Abstract
The development of new antiviral compounds active against hepatitis C virus (HCV) has surged in recent years. In order for these new compounds to be efficacious in humans, optimal dosage regimens for each compound must be elucidated. We have developed a novel in vitro pharmacokinetic/pharmacodynamic system, the BelloCell system, to identify optimal dosage regimens for anti-HCV compounds. In these experiments, genotype 1b HCV replicon-bearing cells (2209-23 cells) were inoculated onto carrier flakes in BelloCell bottles and treated with MK-4519, a serine protease inhibitor. Our dose-ranging studies illustrated that MK-4519 inhibited replicon replication in a dose-dependent manner, yielding a 50% effective concentration (EC50) of 1.8 nM. Dose-fractionation studies showed that shorter dosing intervals resulted in greater replicon suppression, indicating that the time that the concentration is greater than the EC50 is the pharmacodynamic parameter for MK-4519 linked with inhibition of replicon replication. Mutations associated with resistance to serine protease inhibitors were detected in replicons harvested from all treatment arms. These data suggest that MK-4519 is highly active against genotype 1b HCV, but monotherapy is not sufficient to prevent the amplification of resistant replicons. In summary, our findings show that the BelloCell system is a useful and clinically relevant tool for predicting optimal dosage regimens for anti-HCV compounds.
INTRODUCTION
Hepatitis C virus (HCV) infects an estimated 3% of the world's population. For about 70% of the infected people, this results in a chronic infection leading to cirrhosis of the liver, liver cancer, and eventually death. There is no vaccine to prevent this infection, and until recently, the only treatment approved for clinical use is a combination of ribavirin and pegylated alpha interferon. This therapy is often associated with adverse side effects and is effective in less than 50% of patients infected with genotype 1a or 1b HCV (7). These important issues highlight the pressing need to develop more effective therapies for the treatment of HCV infections.
The development of novel compounds that inhibit specific processes in the viral replication cycle, often referred to as direct-acting antiviral agents (DAAs), has been brought to the forefront of HCV antiviral research. The HCV NS3-NS4 serine protease, NS5B polymerase, and NS5A proteins have all been recognized to be potential targets for antiviral drugs, but serine protease inhibitors are the most clinically advanced. The serine protease inhibitors boceprevir (Victrelis; Merck) and telaprevir (Incivek; Vertex), for example, have just received approval from the Food and Drug Administration (FDA) for the treatment of chronic genotype 1 HCV infections (17, 23). Several additional protease inhibitors have been described to be at early stages of clinical development and show promising anti-HCV activity in vitro (19, 26). For our studies, we employed MK-4519 (Merck), a serine protease inhibitor in the preclinical phase of development.
In order for new anti-HCV compounds, such as MK-4519, to be effective in a clinical setting, it is imperative to determine the optimal dose and schedule of administration (pharmacodynamics [PDs]) for each compound. We have developed a new in vitro model system to study the PDs of antiviral compounds active against HCV. This system, the BelloCell system, utilizes a Huh-7 human hepatoma cell line that carries a subgenomic replicon for genotype 1b HCV (2209-23 cells). The 2209-23 cells are adherent and do not export a product that can be measured in the supernatant. Hence, the classical hollow-fiber infection model (HFIM) system approach, used to study PDs of antiviral compounds active against influenza, HIV, and vaccinia virus (2–6, 15, 16), cannot be employed if serial sampling is desired. The major advantages of the BelloCell system are that it permits serial sampling of the replicon-containing cells and allows the simulation of human pharmacokinetic (PK) profiles for any compound. Both experimental conditions are paramount for the identification of optimal dosage regimens for anti-HCV agents.
In this work, we employed the BelloCell system to identify the pharmacodynamic parameter for MK-4519 that is linked to inhibition of replication and suppression of emergence of resistance for a genotype 1b replicon. Additionally, we used a mathematical model to describe the effect of MK-4519 on replicon populations over time. Our findings indicate that the BelloCell system is an effective in vitro PD model system which can be used for predicting optimal dosage regimens for antiviral compounds active against HCV. To our knowledge, the BelloCell system is the only in vitro model system available for PD investigations of anti-HCV drugs.
MATERIALS AND METHODS
Cells.
The genotype 1b HCV subgenomic replicon cell line 2209-23 (a kind gift from Roche, Palo Alto, CA) has been described previously (9). Briefly, the replicon contains a Renilla luciferase (rLUC) reporter gene and neomycin resistance (Neor) selection gene in the first open reading frame (ORF), which is driven by the HCV internal ribosomal entry site (IRES). Gene expression for the second ORF is controlled by the IRES derived from encephalomyocarditis virus (EMCV) and includes HCV nonstructural (NS) proteins NS3 through NS5B. An illustration of the HCV replicon used in these studies is depicted in Fig. 1. Cells were maintained in high-glucose Dulbecco's modified Eagle's medium (DMEM; HyClone, Logan, UT) containing 4.0 mM l-glutamine and sodium pyruvate and supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin solution, and 0.5 mg/ml of G418 (Geneticin; Invitrogen, Carlsbad, CA) at 37°C in 5% CO2. Cells were passaged twice weekly to maintain subconfluency.
Fig 1.
Organization of HCV replicon in the 2209-23 cell line. The bicistronic replicon genome is flanked by noncoding regions (NCR) at both the 5′ and 3′ ends. An HCV IRES, located in the 5′ noncoding region, drives translation of the first ORF, which contains the Renilla luciferase (rLUC) and neomycin resistance (Neor) genes. HCV nonstructural (NS) proteins NS3 to NS5B, derived from a genotype 1b virus, are located in the second ORF. Gene expression for this ORF is controlled by the IRES from EMCV. The replicon was transfected into Huh-7 cells to produce the 2209-23 HCV replicon cell line.
