Abstract
The multiresistance gene cfr was identified for the first time in an Enterococcus faecalis isolate of animal origin. The 32,388-bp plasmid pEF-01, which carried the cfr gene, was sequenced completely. Three copies of the insertion sequence IS1216 were identified in pEF-01, and the detection of a cfr- and IS1216-containing amplicon by inverse PCR suggests that IS1216 may play a role in the dissemination of cfr by a recombination process.
TEXT
The cfr gene encodes a methyltransferase that modifies A2503 in bacterial 23S rRNA (12) and confers resistance to five chemically unrelated antimicrobial classes, including phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A (15), and decreased susceptibility to the 16-membered macrolides spiramycin and josamycin (26). Since its initial identification on the multiresistance plasmid pSCFS1 from Staphylococcus sciuri in 2000 (21), several studies have reported the cfr gene in staphylococcal isolates of animal and human origin. In most reports, the cfr gene was located on different plasmids (9, 10, 16), while in a few cases, it was also detected in the chromosomal DNA (9, 14, 27). Recently, the first outbreak case of linezolid-resistant, cfr-carrying Staphylococcus aureus was reported in Spain (17), and the cfr gene has also been identified in Panton-Valentine leukocidin-positive sequence type 8 (ST8) methicillin-resistant S. aureus IVa (USA300) (23). These observations underline the increasing threat of this resistance determinant to public health. In recent studies, the cfr-harboring plasmids pBS-01 and pBS-02 have also been identified in Bacillus strains from swine feces (5, 29). In addition, a poster presented by Cercenado and coworkers described two human clinical isolates of cfr-carrying enterococci, one Enterococcus faecalis and one Enterococcus faecium (2). To date, there has been no report of cfr in Enterococcus species of animal origin.
During a surveillance study on bacterial susceptibility to commonly used antibiotics on cattle farms in Sichuan province, China, in 2009, an enterococcal isolate from bovine feces exhibited elevated MICs of florfenicol and chloramphenicol, as determined by broth microdilution according to CLSI recommendations (3). This isolate, designated EF-01, was initially identified by Gram staining and Enterococcus-specific PCR (6) and confirmed as E. faecalis by the Rapid ID 32 Strep system (bioMérieux, Craponne, France).
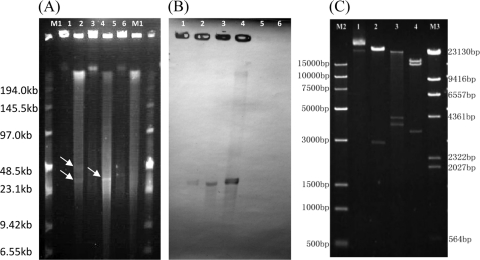
Isolate EF-01 was screened for the genes cfr and fexA using previously described primers (5). A cfr-specific PCR product was obtained and confirmed by sequence analysis. To determine the location of the cfr gene, whole-cell DNA in agarose gel plugs from EF-01 was treated with S1 nuclease (TaKaRa, Shiga, Japan) and then separated by pulsed-field gel electrophoresis (PFGE) as described previously (1). Two plasmids were observed in EF-01, and their sizes were approximately 32 kb and 48 kb, as estimated by using the standard low-range PFG markers (NEB, United Kingdom) (Fig. 1A). In a Southern blot analysis, a cfr-specific digoxigenin-labeled probe hybridized to the ca. 32-kb plasmid, designated pEF-01 (Fig. 1B).
Fig 1.
