass flasks in October 2008 and 2009 and were analyzed within 1 to 2 days after collection. From each sample, 1 to 5 g was taken for analysis, and the remainder was stored at 8°C.
Quantification and characterization of microorganisms.
The following three tests were performed to quantify and characterize the culturable microorganisms in soils and compost. The number of viable culturable microorganisms was evaluated by inoculating nutrient broth (Difco, Detroit, MI) with serial dilutions of soil samples and plating them onto nutrient agar (Difco). The number of endospore-forming bacterial cells was determined by plating samples that had been heated at 70°C for 10 min onto nutrient agar. The various physiological groups of microorganisms present in soil samples were identified by inoculating selective agar or liquid medium. McCrady's most-probable-number (MPN) method was used to determine the CFU/ml (40). The numbers of amylolytic, proteolytic, lipolytic, sulfate-reducing, denitrifying, ammonifying, nitrifying, actinomycetous, and urea-hydrolyzing bacteria were determined using specific media, as described previously (48). After sterilization, all media were supplemented with cycloheximide (50 μg/liter) and nystatin (10 μg/liter). Liquid cultures or plates were incubated at 26°C for 3 days (for proteolytic, amylolytic, ammonifying, and ureolytic bacteria), 7 days (for actinomycetous, denitrifying, lipolytic, and sulfate-reducing bacteria), or 14 days (for nitrifying bacteria). The number of bacteria was calculated per gram (wet weight) of soil.
Isolation and identification of antibiotic-resistant soil bacteria.
Antibiotic-resistant bacteria were isolated from each soil by suspending a soil sample in saline and plating it onto R2A agar complete medium (Graso Biotech, Starogard Gdański, Poland) supplemented with 10 μg/ml of tetracycline, streptomycin, or erythromycin. The plates were incubated at room temperature for 3 to 4 days. Strains were stored at 4°C on antibiotic agar plates and in LB supplemented with 10% glycerol at −70°C.
The complete 16S rRNA gene was used to identify bacterial isolates. The colony PCR method was used for the amplification of 16S rRNA genes. Amplification reaction mixtures of 50 μl were composed of 1 to 4 μl of lysed cell sample, buffer for DreamTaq polymerase with 1.5 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate (dNTP), 1 mM (each) primers 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT), and 1 U of DreamTaq polymerase (Fermentas) (33). PCR was performed using a Mastercycler EP S gradient thermocycler (Eppendorf, Hamburg, Germany) under the following conditions: 5 min at 94°C followed by 20 cycles of 30 s at 94°C, 50 s at 53°C, and 1 min 20 s at 72°C, 15 cycles of 30 s at 94°C, 30 s at 46°C, and 1 min 20 s at 72°C, and 1 cycle of 10 min at 72°C. The PCR products were separated by 0.8% agarose gel electrophoresis and purified using a PCR purification kit (Qiagen, Hilden, Germany) or a Gel Out kit (DNA Gdansk II) according to the manufacturer's instructions. The PCR amplicons were sequenced at Genomed (Warsaw, Poland). Sequence analysis was performed using the Clone Manager-9 program (a commercial bioinformatic software work suite of Sci-Ed that supports molecular biologists with data management). Identification to the species level was performed by comparison with the Ribosomal Database Project database (http://rdp.cme.msu.edu/) and by using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Genus-level and species-level identifications were assigned using the following criteria: ≥99% identity of a 16S rRNA gene sequence to a reference entry identified a bacterium to the species level, while 95 to 98.9% identity identified a bacterium to the genus level.
Antibiotic resistance screening.
The susceptibility of the bacterial isolates to tetracycline, streptomycin, and erythromycin, at concentrations of 30 μg/ml, 25 μg/ml, and 15 μg/ml, respectively, was determined using the standard EUCAST disk diffusion method (19). The plates were incubated at room temperature for 24 to 48 h. The diameters (in millimeters) of the zones of growth inhibition around the disks were measured using precision calipers.
Determination of MICs.
The MICs of tetracycline, streptomycin, and erythromycin against the soil isolates were determined using Etest (bioMérieux, Marcy l'Etoile, France) on Mueller-Hinton agar plates (Oxoid, Basingstoke, United Kingdom) according to the manufacturers' instructions.
qPCR amplification of streptomycin and tetracycline resistance genes.
Total soil DNA was extracted from soil samples by using a MoBio Power soil DNA isolation kit (Süd-Laborbedarf GmbH, Gauting, Germany) according to the manufacturer's instructions. The relative quantities of the streptomycin resistance genes aad(A), str(A), and str(B), as well as IS1133, and the tetracycline resistance genes tet(B), tet(M), and tet(W) (see Table S1 in the supplemental material) in soil DNA were determined using multiplex quantitative real-time PCR (qPCR) as previously described (41, 47, 63).
Identification of resistance genes.
Tetracycline, streptomycin, and erythromycin resistance genes and associated transposon (Tn916, TnB1230, Tn1549, Tn5397, and Tn4451) sequences were identified by PCR amplification using primers specific for each gene, as previously described (46) (see Tables S2 to S5 in the supplemental material). Positive and negative controls were used in each run. For the design of specific PCR primers for the resistance genes, reference resistance gene nucleotide sequences were extracted from the NCBI (http://www.ncbi.nlm.nih.gov/) and ARDB (http://arpcard.mcmaster.ca/) databases. The reference nu-cleotide sequences AF321548, AJ862840, AY602212, AY602406, and EF031554 were used to design primers for PCR amplification of the streptomycin resistance genes str(A), str(B), aad(K), aad(A), and aac, respectively. Escherichia coli strains containing the aad(A) gene and the plasmid RSF1010 [carrying the str(A) and str(B) genes] were used as positive controls (strain Se 131 [GenBank accession no. AJ238350] and strain CB613 [GenBank accession no. AF027768], respectively), and E. coli DH5α was used as the negative control. PCR was performed under the following conditions: 5 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 49 or 60°C, and 1 min at 72°C, with one final cycle of 7 min at 72°C. Amplicons were separated in 0.8% or 1% agarose gels.
RESULTS AND DISCUSSION
Characterization of total microbial populations in soil.