Anti-HCV compounds.
MK-4519 (Merck & Co., Inc. West Point, PA) was reconstituted in dimethyl sulfoxide (DMSO) to a final concentration of 10 mg/ml. It was then aliquoted and stored at −80°C prior to use. The chemical structure of MK-4519 is shown in Fig. 2 and has been previously described under the name compound 25a (13).
Fig 2.

Structure of MK-4519.
Determining antiviral effect of MK-4519 in tissue culture plates.
The antiviral activity of MK-4519 against the HCV genotype 1b replicon-bearing 2209-23 cell line was assessed in tissue culture cells prior to dose-ranging studies in the BelloCell system. The 50% effective concentration (EC50) of MK-4519 was determined using two different methods.
(i) Renilla luciferase drug assay.
2209-23 cells were removed from tissue culture flasks, counted, and resuspended in assay medium. Cells were seeded into a 96-well opaque white plate at a final concentration of 5,000 cells per well and incubated at 37°C in 5% CO2. Twenty-four hours later, serial 2-fold dilutions of MK-4519 were made in assay medium. Each dilution, including a no-treatment control, was added to the 96-well plate in triplicate and incubated at 37°C in 5% CO2 for 3 days. Luciferase activity was measured at the end of the 3 days using the Renilla luciferase assay kit, as per the manufacturer's instructions.
The EC50 was calculated by fitting to the data an inhibitory sigmoid-maximum effect (Emax) model defined by the equation effect = control effect − (maximal effect · exposureH/(exposureH + EC50H). Control effect is the measured output (luciferase activity) in the absence of MK-4519, maximal effect is the greatest reduction in output produced by MK-4519 exposure, EC50 is the MK-4519 exposure that results in 50% of maximal effect, and H is Hill's constant. The model was fit to the data by nonlinear regression analysis using S-ADAPT (version 1.56) software.
(ii) Quantitative real-time RT-PCR drug-response assay.
2209-23 cells were removed from tissue culture flasks, counted, and resuspended in assay medium. Cells were seeded into 6-well plates (Corning Inc., Corning, NY) at a final concentration of 1.5 × 105 cells per well and placed at 37°C in 5% CO2 overnight. A no-treatment control and serial 2-fold dilutions of MK-4519 were added in triplicate to the 6-well plates and incubated at 37°C in 5% CO2 for 3 days. After the incubation period, total cellular RNA was isolated from each sample using an RNeasy minikit (Qiagen, Valencia, CA) and extracts were frozen at −80°C until further processing. Antiviral activity of MK-4519 was determined by performing quantitative real-time reverse transcription-PCR (RT-PCR) on extracted RNA samples. RNA was amplified using one-step TaqMan EZ RT-PCR core reagents (Applied Biosystems, Foster City, CA) and a primer-probe set targeting the 5′ untranslated region of the HCV replicon (Integrated DNA Technologies, Coralville, IA): primers 5′-CCAAATCTCCAGGCATTGAGC-3′ (forward, 500 nM) and 5′-GCCATAGTGGTCTGCGGAAC-3′ (reverse, 500 nM) and probe 5′-6-FAM-CACCGGAATTGCCA GGACGACCGG–3′-Iowa Black fluorescein quencher (250 nM; FAM is 6-carboxyfluorescein). HCV RT-PCR assays were multiplexed with primers and a probe specific for the constitutively expressed housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH; TaqMan GAPDH control reagents [human] kit; Applied Biosystems) for normalization. The thermal cyclic parameters were 50°C for 2 min, 60°C for 30 min, and 95°C for 5 min, followed by 40 cycles of 20 s at 94°C and 1 min at 61°C. Reactions were analyzed using an ABI Prism 7700 sequence detector system (Applied Biosystems). The amount of HCV RNA was normalized to the amount of GAPDH RNA for each sample. EC50s were calculated using an inhibitory sigmoid Emax model in the S-ADAPT program, as described above.
Cytotoxicity assay.
The toxicity of MK-4519 was determined in parallel with the 96-well-plate drug assays. 2209-23 cells suspended in assay medium were seeded into a clear tissue culture-treated 96-well plate at a final concentration of 5,000 cells per well. Cells were incubated overnight at 37°C in 5% CO2. Twenty-four hours later, a no-treatment control and serial 2-fold dilutions of MK-4519 were added in triplicate to the cells and incubated for an additional 3 days. Cell viability was measured using the cell proliferation reagent WST-1 (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. Samples were read on a Synergy HT multidetection microplate reader (BioTek, Winooski, VT). Cytotoxicity was assessed by comparing the metabolic activity of compound-treated cells to that of untreated control cells.
Growth kinetics of 2209-23 cells in BelloCell system.