Identification and characterization of cfr-carrying pEF-01. (A) Plasmid size determination by S1 nuclease treatment coupled with pulsed-field gel electrophoresis. Lane M1, low-range PFG marker (NEB); lanes 1 and 2, EF-01; lanes 3 and 4, the transformant JH2-2+pEF-01; lanes 5 and 6, E. faecalis JH2-2 (control for lanes 3 and 4). The samples (gel plug) in lanes 1, 3, and 5 were not treated with S1 nuclease, while the samples in lanes 2, 4, and 6 were incubated with S1 nuclease prior to electrophoresis. Arrows indicate the locations of plasmids. (B) Southern blot hybridization with the cfr probe. The lane numbers correspond to those in panel A. (C) Restriction digestion analysis of plasmid pEF-01, extracted from E. faecalis JH2-2+pEF-01. Lane 1 contains the nondigested plasmid pEF-01, while lanes 2, 3, and 4 contain the plasmid DNA digested with StuI, EcoRI, and XbaI, respectively. Lanes M2 and M3 contain the Trans15K (TransGen) and λ-HindIII digest (TaKaRa) DNA markers, respectively.
To investigate the transferability of plasmid pEF-01, conjugation and transformation assays were performed. Plasmid DNA was extracted by using the Qiagen plasmid extraction midi kit (Qiagen, Hilden, Germany) with the following modification: after the enterococci were suspended in buffer P1, lysozyme was added at a final concentration of 20 μg/ml, and the mixture was incubated for 2 h at 37°C before adding buffer P2. Transfer of the purified plasmid DNA was attempted with E. faecalis JH2-2 and protoplasts of S. aureus RN4220 by electrotransformation (4, 19). The transformants were selected on brain heart infusion (BHI) agar supplemented with 10 μg/ml florfenicol. Additionally, conjugative mating into E. faecalis JH2-2 was attempted as described elsewhere (8). Although the conjugation was not successful, pEF-01 was successfully transferred into strains JH2-2 (JH2-2+pEF-01) and RN4220 (RN4220+pEF-01) by electrotransformation, as confirmed by a Southern blot analysis (Fig. 1A and B). Compared to the recipient strains, the transformants JH2-2+pEF-01 and RN4220+pEF-01 exhibited elevated MICs of phenicols, clindamycin, linezolid, and tiamulin (Table 1), which indicated the functionality of the cfr gene in the new host bacteria.
Table 1.
Impact of pEF-01 on antimicrobial susceptibility in E. faecalis and S. aureus
| Antimicrobial agent | MIC (μg/ml) fora: |
||||
|---|---|---|---|---|---|
|
E. faecalis strain |
S. aureus strain |
||||
| EF-01 | JH2-2 | JH2-2+pEF-01 | RN4220 | RN4220+pEF-01 | |
| Chloramphenicol | 64 | 8 | 32 | 4 | 64 |
| Florfenicol | 128 | 8 | 64 | 4 | 128 |
| Linezolid | 4 | 2 | 8 | 2 | 8 |
| Tiamulin | – | – | – | ≤0.5 | 32 |
| Clindamycin | – | – | – | ≤0.0625 | 8 |
–, MIC was not measured.
The cfr gene of pEF-01 encodes a 349-aa protein which differs from the Cfr proteins of pSCFS1 and pSCFS3 by only two amino acid (aa) substitutions (K88E and N123D) (11, 21). To determine the genetic environment of the cfr gene, pEF-01 DNA purified from the transformant JH2-2+pEF-01 was sequenced by shotgun sequencing combined with primer walking for gap closure, with both performed by the Beijing Genomics Institute (BGI; China). Sequences were annotated using the VectorNTI program (Invitrogen), and the predicted coding sequences (CDSs) were identified via Glimmer software and manually by correlation scores of the open reading frames (ORFs) with ≥50 amino acids. Sequence comparison was performed using the BLAST system (http://www.ncbi.nlm.nih.gov/BLAST/). The final assembly of the whole plasmid was verified by two methods. Based on the assembled plasmid sequence, we designed multiple pairs of PCR primers, and by using these primers, we obtained amplicons of the expected sizes from JH2-2+pEF-01. The amplicons were sequenced, and they confirmed the assembled plasmid sequence. Second, we conducted restriction analyses of the plasmid using EcoRI, HindIII, MluI, NheI, NdeI, StuI, XbaI, and XhoI. All obtained fragment patterns were consistent with the assembled plasmid sequence. Some of the restriction patterns are shown in Fig. 1C.