The total numbers of heterotrophic bacteria determined by growth in nutrient broth and on nutrient agar were higher for compost (9.5 × 107 CFU/g and 3.6 × 106 CFU/g, respectively), vegetable garden soil (6.7 × 107 CFU/g and 4.5 × 106 CFU/g, respectively), and arable farmland soil (2.4 × 107 CFU/g and 6.5 × 105 CFU/g, respectively) than for forest soil (2.0 × 105 CFU/g and 5.0 × 104 CFU/g, respectively) and apple orchard soil (6.5 × 105 CFU/g and 3.5 × 104 CFU/g, respectively) or mixed fruit orchard soil (2.0 × 106 CFU/g and 6.4 × 105 CFU/g, respectively). Forest soil contains fewer easily assimilable organic compounds required to support the growth of a heterotrophic microbiota (61). The largest numbers of spore-forming bacteria were found in compost and in apple orchard soil (4.1 × 105 and 1.3 × 105 cells/g, respectively). This was probably due to the small amount of easily accessible organic matter in mature compost or orchard soils. The depletion of easily degradable organic compounds is known to result in an increase in the number of spore-forming bacteria (61). Amylolytic, proteolytic, lipolytic, ammonifying, nitrifying, denitrifying, sulfate-reducing, actinomycetous, and urea-hydrolyzing bacteria were quantified in the soil samples (see Table S6 in the supplemental material). The number of bacteria in the individual physiological groups (e.g., amylolytic, proteolytic, ammonifying, or nitrifying) was significant in many cases. The mixed fruit orchard soil contained a smaller number of specific groups of bacteria (especially the lipolytic and denitrifying groups) than the other soils. All of the soils contained bacteria that are potentially capable of degrading proteins, hydrocarbons, and fats formed during the biodegradation of organic matter. Microorganisms that contribute to conversion of carbon, nitrogen, and sulfur were also present in large numbers in all soils (Table S6).
Species of bacteria isolated from studied soils under the pressure of the antibiotics tested.
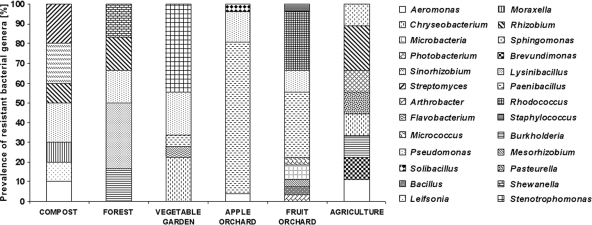
Bacterial isolates showed identity to the typical genera of soil bacteria (27), including Arthrobacter, Bacillus, Chryseobacterium, Moraxella, Paenibacillus, Pseudomonas, Rhizobium, Shewanella, Sphingomonas, Stenotrophomonas, and Streptomyces (Fig. 1; see Table S7 in the supplemental material). Bacteria from taxa reported to include opportunistic human and/or animal pathogens (i.e., Aeromonas salmonicida, Burkholderia cepacia, Chryseobacterium meningosepticum, Moraxella sp., Photobacterium damselae, Pseudomonas fluorescens, Sphingomonas multivorum, and Stenotrophomonas maltophilia) were isolated from all soils (14, 25, 29).
Fig 1.
Prevalences of resistant bacteria belonging to different genera identified in the studied soils.
Relative abundances of streptomycin and tetracycline resistance genes.
qPCR analysis of tetracycline and streptomycin resistance genes was performed on DNAs extracted directly from soil and compost samples. There was little variation in the relative quantities of the tet(M), tet(W), and str(B) genes in DNAs extracted from the six soils and from compost (see Table S8 in the supplemental material). The DNA extracted from the apple orchard soil contained a high relative quantity of aad(A). The relative quantities of the tet(B) and str(A) genes were low in the mixed fruit orchard soil and compost DNAs, respectively, but higher than in the other soil DNAs (Fig. 2; see Table S8). The tet(M) and tet(W) genes were detected at almost identical relative quantities in the DNAs extracted from the six soils. However, they were not detected by PCR in bacteria isolated from three soils. The streptomycin resistance genes aad(A) and str(A) were detected by PCR in bacteria isolated from four and five soils, respectively, but were detected in only one soil by qPCR. The relative quantities of str(B) were similar in all soil DNAs, but it was detected in the isolates from only three soils. The insertion sequence IS1133, associated with the plasmid-mediated str(A) and str(B) genes, was not detected.
Fig 2.
Quantities of antibiotic resistance genes in soils. (A) Forest soil, compost, and agricultural soil; (B), vegetable garden soil, mixed fruit orchard soil, and apple orchard soil.
The methods of resistance detection (direct culture and PCR amplification of resistance genes) and quantification (qPCR) complement each other. The str(B) gene was detected only by qPCR in three soils, tet(M) was detected only by qPCR in two soils, and tet(W) was detected only by qPCR in two soils. In these soils, these genes were not detected in the culturable bacteria by use of PCR. When the genes were present in sufficient quantities in both the soil and culturable bacteria, they were detected using both assays. These results therefore identified that the str(B), tet(M), and tet(W) genes were also present in nonculturable bacteria or in bacteria not phenotypically resistant to the antibiotics. The qPCR method may not be able to detect genes present at levels lower than the detection limits of the assay (between 10 fg and 1 pg), but unlike PCR or culture methods, it provides a quantification of the resistance genes detected. However, culture of bacteria with antibiotic-specific selection ensures a sufficient quantity of DNA for PCR analysis but is limited to the detection of resistance in culturable bacteria. Antibiotic resistance genes have not previously been quantified in environmental samples where streptomycin has been applied or in soil samples without previous fertilization or treatment. Previous studies investigating antibiotic resistance genes in the environment by use of PCR have been limited to qualitative detection (9, 10, 24, 42, 62).
Susceptibility to antimicrobial agents and determination of MIC.