Studies were performed to determine if 2209-23 cells would maintain consistent cell growth kinetics while retaining stable replicon levels in the BelloCell system. A detailed description of the BelloCell system (BelloCell HD continuous cell culture system; catalog no. BS7000; Cesco Bioengineering Inc., Taichung, Taiwan) is described elsewhere (8, 22). Briefly, 2209-23 cells were removed from tissue culture flasks, counted, and resuspended in assay medium (DMEM supplemented with 5% FBS and 1% penicillin-streptomycin solution) to concentrations of 2 × 106 cells per ml. BelloCell-500P perfusion bottles (Cesco Bioengineering Inc., Taichung, Taiwan) were loaded with 470 ml of assay medium in the bottom of the bottle and 30 ml of cell suspension (∼6 × 107 cells) into the carrier basket, for a total working volume of 500 ml per bottle. BelloCell bottles were incubated at 37°C in 5% CO2 for 24 h to allow optimal cell attachment to carrier flakes (catalog no. BC1000; Cesco Bioengineering Inc.). During the attachment period, the BelloStage maintained an up/down speed of 2 mm/s, an upper holding time of 2 min, and a bottom holding time of 0 min. On the following day, BelloStage parameters were altered to sustain an up/down speed of 1.5 mm/s, an upper holding time of 2 min, and a bottom holding time of 1.5 min. These parameters were maintained for the duration of the study. Additionally, Masterflex pumps (Cole-Parmer, Vernon Hills, IL) were employed to constantly perfuse fresh assay medium into the BelloCell bottles (0.6 ml/min) and to remove waste. Assay medium was made fresh and added to the system daily. FBS concentrations in the assay medium were decreased to 2.5% beginning at day 5 to maintain logarithmic cell growth.
To monitor cell growth and replicon stability, the BelloCell system was sampled at various time points (Fig. 3). To sample, three to six sets of six carrier flakes were removed from the carrier basket of each BelloCell bottle using sterile forceps. Sampling was always conducted inside a class II biosafety cabinet to prevent contamination. Carrier flakes were washed twice with phosphate-buffered saline (PBS) to remove the assay medium. Three sets of flakes were placed directly into 1× Renilla luciferase lysis buffer (Promega, Madison, WI), vortexed thoroughly, incubated at room temperature for 30 min, and frozen at −80°C for future analysis. 2209-23 cells were removed from the remaining set(s) of flakes using 0.05% trypsin with EDTA (Invitrogen, Carlsbad, CA), and viable cells were counted using the trypan blue exclusion test.
Fig 3.
2209-23 cell growth and HCV replicon levels in the BelloCell system. (A) Cell growth was measured daily. 2209-23 cells were removed from carrier flakes with trypsin-EDTA, and viable cells were enumerated using the trypan blue exclusion method. Data points represent the average cell count from three independent samples, and error bars correspond to 1 standard deviation. (B) Replicon stability was assessed by quantifying the luciferase activity of 2209-23 cells from 6 carrier flakes harvested each day from the BelloCell bottle.
Replicon stability was monitored by measuring the luciferase activity of the 2209-23 cells grown in the BelloCell system using the Renilla luciferase assay kit, as per the manufacturer's instructions. Briefly, all samples were assayed on the same day to avoid assay-to-assay variability. Carrier flakes frozen in Renilla luciferase lysis buffer were thawed completely at room temperature and vortexed thoroughly. Twenty microliters of each sample was transferred in triplicate into a white opaque 96-well plate (Dynex Technologies, Chantilly, VA) and analyzed on a Glomax 20/20 luminometer (Promega).
BelloCell system experimental design.
MK-4519 dose-ranging studies consisted of four BelloCell bottles each loaded with approximately 6 × 107 2209-23 cells, as described above. Following the 24-h cell attachment period, three BelloCell bottles received a bolus injection of either 6 nM, 10 nM, or 30 nM MK-4519 and one bottle served as a no-treatment control. Drug was delivered into the system via Masterflex pumps which continuously perfused assay medium containing the appropriate concentration of compound into the BelloCell bottles to maintain the original concentration. Each day, assay medium with MK-4519 was made fresh and added to the system. PK samples were harvested from the BelloCell bottles several times during the first 48 h of the study, and MK-4519 concentrations were quantified by high-pressure liquid chromatography–tandem mass spectrometry (LC/MS/MS) to ensure that proper drug concentrations were achieved. Studies were conducted for up to 14 days. At various times after drug exposure, six sets of six carrier flakes were harvested from each BelloCell bottle using sterile forceps. Flakes were washed twice with PBS to remove all traces of assay medium. Three sets of flakes were completely immersed in 1× Renilla luciferase lysis buffer, vortexed, incubated at room temperature for at least 30 min, and frozen at −80°C until the end of the study. The remaining three sets of flakes were submerged in RLT buffer (Qiagen), vortexed, incubated at room temperature for 15 min, and frozen at −80°C. At various times throughout the course of the study, samples frozen in RLT buffer were thawed at room temperature and RNA extractions were performed according to the manufacturer's instructions (Qiagen). RNA extracts were stored at −80°C until the end of the study. Renilla luciferase and RT-PCR assays were performed on the appropriate samples of flakes, as described above. All samples for each assay were run simultaneously at the end of the study to avoid assay-to-assay variability.
Dose-fractionation studies were conducted in the BelloCell system to identify the pharmacodynamic index (indices) linked with inhibition of replicon turnover and suppression of resistance. Dose-fractionation experiments were conducted in parallel with dose-ranging experiments. In these studies, four BelloCell bottles were inoculated with 2209-23 cells as described above. One arm served as a no-treatment control, and the remaining three arms were treated with MK-4519. For the treatment arms, the total daily dose of 10 nM was administered as a continuous infusion or as an injection over a 1-h period via computer-controlled syringe pumps on a schedule of a whole dose every 24 h (q24h) or one-half dose every 12 h (q12h). Following the injection, fresh medium without drug was pumped into the q24h and q12h study arms at a specific rate to simulate the species scale-up estimate of a human half-life (t1/2) of 3.6 h (provided by Merck & Co.). All three treatment arms received the same 24-h area under the concentration-time curve (AUC) exposure of MK-4519 (240 nM · h). PK samples were harvested from the BelloCell bottles during the first 48 h of the study and assayed by LC/MS/MS. Flakes were sampled and processed as indicated for dose-ranging studies.