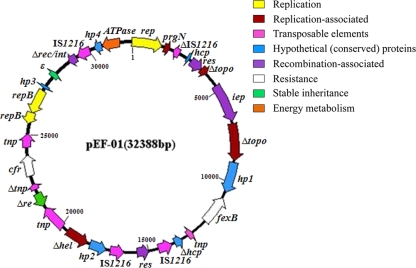
Plasmid pEF-01 consists of 32,388 bp and contains 30 potential coding sequences (CDSs) for proteins of ≥50 aa. The putative functions of 28 CDSs were predicted on the basis of their sequence homology to previously characterized proteins (see Table S1 in the supplemental material). The G+C content of pEF-01 is 35.3%, which is similar to that of E. faecalis genomic DNA (13). The G+C contents of individual CDSs ranged from 27.7% to 47.3%, suggesting that pEF-01 is composed of DNA regions which originated from multiple sources. A circular genetic map of pEF-01 is shown in Fig. 2. In addition to cfr, the phenicol exporter gene fexB (CDS10), recently described in E. faecium and Enterococcus hirae, was identified in pEF-01 (13).
Fig 2.
Genetic map of pEF-01. Coding regions larger than 50 amino acids are represented by arrows indicating the direction of transcription. All CDSs are colored according to their predicted functions. Truncated CDSs are indicated with a Greek delta symbol (e.g., ΔIS1216).
Comparative analyses of the sequences of pEF-01 and the vanA-carrying, vancomycin-resistant plasmids pVEF1/pVEF2/pVEF4 from Enterococcus faecium revealed three interesting observations. First, a region of 9,771 bp of pEF-01 spanning from CDS 28 to CDS 7 (Fig. 2; see also Table S1 in the supplemental material) showed 98.4% identity to the corresponding region of pVEF4 from an E. faecium strain derived from poultry in Norway (25). A smaller region (6,925 bp; from CDS 28 through part of CDS 6) of pEF-01 also showed 99.0% identity to the corresponding regions of pVEF1 and pVEF2 from E. faecium strains isolated from poultry and a poultry farmer in Norway (24). The common DNA segments in these four plasmids suggest recombination between plasmids and exchange among Enterococcus strains of different origins. Second, multiple replication genes were identified in these plasmids (pVEF1, pVEF2, pVEF4, and pEF-01). Three replication genes in pEF-01 showed similarities to those from three different bacteria. Specifically, CDS 1 is identical to the rep gene in pVEF1/pVEF2/pVEF4 from E. faecium, while the deduced aa sequences of CDS 23 and CDS 24 showed identities of 45% and 87% to the RepB proteins of pWV05 from Lactococcus lactis and Leuconostoc mesenteroides subsp. cremoris, respectively (22). The presence of three different rep genes in pEF-01 suggests that this plasmid might have the ability to replicate in different bacteria—as also confirmed by its ability to replicate in S. aureus RN4220—and thus contribute to the dissemination of the multiresistance gene cfr. Third, all of these plasmids have multiple copies of the insertion sequence (IS) IS1216 (three intact and one truncated copy in pEF-01) and the same resolvase (CDS 5 in pEF-01). The presence of IS elements, transposases, group II intron reverse transcriptase, integrase, and resolvases (Fig. 2; see also Table S1 in the supplemental material) in these plasmids might facilitate intra- or interplasmid recombination. Additionally, pEF-01 carries a bacterial epsilon antitoxin gene, suggesting that it may possess a toxin-antitoxin system to maintain its stability in bacterial hosts.
