Abstract
We describe the activities of RX-P763, RX-P766, RX-P770, RX-P792, RX-P793, and RX-P808 against strains of resistant Pseudomonas aeruginosa. These compounds target the large subunit of the bacterial ribosome and have broad-spectrum activities against multidrug-resistant pathogens. All compounds demonstrated in vitro activity against P. aeruginosa, with MIC90 values of 4 to 8 μg/ml (range, 0.5 to 64). These novel compounds had narrow MIC distributions and maintained activity despite resistance phenotypes to other commonly utilized agents.
TEXT
Multidrug-resistant (MDR) P. aeruginosa is on the rise, limiting the choice of available anti-infective agents. The National Nosocomial Infections Surveillance System reports that P. aeruginosa resistance rates have increased to available agents (9), increasing morbidity and mortality (1, 6). Further, P. aeruginosa was the second most common type of pathogen isolated from nosocomial pneumonia patients between 1986 and 2003 (7), and MDR P. aeruginosa increased by 10%, from 4% in 1993 to 14% in 2002 (10). Given the high morbidity and mortality and the increased economic burden, new antimicrobial agents are desperately needed to combat highly resistant P. aeruginosa infections.
The RX-04 program at Rib-X Pharmaceuticals has developed a new series of anti-infectives designed to target novel sites of action. These compounds, known as the pyrrolocytosines, bind to the P-loop of the large ribosomal subunit and inhibit translation by stabilizing a distorted mode of P-tRNA binding, a new and unique mechanism of inhibition among antibiotics in clinical use (11). RX-P763, RX-P766, RX-P770, RX-P792, RX-P793, and RX-P808 are fruits of the RX-04 program (Fig. 1) (2). Previous in vitro work demonstrated activities against many Gram-positive pathogens and Gram-negative pathogens, including MDR P. aeruginosa, Enterobacteriaceae including carbapenemase-producing Klebsiella pneumoniae, and Acinetobacter baumanii (2). In this study, we evaluated the potency of six novel compounds, RX-P763, RX-P766, RX-P770, RX-P792, RX-P793, and RX-P808, against a collection of highly resistant P. aeruginosa isolates.
Fig 1.
Chemical structures of the novel compounds.
A total of 200 P. aeruginosa isolates were selected from a population of 1,788 nonduplicate, nonurine isolates collected from 41 hospitals distributed throughout the United States over the period of October 2005 to June 2010 (4, 5). The isolates were selected to represent the following drug-resistant phenotypes: imipenem resistant (R-IMI; MIC, ≥16 μg/ml); ceftazidime resistant (R-TAZ; MIC, ≥32 μg/ml); piperacillin-tazobactam-nonsusceptible (R-TZP; MIC, ≥32 μg/ml), ciprofloxacin resistant (R-CIPRO; MIC, ≥4 μg/ml), tobramycin resistant (R-TOB; MIC, ≥16 μg/ml), and MDR (resistant to ≥3 classes of antimicrobials). Of the 200 isolates selected, 181 had accompanying patient information. The mean ± standard deviation age was 54 ± 23 years, and 54% of patients were males. Further, 50% of isolates were collected in the intensive care unit (ICU), and the respiratory tract was the most common collection site (61%), followed by blood (11%) and bodily fluids (11%), skin wounds (9%), and other sites (8%).
MICs were determined in duplicate with the reference Clinical and Laboratory Standards Institute broth microdilution method (3). MICs for the antimicrobials imipenem, meropenem, piperacillin-tazobactam, cefepime, ceftazidime, ciprofloxacin, levofloxacin, and tobramycin were also determined by broth microdilution. P. aeruginosa ATCC 27853 was used as the quality control organism for all six novel compounds. Acceptable ranges for all six compounds were 2 to 8 μg/ml. Of the 200 isolates, 67% were R-TZP, 42% were R-TAZ and R-IMI, 32% were R-CIPRO, and 21% were R-TOB. Further, 21% were considered MDR, with resistance to 3 or more drug classes.