NS3/4A population and clonal sequencing.
Reverse transcription and PCR amplification of the NS3/4A gene were performed on RNA isolated from HCV Con1b replicon cells from BelloCell experiments using a Superscript III One Step RT-PCR system with Platinum Taq High Fidelity (Invitrogen) and the following amplification primers: 377IRES1F (5′-TCTCCTCAAGCGTATTCAACAAGG) and NS4AR4 (5′-AGGCAGAGTGGACAAGCCTGCTAA). Population sequence analysis of the NS3/4A region was performed using 10 sequencing primers spanning the NS3/4A region. For clonal sequencing, the NS3/4A amplicons were purified by gel electrophoresis (Clonewell gel purification system; Invitrogen). The purified NS3/4A DNA was cloned using a TOPO TA cloning kit for sequencing (Invitrogen). DNA was extracted from 96 clones per time point and amplified using a TempliPhi amplification system (GE Healthcare). The NS3 protease region was sequenced from each clone following duplicate sequencing reactions run with the primer NS3_1B-4013 (5′-TTGRAAKGTYTGCGGTAC). All sequencing was performed using a BigDye Terminator cycle sequencing kit (version 3.1; Applied Biosystems) and analyzed using SeqScape (version 2.5) software (Applied Biosystems). All commercially available products were used as per the manufacturer's instructions. The number of sequence determinations used to determine the percentage of each mutant is shown in Table 1.
Table 1.
Total number of clones analyzed from each BelloCell treatment arm
| Treatment arm | No. of clones analyzed |
|
|---|---|---|
| Day 6 | Day 13 | |
| 0 nM | 86 | 84 |
| 6 nM CIa | 87 | 82 |
| 10 nM CI | 72 | 92 |
| 30 nM CI | 88 | 70 |
| 10 nM q24h | 86 | 92 |
| 10 nM q12h | 89 | 85 |
CI, continuous infusion.
MK-4519 concentrations in BelloCell system.
Medium samples were diluted with HPLC-grade water (0.050 ml sample into 49 .0 ml 50:50 methanol-water) and were analyzed by LC/MS/MS for MK-4519 concentrations. The LC/MS/MS system was comprised of a Shimadzu Prominence HPLC system and an Applied Biosystems/MDS Sciex API5000 LC/MS/MS.
Chromatographic separation was performed using a Thermoscientific Hypersil Gold C18 column (150 by 4.6 mm, 5 μm) and a mobile phase consisting of 25% 0.1% formic acid in water and 75% methanol at a flow rate of 0.75 ml/min.
MK-4519 concentrations were obtained using TurboIonSpray LC/MS/MS monitoring of the MS/MS transition m/z 694.7 → m/z 341. Analysis run time was 8.0 min. The assay was linear over a range of 0.5 nM to 50.0 nM (r2 > 0.996). The interday coefficients of variation for the quality control samples analyzed in replicates of three at three concentrations on each analysis day (0.100, 1.00, and 10.0 nM) were 68.02% or less, with accuracies (percent recovery) ranging from 98.6% to 102%.
Mathematical model describing the exposure-response relationship of total replicon population and subpopulation resistant to MK-4519 exposure.
A mathematical model was fit to the Renilla luciferase data generated by dose-ranging and dose-fractionation studies to determine both the time course of HCV replicon turnover and the effect of MK-4519 on the turnover. Three simultaneous parallel inhomogeneous differential equations, shown below, were used to describe the time course of MK-4519 concentrations and the total and resistant replicon subpopulations. The model was evaluated using the Big Non-Parametric Adaptive Grid (BigNPAG) approach. Bayesian estimates were generated for each dosing regimen.
| (1) |
for initial condition 2 (IC2):
| (2) |
for IC3:
| (3) |
Equation 1 is a standard one-compartment model with zero-order input and first-order elimination that describes the pharmacokinetics of MK-4519 in the BelloCell system. X1 is the amount of MK-4519 (in mg) in the central compartment (BelloCell bottle). R(1) is the piecewise constant infusion rate of MK-4519 (in nM/h) into the central compartment. B(1) is the bolus injection of MK-4519 (in nM) into the central compartment. CL (in liters/h) is the rate of clearance of MK-4519 from the central compartment. V (in liters) is the volume of the central compartment.
Equations 2 and 3 describe luciferase activity of the susceptible population (equation 2) and the mutant population (equation 3), where X2 and X3 represent wild-type luciferase activity and mutant luciferase activity, respectively. Kturn-s and Kturn-r are the first-order turnover rate constants for wild-type (sensitive) and mutant (resistant) replicons, respectively. C50-s and C50-r are the concentrations of MK-4519 for wild-type and mutant replicons at which the turnover rate constants are reduced by half, respectively. Hs and Hr are Hill's constants for the wild-type and mutant populations, respectively. POPMAX is the maximal amount of total luciferase activity and is part of the logistic carrying function. Kloss is the first-order rate of loss of the replicon from the cells.
System outputs.
System outputs are Y(1), which is the measured MK-4519 concentrations in the in vitro system; Y(2), which is log10 luciferase activity of the total replicon population; and Y(3), which is log10 luciferase activity of the mutant replicon subpopulation.
RESULTS
Susceptibility of HCV replicon RNA to MK-4519.