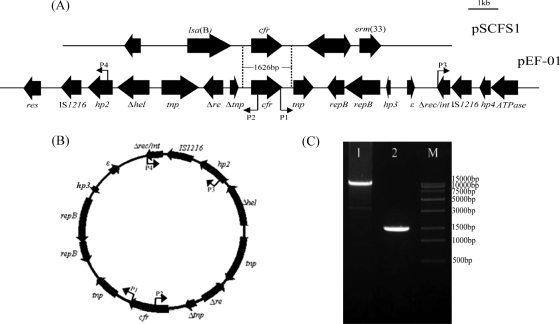
In pEF-01, two direct repeat copies of the IS1216 element flanked a 12.4-kb segment that contains the cfr gene (Fig. 3A). It has been reported that IS1216 plays an important role in the dissemination of antimicrobial resistance determinants (28) and in plasmid recombination (20). Since IS-mediated transpositions involve a simple cut-and-paste mechanism and the formation of circular forms (7, 9), we performed inverse PCR to detect whether IS1216 in pEF-01 mediated the formation of a cfr-carrying circular form. The inverse PCR assay was first performed using primers P1 (5′-TGAAGTCTGCTGGTATCCATGT-3′) and P2 (5′-TTTGCTCTGCTAAGAGCTTGAT-3′) and total DNA purified from EF-01 as the template. Indeed, a ca. 13-kb PCR product was obtained, and sequence analysis of the PCR product confirmed that it contained one copy of IS1216 and the intact region between the two direct repeat copies of IS1216, as shown in Fig. 3B and C. To further confirm the formation of a circular form, a second inverse PCR assay was conducted using primers P3 (5′-GACTGGCTTGATCTAAACCG-3′) and P4 (5′-GAGTCCTAATGAACCCAATACAG-3′), which are located near the two direct repeat copies of IS1216 (Fig. 3A). This inverse PCR yielded a ca. 1.5-kb product that contained one intact copy of IS1216 and partial sequences of hp2 and Δrec/int (Fig. 3B and C). Moreover, identical inverse PCR products were amplified using either plasmid or whole-cell DNA of the original strain EF-01 and its transformant JH2-2+pEF-01. These results might suggest that intraplasmid recombination occurred between the two direct repeat copies of IS1216 in pEF-01 (Fig. 3B). Thus, the association of cfr with IS1216 may facilitate its dissemination; however, the definitive role of IS1216 in mobilizing cfr remains to be confirmed in future studies.
Fig 3.
Genetic features flanking the cfr gene and detection of IS1216-mediated formation of a circular product by inverse PCR. (A) Comparison of plasmids pEF-01 (from E. faecalis EF-01) and pSCFS1 (from S. sciuri) in the regions flanking cfr. The positions and orientations of the genes are indicated by solid box arrows. The area of homology (1,626 bp) between the two plasmids is indicated by two dashed lines. Arrows P1, P2, P3, and P4 indicate the locations and orientation of the primers used for inverse PCR. (B) A circular product detected by inverse PCR and confirmed by sequence analysis. The locations and orientation of the primers (P1 to P4) used for inverse PCR are indicated by arrows. (C) Agarose gel analysis of the inverse PCR products amplified with primers P1 and P2 (lane 1) or primers P3 and P4 (lane 2). Lane M contains the Trans15K DNA marker.
In conclusion, we report the first identification of a cfr-carrying plasmid in E. faecalis of animal origin and present evidence for the involvement of IS1216 in the mobility of the cfr gene. Given that enterococci are widely distributed in the environment and are known to be able to transmit antibiotic resistance determinants to other pathogens (18), a mobile cfr gene in this Gram-positive organism will likely increase the spread of this multiresistance determinant. Thus, an enhanced surveillance effort is needed to monitor the emergence and spread of cfr in enterococci and other pathogens in clinical settings.
Nucleotide sequence accession number.
The sequence of a 32,388-bp sequence of the cfr-carrying plasmid pEF-01 has been deposited in the GenBank database under accession no. NC_014508.