The antibiotic susceptibility of pure cultures of bacterial strains to tetracycline, streptomycin, and erythromycin was initially examined using the disk diffusion method (data not shown). The MICs of tetracycline, streptomycin, and erythromycin against pure cultures of bacteria isolated from compost (Table 1), forest soil (Table 2), vegetable garden soil (Table 3), apple orchard soil (Table 4), mixed fruit orchard soil (Table 5), and arable farmland soil (Table 6) reflected the results from the disk diffusion method (19). We observed no growth inhibition zones or high MICs of tetracycline, streptomycin, and erythromycin for Aeromonas salmonicida, Brevundimonas vesicularis, and Chryseobacterium jejuense. High-level resistance to two antibiotics was observed for Chryseobacterium meningosepticum, Lysinibacillus sphaericus, Moraxella spp., Photobacterium damselae, Rhizobium radiobacter, Shewanella putrefaciens, Sinorhizobium meliloti, Sphingomonas multivorum, and Streptomyces griseus. The diversity of antibiotic-resistant bacterial species was highest in the vegetable garden soil amended with manure and in orchard soils from beneath trees that had been treated in the past with foliar sprays of streptomycin and tetracycline for disease protection. The compost, forest soil, and arable farmland soils yielded a lower species diversity, in contrast to a previous study that suggested that a reduction in bacterial species variation was due to the selection pressure of antibiotics or manure (18, 20).
Table 1.
Susceptibility profiles and corresponding antibiotic resistance genes of strains isolated from composta
| Strain | TET MIC (μg/ml) | TET resistance gene(s) | STR MIC (μg/ml) | STR resistance gene(s) | E MIC (μg/ml) | E resistance gene(s) | Transposon gene |
|---|---|---|---|---|---|---|---|
| Aeromonas salmonicida T-C5 | 128 | tet(B), tet(T) | ND | ND | 32 | erm(C) | I |
| Leifsonia xyli subsp. xyli T-C3 | 32 | tet(B), tet(O) | ND | ND | 32 | erm(V), erm(X), vga | ND |
| Moraxella S-C9 | ND | ND | 512 | ND | 48 | ole(B) | I |
| Pseudomonas luteola T-C1 | 32 | tet(D), tet(O), tet(T), tet(W) | ND | ND | 32 | erm(V), vga | I |
| Pseudomonas luteola S-C6 | ND | ND | 512 | str(A) | 32 | ND | ND |
| Rhizobium radiobacter S-C7 | ND | ND | 512 | str(A) | 48 | erm(C) | ND |
| Sinorhizobium meliloti S-C8 | ND | ND | 512 | str(A) | 48 | ole(B) | I |
| Sinorhizobium meliloti S-C10 | ND | ND | 512 | str(A) | 48 | erm(C), ole(B) | ND |
| Streptomyces griseus subsp. griseus T-C4 | 128 | tet(D) | ND | ND | 32 | erm(C), erm(X), msr(A) | ND |
TET, tetracycline; STR, streptomycin; E, erythromycin; I, Tn916 or Tn1549 integrase; ND, not detected.
Table 2.
Susceptibility profiles and corresponding antibiotic resistance genes of strains isolated from forest soila
| Strain | TET MIC (μg/ml) | TET resistance gene(s) | STR MIC (μg/ml) | STR resistance gene(s) | E MIC (μg/ml) | E resistance gene(s) | Transposon gene |
|---|---|---|---|---|---|---|---|
| Burkholderia pseudomallei T-F2 | 16 | tet(D) | ND | ND | 48 | msr(A) | ND |
| Lysinibacillus sphaericus S-F5 | ND | ND | 512 | str(A), str(B), aac | 256 | msr(A) | I |
| Lysinibacillus sphaericus S-F6 | ND | ND | 512 | str(A), aac | 48 | ole(B) | |
| Pseudomonas fluorescens T-F1 | 32 | tet(B), tet(D), tet(O), tet(T), tet(W) | ND | ND | 256 | erm(C), erm(X) | I |
| Rhizobium radiobacter T-F3 | 128 | tet(A), tet(B), tet(D), tet(M), tet(O), tet(T), tet(W) | ND | ND | 256 | erm(C), erm(V), erm(X) | I, RE, RS |
| Shewanella putrefaciens T-F4 | 128 | tet(B), tet(D), tet(M), tet(O), tet(T), tet(W) | ND | ND | 256 | erm(V), erm(X), msr(A) | I, RE, RS |
TET, tetracycline; STR, streptomycin; E, erythromycin; I, Tn916 or Tn1549 integrase; RE, Tn4451 recombinase; RS, Tn5397 resolvase; ND, not detected.
Table 3.
Susceptibility profiles and corresponding antibiotic resistance genes of strains isolated from vegetable garden soila
| Strain | TET MIC (μg/ml) | TET resistance gene(s) | STR MIC (μg/ml) | STR resistance gene(s) | E MIC (μg/ml) | E resistance gene(s) | Transposon gene |
|---|---|---|---|---|---|---|---|
| Chryseobacterium piscium T-VG4 | 8.0 | tet(T) | 12 | ND | 2 | ND | I, RS |
| Chryseobacterium jejuense TES-VG6 | >256 | tet(D) | >1,024 | aad(K) | >256 | ND | RS |
| Chryseobacterium ginsengisoli ES-VG9 | 16 | tet(W) | >1,024 | ND | 32 | ND | RE, RS |
| Chryseobacterium ginsengisoli TES-VG11 | 16 | tet(D) | 64 | ND | 4.0 | ND | I, RE |
| Flavobacterium sp. TES-VG14 | ND | ND | 64 | ND | 4.0 | ND | ND |
| Paenibacillus pabuli TES-VG13 | ND | ND | >1,024 | ND | 0.125 | ND | I |
| Pseudomonas jessenii TES-VG12 | ND | ND | 96 | ND | >256 | erm(V), erm(X) | I |
| Pseudomonas mandelii TE-VG10 | 16 | tet(D), tet(O), tet(T) | 0.064 | ND | >256 | erm(C), erm(X) | RE |
| Pseudomonas putida ES-VG7 | ND | ND | 8.0 | aad(A) | >256 | erm(C) | ND |
| Pseudomonas putida TES-VG8 | 16 | tet(T), tet(W) | 12.0 | aad(A) | >256 | erm(X) | I |
| Stenotrophomonas maltophilia TE-VG1 | 12 | tet(M) | 64 | ND | >256 | ND | RE |
| Stenotrophomonas maltophilia TE-VG2 | 48 | tet(O) | 96 | aac | >256 | ND | ND |
| Stenotrophomonas maltophilia TE-VG3 | >256 | ND | 126 | ND | >256 | ND | ND |
| Stenotrophomonas maltophilia TE-VG5 | 32 | tet(T) | 192 | aac | >256 | erm(C) | RE |
| Stenotrophomonas rhizophila TE-VG15 | 32 | ND | 16 | ND | >256 | erm(C) | ND |
| Stenotrophomonas rhizophila TES-VG16 | 64 | tet(T) | 24 | ND | >256 | erm(C) | I |
| Stenotrophomonas rhizophila TES-G17 | 32 | ND | 12 | ND | >256 | erm(X) | I |
| Stenotrophomonas rhizophila TES-VG18 | >256 | ND | 0.38 | aad(A) | 1.5 | ND | ND |
TET, tetracycline; STR, streptomycin; E, erythromycin; I, Tn916 or Tn1549 integrase; RE, Tn4451 recombinase; RS, Tn5397 resolvase; ND, not detected.