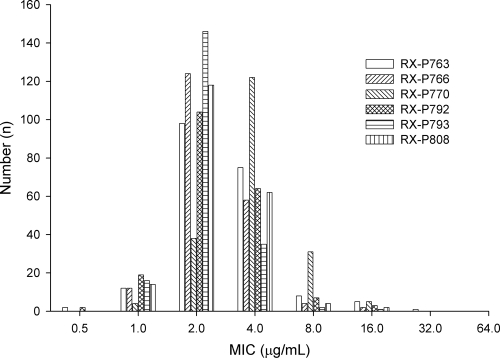
The MIC distributions of all six compounds are shown in Fig. 2. The MIC profiles of each of the six novel compounds were narrow, and modal MICs of 2 μg/ml were observed for all compounds except RX-P770, which had a modal MIC of 4 μg/ml. RX-P763, RX-P766, RX-P792, RX-P793, and RX-P808 had similar MIC distributions, while RX-P770 seemed to be 1 dilution less potent than its fellow novel compounds, as illustrated in Fig. 2. Marketed compounds tended to have higher modal MICs (in μg/ml, with ranges shown in parentheses): 16 (0.25 to ≥128) for imipenem, 32 (≤0.5 to ≥128) for piperacillin-tazobactam, 32 (1 to ≥128) for ceftazidime, 0.125 (≤0.06 to ≥128) for ciprofloxacin, and 1 (0.25 to ≥128) for tobramycin. Imipenem and piperacillin-tazobactam had elevated MIC50 and MIC90 values of 4 and 32 and of 32 and 256 μg/ml, respectively. Ceftazidime MIC50 and MIC90 values were 16 and 128 μg/ml, respectively. The final two marketed compounds, ciprofloxacin and tobramycin, had MIC50 and MIC90 values of 0.5 and 1 and of 16 and 128 μg/ml, respectively. The MIC distributions of each resistant phenotype are shown in Table 1. The MIC profile of the six novel compounds did not change when R-IMI was broken out, with the exception of the RX-P763 MIC50, which increased by one dilution to 4 μg/ml. None of the novel compounds' MIC profiles changed based on R-TAZ isolates, while the potency of ceftazidime decreased by approximately 2 dilutions. Similar results were seen for all agents against R-CIPRO isolates. MIC50 and MIC90 values for the novel compounds changed by ≤1 dilution, while the MIC50 for ciprofloxacin increased by 5 dilutions and the MIC90 increased by 1 dilution, to 16 and 32 μg/ml, respectively. After separating those isolates that were multidrug resistant, the MIC50 values for RX-P763 and RX-P792 changed by 1-fold dilutions from 2 to 4 μg/ml, while the other four compounds did not show any changes in MIC profiles. Of note, six isolates contributed to all of the MICs that were ≥16 μg/ml for the novel compounds. The isolates were collected from geographically diverse locations (two isolates from Ohio and one each from Alabama, the District of Columbia, Georgia, and Florida), were generally not MDR, and were obtained from different sites of infection. Of these six isolates, three were R-IMI, three were R-TAZ, four were R-TZP, one was R-TOBRA, and two were R-CIPRO. The modal MICs for each of the compounds against these six isolates were 4 μg/ml for RX-P793 (range, 4 to 16 μg/ml), 8 μg/ml for RX-P766 (range, 4 to 16 μg/ml) and RX-P808 (range, 4 to 16 μg/ml), and 16 μg/ml for RX-P763 (range, 4 to 32 μg/ml), RX-P770 (range, 8 to 16 μg/ml), and RX-P792 (range, 4 to 64 μg/ml).
Fig 2.
MIC distributions for the six novel compounds, RX-7957, RX-7960, RX-7973, RX-7999, RX-8000, and RX-8015, against 200 clinical P. aeruginosa isolates.
Table 1.
MIC distributions of novel compounds and comparator agents against 200 P. aeruginosa isolates, by resistance phenotype
| Resistance phenotype (MIC; n) | Agent | No. of isolates (cumulative %) inhibited at MIC (μg/ml) of: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ≥128 | ||
| R-IMI (≥16 μg/ml; 84) | RX-P763 | 2 (2) | 5 (8) | 33 (48) | 38 (93) | 3 (96) | 3 (100) | 0 (100) | 0 (100) | 0 (100) |
| RX-P766 | 0 (0) | 7 (8) | 48 (65) | 26 (96) | 2 (99) | 1 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P770 | 0 (0) | 3 (4) | 12 (18) | 54 (82) | 13 (98) | 2 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P792 | 2 (2) | 8 (12) | 35 (54) | 34 (94) | 3 (98) | 2 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P793 | 0 (0) | 7 (8) | 62 (82) | 13 (98) | 2 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P808 | 0 (0) | 8 (10) | 42 (60) | 30 (95) | 3 (99) | 1 (100) | 0 (100) | 0 (100) | 0 (100) | |
| Imipenem | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 53 (63) | 22 (89) | 5 (95) | 4 (100) | |
| R-TAZ (≥32 μg/ml; 83) | RX-P763 | 2 (2) | 6 (10) | 39 (57) | 30 (93) | 3 (96) | 2 (99) | 1 (100) | 0 (100) | 0 (100) |
| RX-P766 | 0 (0) | 8 (10) | 51 (71) | 20 (95) | 2 (98) | 2 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P770 | 0 (0) | 3 (4) | 18 (25) | 48 (83) | 12 (98) | 2 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P792 | 2 (2) | 9 (13) | 41 (63) | 24 (92) | 5 (98) | 1 (99) | 1 (100) | 0 (100) | 0 (100) | |