Supplementary Material
ACKNOWLEDGMENTS
We thank Don B. Clewell (University of Michigan) for providing Enterococcus faecalis reference strain JH2-2. We also thank Anne-Kathrin Schink (Friedrich-Loeffler-Institut) for expert assistance in the S1 nuclease PFGE assay.
This work was supported by grants from the National Natural Science Foundation of China (no. 31001087 and no. U1031004), the Program for Chang Jiang Scholars, and the Innovative Research Team at the University of China (no. IRT0866).
Footnotes
Published ahead of print 27 December 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 2. Cercenado E, Marín M, Insa R, Bouza E. 2010. Abstr. Eur. Congr. Clin. Microbiol. Infect. Dis., poster A-869. European Society of Clinical Microbiology and Infectious Diseases, Munich, Germany [Google Scholar]
- 3. Clinical and Laboratory Standards Institute 2008Performance standards for antimicrobial susceptibility testing; 18th informational supplement. CLSI M31-A3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. Cruz-Rodz AL, Gilmore MS. 1990. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224:152–154 [DOI] [PubMed] [Google Scholar]
- 5. Dai L, et al. 2010. First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob. Agents Chemother. 54:3953–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dutka-Malen S, Evers S, Courvalin P. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franco AA. 2004. The Bacteroides fragilis pathogenicity island is contained in a putative novel conjugative transposon. J. Bacteriol. 186:6077–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huys G, D'Haene K, Collard JM, Swings J. 2004. Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl. Environ. Microbiol. 70:1555–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kehrenberg C, Aarestrup FM, Schwarz S. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51:483–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kehrenberg C, Cuny C, Strommenger B, Schwarz S, Witte W. 2009. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 53:779–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57:1064–1073 [DOI] [PubMed] [Google Scholar]
- 13. Liu H, et al. 2012. A novel phenicol exporter gene, fexB, found in enterococci of animal origin. J. Antimicrob. Chemother. 67:322–325 [DOI] [PubMed] [Google Scholar]
- 14. Locke JB, Rahawi S, LaMarre J, Mankin AS, Shaw KJ. 2012. Genetic environment and stability of cfr in methicillin-resistant Staphylococcus aureus CM05. Antimicrob. Agents Chemother. 56:332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mendes RE, et al. 2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob. Agents Chemother. 52:2244–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morales G, et al. 2010. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 50:821–825 [DOI] [PubMed] [Google Scholar]
- 18. Paulsen IT, et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 19. Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 94:133–138 [DOI] [PubMed] [Google Scholar]
- 20. Schwarz FV, Perreten V, Teuber M. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46:170–187 [DOI] [PubMed] [Google Scholar]
- 21. Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seegers JF, Bron S, Franke CM, Venema G, Kiewiet R. 1994. The majority of lactococcal plasmids carry a highly related replicon. Microbiology 140:1291–1300 [DOI] [PubMed] [Google Scholar]
- 23. Shore AC, et al. 2010. Identification and characterization of the multidrug resistance gene cfr in a Panton-Valentine leukocidin-positive sequence type 8 methicillin-resistant Staphylococcus aureus IVa (USA300) isolate. Antimicrob. Agents Chemother. 54:4978–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sletvold H, et al. 2007. Comparative DNA analysis of two vanA plasmids from Enterococcus faecium strains isolated from poultry and a poultry farmer in Norway. Antimicrob. Agents Chemother. 51:736–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sletvold H, et al. 2010. Tn1546 is part of a larger plasmid-encoded genetic unit horizontally disseminated among clonal Enterococcus faecium lineages. J. Antimicrob. Chemother. 65:1894–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith LK, Mankin AS. 2008. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 52:1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Toh SM, et al. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu X, et al. 2010. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob. Agents Chemother. 54:4643–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang W, et al. 2011. The new genetic environment of cfr on plasmid pBS-02 in a Bacillus strain. J. Antimicrob. Chemother. 66:1174–1175 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.