Table 4.
Susceptibility profiles and corresponding antibiotic resistance genes of strains isolated from apple orchard soila
| Strain | TET MIC (μg/ml) | TET resistance gene(s) | STR MIC (μg/ml) | STR resistance gene(s) | E MIC (μg/ml) | E resistance gene(s) | Transposon gene |
|---|---|---|---|---|---|---|---|
| Aeromonas salmonicida S-AO16 | 24 | tet(O) | >1,024 | str(B), aac | 8.0 | ND | ND |
| Paenibacillus amylolyticus S-AO1 | 12 | ND | >1,024 | ND | 6.0 | ND | ND |
| Paenibacillus sp. S-AO2 | 8 | ND | >1,024 | ND | 0.25 | ND | ND |
| Paenibacillus amylolyticus S-AO3 | 12 | ND | >1,024 | ND | 3.0 | ND | ND |
| Paenibacillus amylolyticus S-AO4 | 16 | ND | >1,024 | ND | 8.0 | ND | ND |
| Paenibacillus amylolyticus S-AO6 | 12 | ND | >1,024 | ND | 2.0 | ND | ND |
| Paenibacillus amylolyticus S-AO8 | 12 | ND | >1,024 | ND | 6.0 | ND | ND |
| Paenibacillus amylolyticus S-AO9 | 12 | ND | >1,024 | ND | 1.5 | ND | ND |
| Paenibacillus amylolyticus S-AO10 | 12 | ND | >1,024 | aac | 6.0 | ND | ND |
| Paenibacillus amylolyticus S-AO11 | 12 | ND | >1,024 | ND | 6.0 | ND | ND |
| Paenibacillus sp. S-AO12 | 8 | ND | >1,024 | str(B) | 8.0 | ND | ND |
| Paenibacillus amylolyticus S-AO13 | 12 | ND | >1,024 | aac | 8.0 | ND | ND |
| Paenibacillus amylolyticus S-AO14 | 12 | ND | >1,024 | str(B) | 4.0 | ND | ND |
| Paenibacillus amylolyticus S-AO15 | 12 | ND | >1,024 | ND | 12.0 | ND | ND |
| Paenibacillus sp. S-AO17 | 8 | ND | >1,024 | str(B) | 2.0 | ND | ND |
| Paenibacillus amylolyticus S-AO18 | 12 | ND | >1,024 | ND | 1.5 | ND | ND |
| Paenibacillus sp. S-AO19 | 8 | ND | >1,024 | aad(A) | 4.0 | ND | ND |
| Paenibacillus xylanilyticus S-AO20 | 16 | tet(O) | >1,024 | ND | 3.0 | ND | ND |
| Paenibacillus sp. S-AO21 | 12 | ND | >1,024 | ND | 1.5 | ND | ND |
| Paenibacillus xylanilyticus S-AO22 | 16 | tet(O) | >1,024 | str(B), aac | 1.5 | ND | ND |
| Pseudomonas sp. E-AO23 | 12 | ND | 12.0 | ND | >256 | erm(V) | I, RE |
| Pseudomonas putida E-AO24 | 24 | tet(T) | 12.0 | ND | >256 | erm(C), erm(V) | I, RE |
| Pseudomonas sp. E-AO25 | 24 | tet(T) | 16.0 | ND | >256 | erm(V) | I |
| Pseudomonas sp. SE-AO26 | 24 | tet(T) | 96.0 | aad(K) | 24.0 | erm(V) | ND |
| Solibacillus silvestris S-AO7 | 8 | ND | >1,024 | str(A), aad(A) | 1.5 | ND | ND |
TET, tetracycline; STR, streptomycin; E, erythromycin; I, Tn916 or Tn1549 integrase; RE, Tn4451 recombinase; ND, not detected.
Table 5.