We evaluated the susceptibility of HCV replicon RNA to MK-4519 by measuring the inhibitory effect of the compound on replicon RNA replication using Renilla luciferase and RT-PCR assays. The EC50s for MK-4519 were 1.58 ± 0.43 nM (mean ± standard deviation) by the Renilla luciferase assay and 1.73 ± 0.18 nM by the real-time RT-PCR assay. No signs of cytotoxicity were observed, as 2209-23 cell viability was not altered in the presence of the compound at concentrations up to 144 nM, determined by measurement of the 50% cytotoxic concentration using the cell proliferation reagent WST-1. These findings indicate that MK-4519 actively inhibits replicon output.
Growth kinetics of HCV replicon-bearing 2209-23 cells in BelloCell system.
We evaluated cell growth kinetics and replicon RNA levels of 2209-23 cells in the BelloCell system over time. BelloCell bottles were loaded with 6 × 107 cells and continuously infused with assay medium for 14 days. Carrier flakes were sampled daily from the BelloCell bottle, and viable cells and luciferase activity were quantified. Growth kinetics of 2209-23 cells were robust in the BelloCell system, as the number of cells increased from 106 cells on day 1 to 3.3 × 108 cells on day 10 in the BelloCell bottle (Fig. 3A). Cell numbers then plateaued at about 3 × 108 cells for the remainder of the study. Luciferase activity, indicative of replicon RNA levels, increased steadily with linear kinetics through day 6 and reached peak levels of 6.1 × 107 relative light units (RLU) at day 8 (Fig. 3B). Replicon RNA levels were relatively constant for the duration of the experiment. These data show that 2209-23 cells are able to actively grow and maintain HCV replicon RNA when cultured in the BelloCell system.
Dose-range studies with MK-4519 in BelloCell system.
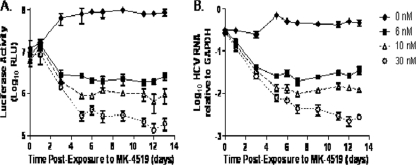
We performed dose-ranging studies with MK-4519 in the BelloCell system to (i) examine the utility of the system as an in vitro PK/PD model for HCV and (ii) assess the antiviral activity of MK-4519 on HCV replicon RNA over time through sequential sampling. Renilla luciferase and RT-PCR assays yielded nearly identical results, showing similar trends in HCV RNA levels over time (Fig. 4). The no-treatment control arm showed consistent amounts of replicon RNA, indicating that the replicon was maintained in 2209-23 cells throughout the duration of the study. MK-4519 inhibited the HCV replicon in a dose-dependent manner, with the 30 nM regimen resulting in the largest amount of suppression and the 6 nM regimen suppressing the least (Fig. 4). Luciferase activity and replicon RNA decreased steadily for all treatment arms through day 5. After day 5, the change in luciferase activity or HCV RNA levels was minimal from day to day in the 6 nM and 10 nM treatment arms, indicating that the maximal extent of inhibition had been achieved. Replicon levels in the 30 nM treatment arms continued to decrease through day 12. The PK results showing the actual MK-4519 concentrations in the BelloCell bottles are illustrated in Fig. 5.
Fig 4.
Dose-dependent inhibition of the HCV replicon by MK-4519 in BelloCell dose-ranging studies. Luciferase activity (A) and HCV RNA relative to GAPDH RNA (B) of 2209-23 cells at various times after MK-4519 exposure. BelloCell bottles were inoculated with 6 × 107 replicon-bearing 2209-23 cells and continuously infused with 0, 6, 10, or 30 nM MK-4519. Data points indicate median values of three independent samples, and error bars correspond to 1 standard deviation.
Fig 5.
MK-4519 concentrations, determined by LC/MS/MS, in the BelloCell system for the dose-ranging study. Triangles, MK-4519 concentrations observed; lines, targeted concentration-time profile.
Since complete inhibition of replicon RNA was never achieved in the dose-ranging studies, we investigated whether replicons resistant to MK-4519 emerged under drug pressure in these experiments. Replicon RNA harvested on days 6 and 13 from dose-range studies was examined for genetic mutations associated with resistance to protease inhibitors. Mutant populations were quantified by clonal sequencing. Replicons harboring resistance mutations were detected in all RNA samples harvested from MK-4519 treatment arms from the dose-range study, and the resulting mutations are shown in Fig. 6. We found that the percentage of wild-type replicons decreased with increasing levels of MK-4519 exposure. Higher exposures also resulted in more diverse mutant populations, as 9 different mutant genotypes were identified in the 30 nM exposure on day 6, whereas 4 different genotypes were found in the 6 nM and 10 nM regimens at the same time point. Finally, detection frequencies of replicon mutants varied with exposure, and this is demonstrated by the F43S mutation (Fig. 6A to C) on day 6 (21% for 6 nM versus 36% for 10 nM versus 18% for 30 nM). In addition to exposure level, the duration of exposure greatly influenced the mutant replicon population. Our results showed that the overall percentage, genotype, and detection frequency of mutant replicons were different on day 6 than on day 13 for samples receiving identical exposures of MK-4519 (Fig. 6). These findings suggest that the level and duration of exposure as monotherapy hold important implications regarding the emergence of drug-resistant replicons.
Fig 6.
Selection of drug-resistant NS3/4A replicon variants as a result of MK-4519 pressure in BelloCell dose-ranging studies. Cellular RNA was extracted from 2209-23 cells that were treated with various concentrations of MK-4519 in the dose-range study. Replicon RNA was assessed for mutations by clonal sequencing as described in Materials and Methods. Pie charts show the genotype and frequency of replicon populations harvested from the 6 nM (A and D), 10 nM (B and E), and 30 nM (C and F) treatment arms at day 6 (top) and day 13 (bottom) after drug exposure.
Dose-fractionation studies in BelloCell system.