| RX-P793 | 0 (0) | 11 (13) | 57 (82) | 13 (98) | 1 (99) | 1 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P808 | 0 (0) | 11 (13) | 45 (67) | 23 (95) | 2 (98) | 2 (100) | 0 (100) | 0 (100) | 0 (100) | |
| Ceftazidime | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 37 (45) | 24 (73) | 22 (100) | |
| R-TZP (≥32 μg/ml; 133) | RX-P763 | 2 (2) | 9 (8) | 59 (53) | 52 (92) | 7 (97) | 3 (99) | 1 (100) | 0 (100) | 0 (100) |
| RX-P766 | 0 (0) | 11 (8) | 77 (66) | 40 (96) | 3 (98) | 2 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P770 | 0 (0) | 4 (3) | 22 (20) | 80 (80) | 24 (98) | 3 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P792 | 2 (2) | 11 (10) | 65 (59) | 45 (92) | 7 (98) | 2 (99) | 1 (100) | 0 (100) | 0 (100) | |
| RX-P793 | 0 (0) | 15 (11) | 88 (77) | 28 (98) | 1 (99) | 1 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P808 | 0 (0) | 13 (10) | 72 (64) | 43 (96) | 3 (98) | 2 (100) | 0 (100) | 0 (100) | 0 (100) | |
| Piperacillin-tazobactam | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 50 (38) | 21 (53) | 62 (100) | |
| R-CIPRO (≥4 μg/ml; 64) | RX-P763 | 1 (2) | 4 (8) | 22 (42) | 31 (91) | 4 (97) | 2 (100) | 0 (100) | 0 (100) | 0 (100) |
| RX-P766 | 0 (0) | 5 (8) | 33 (59) | 24 (97) | 1 (98) | 1 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P770 | 0 (0) | 2 (3) | 9 (17) | 37 (75) | 15 (98) | 1 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P792 | 1 (2) | 6 (11) | 24 (48) | 28 (92) | 4 (98) | 1 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P793 | 0 (0) | 6 (9) | 43 (77) | 14 (98) | 1 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P808 | 0 (0) | 6 (9) | 30 (56) | 25 (95) | 2 (98) | 1 (100) | 0 (100) | 0 (100) | 0 (100) | |
| Ciprofloxacin | 0 (0) | 0 (0) | 0 (0) | 13 (20) | 16 (45) | 14 (67) | 15 (91) | 0 (91) | 6 (100) | |
| R-TOB (≥16 μg/ml; 42) | RX-P763 | 0 (0) | 2 (5) | 17 (46) | 19 (93) | 3 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) |
| RX-P766 | 0 (0) | 3 (7) | 26 (71) | 12 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P770 | 0 (0) | 0 (0) | 10 (24) | 23 (80) | 7 (98) | 1 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P792 | 0 (0) | 5 (12) | 18 (56) | 18 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P793 | 0 (0) | 3 (7) | 31 (83) | 7 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P808 | 0 (0) | 4 (10) | 23 (66) | 14 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | |
| Tobramycin | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 5 (12) | 8 (32) | 6 (46) | 22 (100) | |
| MDR (resistance to ≥3 drug classes; 42) | RX-P763 | 0 (0) | 3 (7) | 15 (43) | 19 (88) | 4 (98) | 1 (100) | 0 (100) | 0 (100) | 0 (100) |
| RX-P766 | 0 (0) | 4 (10) | 23 (64) | 14 (98) | 1 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P770 | 0 (0) | 1 (2) | 6 (17) | 26 (79) | 9 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P792 | 0 (0) | 4 (10) | 15 (45) | 20 (93) | 3 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P793 | 0 (0) | 5 (12) | 29 (81) | 8 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | |
| RX-P808 | 0 (0) | 5 (12) | 20 (60) | 15 (95) | 2 (100) | 0 (100) | 0 (100) | 0 (100) | 0 (100) | |
| Imipenem | 1 (2) | 2 (7) | 6 (21) | 2 (26) | 3 (33) | 12 (62) | 9 (83) | 4 (93) | 3 (100) | |
| Piperacillin-Tazobactam | 1 (2) | 0 (2) | 0 (2) | 1 (5) | 1 (7) | 2 (12) | 2 (17) | 2 (21) | 33 (100) | |
| Ceftazidime | 0 (0) | 0 (0) | 1 (2) | 4 (12) | 2 (17) | 5 (29) | 10 (52) | 6 (67) | 14 (100) | |
| Ciprofloxacin | 0 (0) | 0 (0) | 6 (14) | 7 (31) | 9 (52) | 8 (71) | 9 (93) | 0 (93) | 3 (100) | |
| Tobramycin | 2 (5) | 4 (14) | 2 (19) | 1 (21) | 4 (31) | 5 (43) | 4 (52) | 3 (60) | 17 (100) | |
Respiratory isolates (n = 111) were separated, and three novel compounds, RX-P766, RX-P770, and RX-P793, were compared against this cache of isolates (8). The MIC distributions of the novel compounds were very similar to the whole collection of 200 isolates. When broken out according to those isolates collected in the ward and those collected in the ICU, there were no changes to the MIC distributions of the three compounds (data not shown). Further, the 3 compounds demonstrated greater potencies in vitro than currently utilized antimicrobials for P. aeruginosa respiratory infections.