Susceptibility profiles and corresponding antibiotic resistance genes of strains isolated from mixed fruit orchard soila
| Strain | TET MIC (μg/ml) | TET resistance gene(s) | STR MIC (μg/ml) | STR resistance gene(s) | E MIC (μg/ml) | E resistance gene(s) | Transposon gene |
|---|---|---|---|---|---|---|---|
| Arthrobacter sp. T-MO28 | 12 | ND | 6 | ND | 0.094 | ND | I |
| Bacillus sp. S-MO29 | 12 | ND | 6 | ND | 8 | ND | ND |
| Mesorhizobium sp. S-MO27 | 24 | ND | 12 | ND | >256 | erm(C), erm(V), vga | RE, RS |
| Microbacterium xylanilyticum T-MO13 | 32 | tet(M), tet(O) | 6 | ND | 0.094 | ND | ND |
| Microbacterium xylanilyticum T-MO14 | 32 | tet(M), tet(O) | 6 | ND | 0.094 | ND | ND |
| Micrococcus sp. S-MO8 | 12 | ND | 24 | str(A), str(B), aad(K), aad(A), aac | 4 | ND | ND |
| Paenibacillus sp. S-MO1 | 12 | ND | 24 | str(B), aad(K), aac | 8 | ND | I, RE |
| Paenibacillus sp. S-MO2 | 12 | ND | 12 | aad(K), aac | 6 | ND | I |
| Paenibacillus sp. S-MO3 | 24 | ND | 24 | str(A), str(B), aac | 12 | ND | I, RE, RS |
| Paenibacillus sp. S-MO4 | 8 | ND | 24 | str(A), str(B), aad(K) | 8 | ND | I, RE |
| Paenibacillus sp. S-MO5 | 32 | ND | 12 | str(B), aad(K), aac | >256 | erm(X), vga | I, RS |
| Paenibacillus sp. S-MO6 | 8 | ND | 12 | str(B), aad(K), aad(A), aac | 8 | ND | I, RE |
| Paenibacillus sp. S-MO7 | 24 | ND | 12 | str(A), str(B), aad(K), aac | >256 | erm(C), erm(V) | I, RE, RS |
| Paenibacillus sp. S-MO9 | 32 | ND | 24 | str(A), str(B), aad(A), aac | 4 | ND | I, RE, RS |
| Paenibacillus sp. S-MO10 | 8 | ND | 24 | str(A), str(B), aac | 4 | ND | I, RE, RS |
| Pseudomonas fluorescens S-MO12 | 32 | tet(T) | 12 | aac | >256 | ND | RE, RS |
| Pseudomonas sp. ST-MO24 | 24 | tet(T) | 12 | ND | >256 | ND | RE |
| Pseudomonas fluorescens S-MO25 | 24 | ND | 12 | ND | >256 | vga | RE, RS |
| Rhodococcus erythropolis ST-MO15 | 16 | tet(O), tet(B) | 2 | ND | 4 | ND | RE |
| Rhodococcus erythropolis ST-MO16 | 16 | ND | 2 | ND | 6 | ND | ND |
| Rhodococcus erythropolis ST-MO17 | 16 | ND | 2 | ND | 4 | ND | ND |
| Rhodococcus erythropolis ST-MO19 | 12 | ND | >1,024 | ND | 6 | ND | ND |
| Rhodococcus sp. ST-MO20 | 12 | ND | 12 | aac | 6 | ND | ND |
| Rhodococcus erythropolis ST-MO22 | 16 | ND | 12 | ND | 4 | ND | ND |
| Rhodococcus erythropolis ST-MO23 | 16 | tet(O) | 6 | ND | 4 | ND | ND |
| Rhodococcus erythropolis ST-MO26 | 32 | tet(O) | 12 | ND | 0.094 | ND | ND |
| Staphylococcus sciuri S-MO11 | 32 | ND | 16 | str(B), aac | 2 | ND | ND |
TET, tetracycline; STR, streptomycin; E, erythromycin; I, Tn916 or Tn1549 integrase; RE, Tn4451 recombinase; RS, Tn5397 resolvase; ND, not detected.
Table 6.
Susceptibility profiles and corresponding antibiotic resistance genes of strains isolated from arable farmland soila
| Strain | TET MIC (μg/ml) | TET resistance gene(s) | STR MIC (μg/ml) | STR resistance gene(s) | E MIC (μg/ml) | E resistance gene(s) | Transposon gene |
|---|---|---|---|---|---|---|---|
| Aeromonas salmonicida S-F9 | ND | ND | 512 | str(A), aad(K), aac | 256 | erm(V), vga, ole(B) | I, RS |
| Brevundimonas vesicularis T-F3 | 196 | tet(B), tet(D), tet(O), tet(T), tet(W) | ND | ND | 256 | erm(C), erm(V), erm(X), msr(A) | I, RE, RS |
| Burkholderia cepacia T-F1 | 32 | tet(O) | ND | ND | 32 | erm(C), erm(X) | ND |
| Chryseomonas meningosepticum S-F6 | ND | ND | 512 | str(A), aac | 256 | erm(V), msr(A) | I |
| Pasteurella multocida T-F4 | 32 | tet(D) | ND | ND | 8 | erm(C), erm(X) | ND |
| Photobacterium damselae S-F7 | ND | ND | 512 | str(A), aac, aad(K) | 48 | msr(A), vga | I, RS |
| Rhizobium radiobacter S-F5 | ND | ND | 512 | str(A) | 16 | ND | ND |
| Rhizobium radiobacter T-F2 | 128 | tet(B), tet(D), tet(M), tet(O), tet(T) | ND | ND | 32 | erm(C), erm(V), erm(X) | I, RE, RS |
| Sphingomonas multivorum S-F8 | ND | ND | 512 | str(A), aad(A) | 48 | erm(C), erm(X) | ND |
TET, tetracycline; STR, streptomycin; E, erythromycin; I, Tn916 or Tn1549 integrase; RE, Tn4451 recombinase; RS, Tn5397 resolvase; ND, not detected.
The pure bacterial cultures were most frequently resistant to streptomycin (42.8%), followed by erythromycin (34.7%) and tetracycline (10.2%). The MIC ranges of streptomycin, erythromycin, and tetracycline against isolates from compost and forest soil were 512 μg/ml, 16 to 256 μg/ml, and 16 to 128 μg/ml, respectively, and those against isolates from the cultivated soils (vegetable garden soil, orchard soils, and arable farmland soil) were 2 to >1,024 μg/ml, 8 to >256 μg/ml, and 0.094 to >256 μg/ml, respectively. The strains with the highest tetracycline MICs were isolated from vegetable garden soil (>256 μg/ml) and arable farmland soil (196 μg/ml). Those with the highest streptomycin MICs were from apple orchard soil (>1,024 μg/ml), and those with the highest erythromycin MICs were from vegetable garden and orchard soils (>256 μg/ml), i.e., manure-amended soils from agricultural systems with a history of antibiotic use. These results indicate a higher prevalence of resistance to streptomycin and erythromycin than to tetracycline.
Tetracycline resistance corresponding to an MIC of 196 μg/ml or greater was observed in Brevundimonas, Chryseobacterium, and Stenotrophomonas spp. A streptomycin MIC of 1,024 μg/ml or greater was detected in Chryseobacterium, Paenibacillus, Rhodococcus, and Solibacillus spp., and an erythromycin MIC of >256 μg/ml was detected in Chryseobacterium, Pseudomonas, and Stenotrophomonas spp. Most of these bacteria were further shown to be multidrug resistant. Stenotrophomonas spp. were highly resistant to both tetracycline and erythromycin. Chryseobacterium spp. were multidrug resistant, with high resistance to all three antibiotics. Bacteria with an MIC of ≥196 μg/ml for tetracycline, 1,024 μg/ml for streptomycin, or 256 μg/ml for erythromycin were not detected in compost or forest soil. The strains most resistant to the studied antibiotics were isolated from cultivated agricultural soils that were amended with animal waste (31).