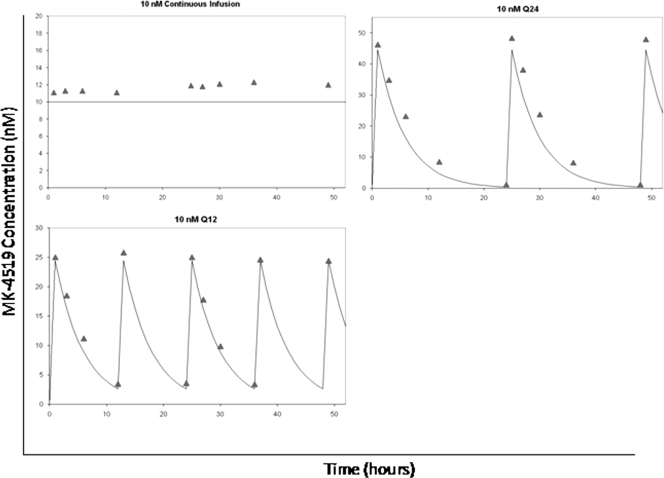
The dose-ranging studies showed that a dose-response relationship exists between MK-4519 and HCV replicon RNA when MK-4519 is administered as a continuous infusion. We also wanted to determine if the schedule of administration of MK-4519 affects inhibition of the replicon. Therefore, we performed dose-fractionation studies in which a 24-h AUC exposure of 240 nM · h was administered as the total exposure once a day (q24h), half the exposure twice daily (q12h), or a continuous infusion. Since dose-range and dose-fractionation studies were performed concurrently, we used the same data for the no-treatment control and continuous-infusion (10 nM) arms from the dose-range study for the dose-fractionation study. Similar to the dose-ranging studies, the results obtained from the luciferase assay and RT-PCR assays correlated well (Fig. 7). All dosage regimens suppressed the HCV replicon relative to the no-treatment control arm as early as day 3. The extent of inhibition was the greatest for the continuous-infusion and q12h treatment arms, as both regimens yielded nearly identical results (Fig. 7). The q24h administration suppressed replicon RNA the least, maintaining at least 3 times more HCV RNA than the other regimens at days 9 through 12. These differences were statistically significant (P < 0.05). Simulated and measured MK-4519 concentrations show that the desired 24-h AUC exposures were attained for all dosage regimens (Fig. 8).
Fig 7.
Inhibition of the HCV replicon by MK-4519 at different dosing intervals in BelloCell dose-fractionation studies. Luciferase activity (A) and relative HCV RNA (B) quantified from 2209-23 cells harvested from the BelloCell system at various times after MK-4519 exposure. BelloCell bottles were inoculated with 6 × 107 2209-23 cells. The 24-h AUC exposure for 10 nM MK-4519 (240 nM · h) was administered q24h, q12h, or as a continuous infusion. Data points indicate median values of three independent samples, and error bars correspond to 1 standard deviation.
Fig 8.
Pharmacokinetic analysis of MK-4519 in the BelloCell system. PK samples were harvested from BelloCell units, and MK-4519 concentrations were determined by LC/MS/MS. Triangles, measured concentrations; lines, target concentration-time profiles.
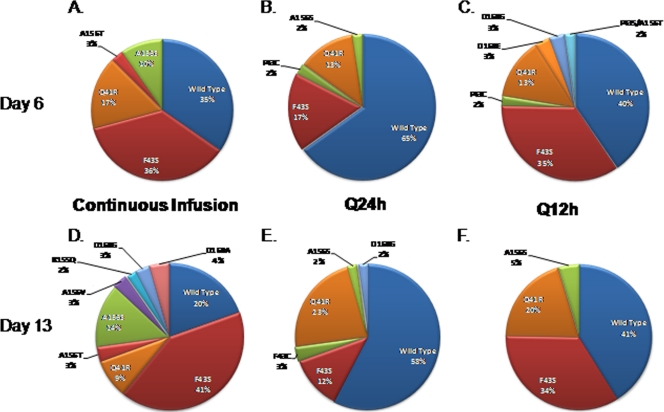
Replicon RNA from dose-fractionation studies was also analyzed for resistance mutations to determine if schedule of administration affects the emergence of drug-resistant replicons. We found that the percentage of wild-type replicons varied between the different dosing intervals, despite the fact that all three regimens received the same level of exposure (Fig. 9). At both time points, the largest percentage of wild-type replicons was found in the q24h regimen (65% on day 6 and 58% on day 13), followed by the q12h (40% on day 6 and 41% on day 13) and continuous-infusion (35% on day 6 and 20% on day 13) regimens. Mutant genotype was also different between the dosing intervals, and so was the frequency of detection of specific replicon mutants (Fig. 9). The duration of exposure did not greatly alter wild-type and mutant replicon populations in the q24h and q12h regimens, as the percentages of mutant genotypes were similar on days 6 and 13 (Fig. 9B, C, E, and F). However, MK-4519 administration by continuous infusion did affect replicon populations by duration, resulting in overall increases in the percentage of mutant replicons and genotypes of detected mutants from day 6 to day 13 (Fig. 9A and D).
Fig 9.
Detection of NS3/4A mutations associated with MK-4519 resistance in replicons harvested from dose-fractionation studies. A 24-h AUC exposure of 240 nM · h of MK-4519 was delivered into the BelloCell system at dosing intervals of q24h or q12h or as a continuous infusion. Cellular RNA was extracted from 2209-23 cells from each treatment arm, and NS3/4a replicon mutants were quantified by clonal sequencing. The pie charts show the replicon populations for the continuous-infusion (A and D), q24h (B and E), and q12h (C and F) dosing intervals at day 6 (top) and day 13 (bottom) after drug exposure.