Overall, these data showed that all six novel compounds demonstrated in vitro activity against a collection of highly resistant P. aeruginosa isolates. All of the novel compounds demonstrated a unimodal MIC distribution with this collection of isolates. However, elevated MICs were observed. These six isolates (3%) contributed to all of the observed MICs that were ≥16 μg/ml, suggesting some type of cross-resistance between the novel compounds. While the genetic profiles of these isolates have not been elucidated and the resistance mechanism(s) has not been identified, it is suspected that multiple resistance mechanisms, possibly including efflux pumps, are involved, given the differing resistance phenotypes (2). With a new mechanism of action and these in vitro data, these novel compounds could provide new options for clinicians against multidrug-resistant P. aeruginosa.
ACKNOWLEDGMENTS
We thank Jennifer Hull for her assistance with the in vitro experimentation.
D.P.N. has received research grants from and is a consultant for Rib-X Pharmaceuticals. C.S. and S.T.H. have no conflicts of interest to disclose.
Footnotes
Published ahead of print 27 December 2011
REFERENCES
- 1. Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. 2006. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob. Agents Chemother. 50:43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhattacharjee A, et al. 2011. Completely novel antibiotics for treating multidrug-resistant Gram-negative infections: the pyrrolocytosines, abstr. F1-1846. Abstr. 51st Intersci. Conf. Antimicrob. Agents Chemother., Chicago, IL, 17 to 20 September 2011 American Society for Microbiology, Washington, DC [Google Scholar]
- 3. Clinical and Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed CLSI publication M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. Eagye KJ, Kuti JL, Sutherland CA, Christensen H, Nicolau DP. 2009 In vitro activity and pharmacodynamics of commonly used antibiotics against adult systemic isolates of Escherichia coli and Pseudomonas aeruginosa at forty US hospitals. Clin. Ther. 31:2678–2688 [DOI] [PubMed] [Google Scholar]
- 5. Eagye KJ, MA Banevicius, Nicolau DP. Pseudomonas aeruginosa is not just in the intensive care unit anymore: implications for empirical therapy. Crit. Care Med., in press [DOI] [PubMed] [Google Scholar]
- 6. Gasink LB, et al. 2006. Fluoroquinolone-resistant Pseudomonas aeruginosa: assessment of risk factors and clinical impact. Am. J. Med. 119:526.e19–526.e25 [DOI] [PubMed] [Google Scholar]
- 7. Gaynes R, Edwards JR, National Nosocomial Infections Surveillance System 2005. Overview of nosocomial infections caused by Gram-negative bacilli. Clin. Infect. Dis. 41:848–854 [DOI] [PubMed] [Google Scholar]
- 8. Housman ST, Sutherland C, Nicolau DP. 2011. In vitro potency of Rib-X novel compounds potency against respiratory Pseudomonas aeruginosa isolates, abstr. 594. Abstr. 49th Annu. Meet. Infect. Dis. Soc. Am., Boston, MA, 20 to 23 October 2011 Infectious Diseases Society of America, Arlington, VA [Google Scholar]
- 9. NNIS System 2003. National Nosocomial Infections Surveillance (NNIS) system report: data summary from January 1992 through June 2003, issued August 2003. Am. J. Infect. Control 31:481–498 [DOI] [PubMed] [Google Scholar]
- 10. Obritsch MD, Fish DN, MacLaren R, Jung R. 2004. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob. Agents Chemother. 48:4606–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wimberly BT, et al. 2011. Crystallographic evidence that TAN-1057 and the blasticidin S family of antibiotics inhibit translation by stabilizing a distorted binding mode of P-site tRNA, abstr. F2-1871. Abstr. 51st Intersci. Conf. Antimicrob. Agents Chemother., Chicago, IL, 17 to 20 September American Society for Microbiology, Washington, DC [Google Scholar]