All standard guidelines used for the classification of bacteria as sensitive or resistant to an antibiotic, such as growth inhibition zone reading and/or MICs (19), apply to typical clinical strains. Therefore, it is difficult to relate them to environmental bacteria. There are no current guidelines for the determination of antibiotic susceptibility of bacterial species isolated from the environment, although many of them are either opportunistic pathogens or emerging pathogens. In the CLSI guidelines, for example, a zone of inhibition of 14 mm or 13 mm and a MIC of 12 or 13 mg/liter are regarded as indicating resistance to tetracycline (30 μg) or erythromycin (15 μg), respectively, for Pseudomonas aeruginosa. All of the tested strains of Pseudomonas spp. in this study had MICs above these standards. Thus, the existence of high-level and multidrug-resistant strains, especially opportunistic pathogens, in the environment poses a serious risk to humans and animals.
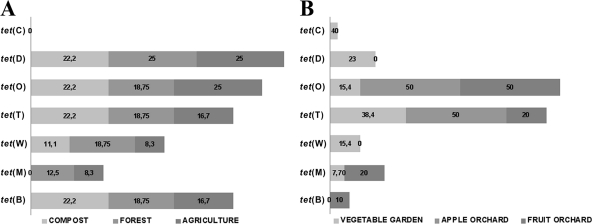
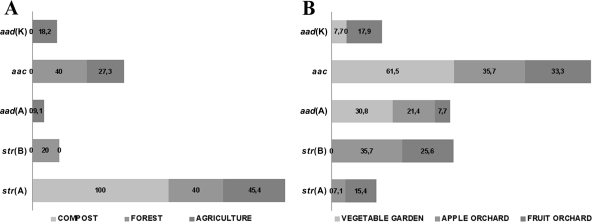
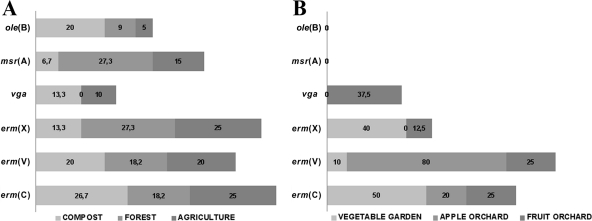
Antibiotic resistance genes detected.
The resistance genes identified by PCR were tet(A), tet(B), tet(D), tet(M), tet(O), tet(T), and tet(W) for tetracycline resistance (Fig. 3), aac, aad(A), str(A), and str(B) for streptomycin resistance (Fig. 4), and erm(C), erm(V), erm(X), msr(A), ole(B), and vga for erythromycin resistance (Fig. 5). The remaining resistance genes investigated were not detected in any of the resistant isolates. Multiple antibiotic resistance genes conferring resistance to the same antibiotic were detected in the same isolates, e.g., tet(B), tet(D), tet(O), tet(T), and tet(W) were detected in Pasteurella multocida; aac, aad(A), str(A), and str(B) were detected in Paenibacillus spp.; and erm(C), erm(V), and erm(X) were detected in Rhizobium radiobacter. Forty-five tetracycline-resistant, 34 streptomycin-resistant, and 8 erythromycin-resistant bacterial strains were negative for all of the tested resistance genes. In compost and forest soils, the frequencies of tetracycline (Fig. 3A) and erythromycin (Fig. 5A) resistance genes were higher than those of streptomycin (Fig. 4A) resistance genes. The farmland soil contained high frequencies of all resistance genes (Fig. 3B, 4B, and 5B). The dominant mechanism of resistance to streptomycin and erythromycin in bacterial isolates from orchard and vegetable garden soils is antibiotic modification. Accordingly, for resistance to streptomycin and erythromycin, the resistance genes aac (chromosomal), aad(A) (plasmid located), str(B) (plasmid located), erm(C), erm(V), and erm(X) (plasmid located) are involved (46). In the case of tetracycline, the resistance mechanism is protection of the ribosome conferred by the tet(O) (plasmid located) and tet(T) (chromosomal) genes (46). This suggests that the resistance we observed in most cases was linked to plasmids or other mobile genetic elements, which theoretically have transfer potential (5, 16, 17, 22, 26, 38, 51, 57, 64, 65).
Fig 3.
Percentages of tetracycline resistance genes identified in bacterial strains isolated from forest soil, compost, and agricultural soil (A) and from vegetable garden soil, mixed fruit orchard soil, and apple orchard soil (B).
Fig 4.
Percentages of streptomycin resistance genes identified in bacterial strains isolated from forest soil, compost, and agricultural soil (A) and from vegetable garden soil, mixed fruit orchard soil, and apple orchard soil (B).
Fig 5.
Percentages of erythromycin resistance genes in bacterial strains isolated from forest soil, compost, and agricultural soil (A) and from vegetable garden soil, mixed fruit orchard soil, and apple orchard soil (B).
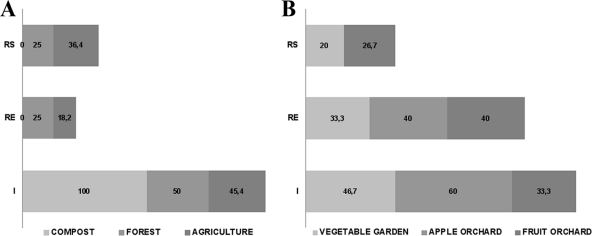
Antibiotic resistance-associated transposons Tn916 and Tn1549 were detected in resistant bacteria isolated from all soils and compost, Tn4451 was detected in resistant bacteria isolated from all soils but not compost, and Tn5397 was detected in resistant bacteria isolated from all soils except for the apple orchard soil and compost (Fig. 6). The presence of tetracycline has previously been shown to increase the incidence of the tet(M)-carrying transposon Tn916 in bacteria (52, 57). Integrase genes were detected in isolates from all soils. The detection of characteristic transposon genes in the antibiotic-resistant soil bacteria, combined with the observed differences in quantity and level of these genes in isolates from nonamended (Fig. 6A) and cultivated (Fig. 6B) soils, suggest that resistance to the tested antibiotics may be associated with transposons (20). It is unclear whether this indicates selection of resistant resident members of the soil community, proliferation of bacteria introduced via manure amendments, and/or dissemination of mobile genetic elements and horizontal gene transfer (5, 6, 12, 16, 17, 28).
Fig 6.