A single-mutation genotype associated with resistance to serine protease inhibitors was detected in the no-treatment control arm. The mutation Q41R was identified on day 6 at a frequency of 2% and on day 13 at a frequency of 4% (data not shown). As there was no drug pressure in this study arm, the mutation is most likely due to the error-prone nature of the replicon polymerase.
Mathematical modeling.
We used a mathematical model to describe the relationship between MK-4519 concentrations and the HCV replicon RNA over time. The mathematical model was simultaneously fit to the PK and Renilla luciferase data generated from the dose-ranging and dose-fractionation studies in the BelloCell system. The mean parameter values and post-Bayesian estimation step for each individual dosage regimen are shown in Table 2. The predicted-versus-observed plots for luciferase activity and MK-4519 concentration show that the model yielded precise and unbiased curve fits (Fig. 10). Linear regression analysis of observed-versus-fitted plots yielded a slope of 1.0, an intercept of −0.008, an r2 value of 0.95, and a P value of less than 10−300 for total luciferase activity (log10 RLU) and a slope of 0.91, an intercept of −0.79, an r2 value of 0.78, and a P value equal to 3.63 × 10−7 for mutant luciferase activity (log10 RLU). An r2 value of 1.0 and a P value of less than 10−300 were obtained for the MK-4519 concentration fits.
Table 2.
Mean and Bayesian estimate parameter values
| Treatment arm | Kturn-s (day−1) | Kturn-r (day−1) | C50-s (nM MK-4519) | C50-r (nM MK-4519) | H-s | H-r | Kloss (day−1) |
|---|---|---|---|---|---|---|---|
| 0 nM | 2.04 | 2.575 | 20.743 | 46.92 | 22.81 | 2.614 | 1.342 |
| 6 nM CIa | 1.975 | 2.736 | 19.4 | 55.51 | 6.998 | 19.25 | 2.199 |
| 10 nM CI | 1.999 | 2.748 | 23.61 | 45.76 | 11.12 | 18.34 | 2.258 |
| 10 nM q24h | 5.612 | 2.086 | 3.076 | 79.53 | 15.83 | 24.65 | 1.569 |
| 10 nM q12h | 3.131 | 4.541 | 15.39 | 14.68 | 14.8 | 3.67 | 2.6 |
| 30 nM CI | 0.242 | 15 | 2.854 | 21.77 | 23.93 | 5.968 | 0.459 |
| Mean ± SD | 2.500 ± 1.63 | 4.947 ± 4.56 | 14.18 ± 8.29 | 44.03 ± 21.5 | 15.91 ± 5.99 | 12.42 ± 8.62 | 1.738 ± 0.71 |
CI, continuous infusion.
Fig 10.

Predicted-versus-observed plots after the Bayesian step for MK-4519 concentration (A), luciferase activity of the total replicon population (B), and luciferase activity of the MK-4519-resistant mutant replicon subpopulation (C).
Figure 11 shows the model fits (simulated) for the PK data and the observed MK-4519 concentrations. The observed luciferase activities for total replicon RNA and mutant replicon RNA (symbols) are shown scattered around the model-predicted luciferase activity (lines) (Fig. 12). These results demonstrate that the luciferase activity detected in the BelloCell arms treated with MK-4519 was largely produced by drug-resistant replicon mutants at later time points. Model predictions showed that mutant replicon populations inevitably replace the total replicon population by the end of the 14-day study in all treatment arms.
Fig 11.
Fitted pharmacokinetics for MK-4519 against a genotype 1b HCV replicon in the BelloCell system. (A) MK-4519 was delivered into BelloCell units as a continuous infusion of 6, 10, or 30 nM. (B) A 24-h AUC exposure of 240 nM · h of MK-4519 was administered q24h or q12h and eliminated from the system with a half-life of 3.6 h. Lines, model-simulated concentration-time profiles (fitted); symbols, MK-4519 concentrations measured in the system.
Fig 12.
Fitted luciferase activities for MK-4519 against a genotype 1b HCV replicon in BelloCell dose-range and dose-fractionation studies. (A to D) Graphs based on the dose-range study where various concentrations of MK-4519 were administered into BelloCell bottles as a continuous infusion. (A, B, D, and F) Graphs based on the dose-fractionation study in which a 24-h AUC exposure of 240 nM · h was fractionated at q24h, q12h, and continuous-infusion dosing intervals. A half-life of 3.6 h was simulated for MK-4519 in the dose-fractionation study. Lines, luciferase activity simulated by the mathematical model; symbols, luciferase activity observed in the BelloCell system at the specified time point; blue lines and symbols, luciferase activity from the total replicon population; red lines and symbols, luciferase activity from the mutant replicon subpopulation.
DISCUSSION
Since the technology for a cell culture-based (genotype 1) HCV system that produces high titers of infectious virus is not yet available (27), we used an HCV replicon-bearing cell line (2209-23 cells) as a surrogate model for HCV infection. As previously discussed, a major obstacle with the replicon system is that it does not secrete a measurable product (e.g., infectious virus). Consequently, serial sampling over time cannot be achieved using the HFIM system (2–6, 15, 16). As a result, we have recently adapted a bioreactor system, the BelloCell system, into a completely novel in vitro system for HCV PK/PD studies. The goal of this study was to elucidate whether the BelloCell system is an effective model system that can be used to predict the optimal dose and schedule of administration for antiviral agents active against HCV.