Percentages of transposon genes identified in bacterial strains isolated from forest soil, compost, and agricultural soil (A) and from vegetable garden soil, mixed fruit orchard soil, and apple orchard soil (B). I, Tn916 or Tn1549 integrase; RE, Tn4451 recombinase; RS, Tn5397 resolvase.
The observations in this soil survey indicate the widespread presence of high-level antibiotic-resistant bacteria. Agricultural soils had more diverse populations of bacteria with resistance. Multidrug-resistant bacteria were detected only in the vegetable garden soil, which also had the highest levels of resistance to three antibiotic classes. For comparison, the composted and forest soils had lower levels of antibiotic resistance, a lower presence of antibiotic resistance genes, with no MDR isolates, and a lower species diversity within the antibiotic-resistant populations.
Supplementary Material
ACKNOWLEDGMENTS
This publication was funded by the State Committee for Scientific Research, Poland (international grant project [not cofinanced]; decision no. 741/N-COST/2010/0). F. Walsh and B. Duffy were funded in part by the Swiss Federal Office for Agriculture and the Swiss Federal Office for the Environment. The research was conducted under the European Co-Operation in the Field of Scientific and Technical (COST) Research Action TD0803 (Detecting Evolutionary Hot Spots of Antibiotic Resistances in Europe [DARE]) (2009–2013).
Footnotes
Published ahead of print 27 December 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Alekshun MN, Levy SB. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128:1037–1050 [DOI] [PubMed] [Google Scholar]
- 2. Allen HK, et al. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8:251–259 [DOI] [PubMed] [Google Scholar]
- 3. Aminov RI. 2009. The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 11:2970–2988 [DOI] [PubMed] [Google Scholar]
- 4. Aminov RI. 2010. A brief history of the antibiotic era: lessons learned and challenges for the future. Front. Microbiol. 1:134 doi:10.3389/fmicb.2010.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aminov RI. 2011. Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aminov RI, Mackie RI. 2007. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271:147–216 [DOI] [PubMed] [Google Scholar]
- 7. Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14:176–182 [DOI] [PubMed] [Google Scholar]
- 8. Benveniste R, Davies J. 1973. Aminoglycoside inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic resistance bacteria. Proc. Natl. Acad. Sci. U. S. A. 70:2276–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Binh CTT, Heuer H, Kaupenjohann M, Smalla K. 2008. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol. Ecol. 66:25–37 [DOI] [PubMed] [Google Scholar]
- 10. Binh CTT, Heuer H, Kaupenjohann M, Smalla K. 2009. Diverse aadA gene cassettes on class 1 integrons introduced into soil via spread manure. Res. Microbiol. 160:427–433 [DOI] [PubMed] [Google Scholar]
- 11. Boxall ABA. 2004. The environmental side effect of medication. EMBO Rep. 12:1110–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boxall ABA, Kolpin D, Halling-Sørensen B, Tolls J. 2003. Are veterinary medicines causing environmental risk? Environ. Sci. Technol. 37:268A–294A [DOI] [PubMed] [Google Scholar]
- 13. Chen J, Michel FC, Sreevatsan S, Morrison M, Yu Z. 2010. Occurrence and persistence of erythromycin resistance genes (erm) and tetracycline resistance genes (tet) in waste treatment systems on swine farms. Microb. Ecol. 60:479–486 [DOI] [PubMed] [Google Scholar]
- 14. Crossman LC, et al. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9:R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74:417–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. D'Costa VM, Griffiths E, Wright GD. 2007. Expanding the soil antibiotic resistome: exploring environmental diversity. Curr. Opin. Microbiol. 10:481–489 [DOI] [PubMed] [Google Scholar]
- 17. D'Costa VM, McGrann KM, Hughes DW, Wright GD. 2006. Sampling the antibiotic resistome. Science 311:374–377 [DOI] [PubMed] [Google Scholar]
- 18. Ding C, He J. 2010. Effect of antibiotics in the environment on microbial populations. Appl. Microbiol. Biotechnol. 87:925–941 [DOI] [PubMed] [Google Scholar]
- 19. EUCAST 2011. Clinical breakpoints. http://www.eucast.org/clinical_breakpoints/
- 20. Fajardo A, Martinez JL. 2008. Antibiotics as signals that trigger specific bacterial responses. Curr. Opin. Microbiol. 11:161–167 [DOI] [PubMed] [Google Scholar]
- 21. Goldman RC, Scaglione F. 2004. The macrolide-bacterium interaction and its biological basis. Curr. Drug Targets Infect. Disord. 4:241–260 [DOI] [PubMed] [Google Scholar]
- 22. Gootz TD. 2010. The global problem of antibiotic resistance. Crit. Rev. Immunol. 30:79–93 [DOI] [PubMed] [Google Scholar]
- 23. Gould IM. 1999. A review of the role of antibiotic policies in the control of antibiotic resistance. J. Antimicrob. Chemother. 43:459–465 [DOI] [PubMed] [Google Scholar]
- 24. Heuer H, Smalla K. 2007. Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9:657–666 [DOI] [PubMed] [Google Scholar]
- 25. Hughes WT, et al. 2002. Guidelines for the use of antimicrobial agents in neutropenic patients with cancer. Clin. Infect. Dis. 34:730–751 [DOI] [PubMed] [Google Scholar]
- 26. Hulscher ME, van der Meer JW, Grol RP. 2010. Antibiotic use: how to improve it? Int. J. Med. Microbiol. 300:351–356 [DOI] [PubMed] [Google Scholar]
- 27. Janssen PH. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kazimierczak KA, Scott KP, Kelly D, Aminov RI. 2009. Tetracycline resistome of the organic pig gut. Appl. Environ. Microbiol. 6:1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kelley St, Theisen U, Angenent LT, St. Aamand Pace NR. 2004. Molecular analysis of shower curtain biofilm microbes. Appl. Environ. Microbiol. 70:4187–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Knapp CW, Dolfing J, Ehlert PA, Graham DW. 2010. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 44:580–587 [DOI] [PubMed] [Google Scholar]
- 31. Kumar K, Gupta SC, Baido SK, Chander Y, Rosen CJ. 2005. Antibiotic uptake by plants from soil fertilized with animal manure. J. Environ. Qual. 34:2082–2085 [DOI] [PubMed] [Google Scholar]
- 32. Kümmerer K. 2008. Pharmaceuticals in the environment: source, fate, effects and risks. Springer, New York, NY [Google Scholar]
- 33. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY [Google Scholar]
- 34. Larsen J, et al. 2010. Porcine-origin gentamicin-resistant Enterococcus faecalis in humans, Denmark. Emerg. Infect. Dis. 16:682–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lesmana M, et al. 2001. In vitro antibiotic susceptibility of Neisseria gonorrhoeae in Jakarta, Indonesia. Antimicrob. Agents Chemother. 45:359–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Madigan MT, Martinko JM, Dunlap PV, Clark DP. 2009. Brock biology of microorganisms, 12th ed Pearson Benjamin-Cummings, San Francisco, CA [Google Scholar]
- 37. Manson JM, Hancock LE, Gilmore MS. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc. Natl. Acad. Sci. U. S. A. 107:12269–12274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martinez JL. 2009. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Poll. 157:2893–2902 [DOI] [PubMed] [Google Scholar]
- 39. Mayers DL. 2009. Antimicrobial drug resistance, vol 1. Mechanisms of drug resistance (infectious disease). Springer, Dordrecht, The Netherlands [Google Scholar]
- 40. McCrady HM. 1915. The numerical interpretation of fermentation-tube results. J. Infect. Dis. 17:183–212 [Google Scholar]
- 41. Nadkarni MA, Martin FE, Jacques NA, Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257–266 [DOI] [PubMed] [Google Scholar]
- 42. Nikolakopoulou T, et al. 2005. PCR detection of oxytetracycline resistance genes otr(A) and otr(B) in tetracycline-resistant streptomycete isolates from diverse habitats. Curr. Microbiol. 51:211–216 [DOI] [PubMed] [Google Scholar]
- 43. Nordmann P, Poirel L. 2005. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56:463–469 [DOI] [PubMed] [Google Scholar]
- 44. O'Connor S, Aga DS. 2007. Analysis of tetracycline antibiotics in soil: advances in extraction, clean-up, and quantification. Trends Anal. Chem. 26:456–465 [Google Scholar]
- 45. O'Connor S, Locke J, Aga DS. 2007. Addressing the challenges of tetracycline analysis in soil: extraction, clean-up, and matrix effects in LC-MS. J. Environ. Monit. 11:1254–1262 [DOI] [PubMed] [Google Scholar]
- 46. Patterson AJ, Colangeli R, Spigaglia P, Scott KP. 2007. Distribution of specific tetracycline and erythromycin resistance genes in environmental samples assessed by macroarray detection. Environ. Microbiol. 9:703–715 [DOI] [PubMed] [Google Scholar]
- 47. Peak N, et al. 2007. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ. Microbiol. 9:143–151 [DOI] [PubMed] [Google Scholar]
- 48. Popowska M, Miernik A, Rzeczycka M, Łopaciuk A. 2010. The impact of environmental contamination with antibiotics on levels of resistance in soil bacteria. J. Environ. Qual. 39:1679–1687 [DOI] [PubMed] [Google Scholar]
- 49. Rayamajhi N, et al. 2010. Antibiotic resistance patterns and detection of blaDHA-1 in Salmonella species isolates from chicken farms in South Korea. Appl. Environ. Microbiol. 76:4760–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rezzonico F, Stockwell VO, Duffy B. 2009. Plant agricultural streptomycin formulations do not carry antibiotic resistance genes. Antimicrob. Agents Chemother. 53:3173–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rice LB. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42:1871–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roberts AP, Mullany P. 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17:251–258 [DOI] [PubMed] [Google Scholar]
- 53. Roe MT, Pillai SD. 2003. Monitoring and identifying antibiotic resistance mechanisms in bacteria. Poult. Sci. 82:622–626 [DOI] [PubMed] [Google Scholar]
- 54. Silbergeld EK, Graham J, Price LB. 2008. Industrial food animal production, antimicrobial resistance, and human health. Public Health 29:151–169 [DOI] [PubMed] [Google Scholar]
- 55. Singer RS, et al. 2003. Antibiotic resistance—the interplay between antibiotic use in animals and human beings. Lancet Infect. Dis. 3:47–51 [DOI] [PubMed] [Google Scholar]
- 56. Stokes HW, Gillings MR. 2011. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram negative pathogens. FEMS Microbiol. Rev. 35:790–819 [DOI] [PubMed] [Google Scholar]
- 57. Su YA, He P, Clewell DB. 1992. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob. Agents Chemother. 36:769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thiele-Bruhn S. 2003. Pharmaceutical antibiotics compounds in soil: a review. J. Plant Nutr. Soil Sci. 166:145–167 [Google Scholar]
- 59. Torres-Cortés G, et al. 2011. Characterization of novel antibiotic resistance genes identified by functional metagenomics on soil samples. Environ. Microbiol. 13:1101–1114 [DOI] [PubMed] [Google Scholar]
- 60. van den Bogaard AE, Stobberingh EE. 2000. Epidemiology of resistance to antibiotics. Links between animals and humans. Int. J. Antimicrob. Agents 14:327–335 [DOI] [PubMed] [Google Scholar]
- 61. van Elsas JD, Trevors JT. 1997. Modern soil microbiology. WM Dekker, New York, NY [Google Scholar]
- 62. van Overbeek LS, et al. 2002. Prevalence of streptomycin-resistance genes in bacterial populations in European habitats. FEMS Microbiol. Ecol. 42:277–288 [DOI] [PubMed] [Google Scholar]
- 63. Walsh F, et al. 2011. Real-time PCR methods for quantitative monitoring of streptomycin and tetracycline resistance genes in agricultural ecosystems. J. Microbiol. Methods 86:150–155 [DOI] [PubMed] [Google Scholar]
- 64. Wright GD. 2007. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5:175–186 [DOI] [PubMed] [Google Scholar]
- 65. Wright GD. 2010. Antibiotic resistance in the environment: a link to the clinic? Curr. Opin. Microbiol. 13:589–594 [DOI] [PubMed] [Google Scholar]
- 66. Yang H, et al. 2010. Presence of antibiotic-resistant commensal bacteria in samples from agricultural, city, and national park environments evaluated by standard culture and real-time PCR methods. Can. J. Microbiol. 56:761–770 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.