In our growth curve studies, we demonstrated that 2209-23 cells grow well in the BelloCell system for up to 14 days. These findings are crucial for the initial validation of the BelloCell system, as active cell growth is required for 2209-23 cells to maintain the subgenomic HCV replicon. We also showed that the replicon, in the absence of the selection pressure of G418, was stably expressed in the system for the entire duration of the experiment. These findings provided early evidence that the BelloCell system is a suitable in vitro model system for studying the PDs of anti-HCV compounds.
For in vitro PK/PD studies in the BelloCell system, we employed a serine protease inhibitor that is currently in preclinical development (MK-4519). MK-4519 was selected on the basis of its high antiviral activity in tissue culture (EC50 = 1.8 nM) and low cytotoxicity. Dose-ranging studies demonstrated that MK-4519 suppresses replicon replication relative to the no-treatment control in a dose-dependent manner. However, complete suppression was never achieved in this study, despite using concentrations up to 17 times the EC50. We hypothesized that replicons resistant to MK-4519 were selected in the BelloCell system under consistent drug pressure for 13 days. To test this hypothesis, we screened replicons from the dose-ranging study for the presence of NS3-NS4A mutations associated with drug resistance.
HCV resistance to serine protease inhibitors has been well described both in vitro and in vivo (11, 12, 20). Resistance occurs through the introduction of specific mutations in the protease by the low-fidelity HCV RNA-dependent RNA polymerase. These specific mutations have been well characterized through clinical trials and in vitro studies with the replicon. The most commonly reported mutations associated with resistance are found at positions V36, Q41, F43, T54, R155, A156, and D168 (14, 20, 21, 25, 28, 29). Clonal sequencing analysis revealed that 4 of the above-mentioned mutations (Q41, F43, A156, and D168) were detected in samples harvested from the dose-ranging study, confirming our hypothesis that MK-4519-resistant replicons were generated in the BelloCell system when placed under consistent drug pressure. Due to the high replication rate of replicon RNA and the error-prone nature of the HCV polymerase, mutant replicons were probably present in the population at levels below the assay limit of detection prior to drug exposure. Drug-resistant HCV variants have been found in treatment-naïve patients with genotype 1 infections (10), illustrating that wild-type and mutant HCV variants naturally exist as a quasispecies. Therefore, replicon mutants were most likely amplified in the BelloCell system under drug pressure, but they may also have been generated de novo. Our results suggest that the overall percentage of replicon mutants and the mutant genotype detected in a given sample were dependent on the level of drug exposure and the duration of exposure (Fig. 6). These findings also indicate that the replicon mutants are relatively biologically fit compared to wild type, as the frequency of detection of various genotypes increased over time. Other replicon mutants, such as the double mutants Q41R/D168E and Q41R/D168G (30 nM treatment arm), were detected at day 6 but were undetectable by day 13, suggestive of a decrease in fitness for these genotypes.
The dose-ranging studies illustrated that the BelloCell system can be used to identify exposure-response relationships between antiviral compounds and HCV replicons. However, the main objective of our experiments was to determine whether this system could be adapted to perform in vitro PK/PD studies using clinically relevant PK profiles. To satisfy this objective, we conducted dose-fractionation studies with MK-4519 in the BelloCell system. For these studies, a 24-h AUC exposure of 240 nM · h for MK-4519 was administered q24h, q12h, or as a continuous infusion, and a half-life of 3.6 h was simulated. We found that the schedule of administration had a significant impact on the suppression of replicon replication, with twice-daily and continuous-infusion administration yielding the largest amount of inhibition (Fig. 7). These results suggest that for MK-4519 the time that the concentration is greater than the EC50 is the PD parameter linked with inhibition of replicon replication and more frequent dosing intervals are necessary for optimal suppression of replication. Similar dosage regimens have been described for the newly FDA-approved compounds boceprevir and telaprevir, which are currently administered three times a day (18, 24). Clonal sequencing analysis from the dose-fractionation study demonstrated that the fewest number of resistant mutants occurred at the q24h dosing interval (Fig. 9). However, this regimen provided the smallest amount of replicon suppression compared to the continuous-infusion and q12h regimens. This finding may be explained by the concentration-time profiles for each dosage regimen, as trough values were 1 ng/ml for the q24h regimen, 3.5 ng/ml for the q12h regimen, and 10 ng/ml for continuous infusion. We suspect that the decreased pressure of once-daily dosing allowed wild-type replicons to replicate. Others have shown through mathematical modeling that active replication of wild-type replicons decreases the capacity for mutant replicons to replicate (1). Thus, the lower frequency of replicon mutants observed in the q24h dosage regimen of our study is likely due to the active replication of the wild-type population. These results may hold important implications on the dosage regimens for the newly FDA approved compounds boceprevir and telaprevir. Our findings suggest that clinical trials analyzing the emergence of resistance using standard dosing intervals of boceprevir and telaprevir compared to using more frequent dosing intervals may be beneficial.
To better describe the effect of MK-4519 on wild-type and resistant replicon populations over time, we employed a mathematical model. The model fit the data well (Fig. 10) and yielded mean EC50s of 14.18 nM for the wild-type (sensitive) replicons and 44.03 nM for the mutant (resistant) replicons (Table 2). Mean turnover rate constants (Kturn-s and Kturn-r) were not significantly different between the wild-type and mutant populations in most treatment arms, suggesting that single resistance mutations did not have a large negative impact on the fitness of these mutant replicons.
Overall, the data presented in these studies suggest that monotherapy with MK-4519 is unlikely to be successful. The EC50s calculated for MK-4519 in BelloCell system dose-ranging studies were 1.1 nM by the luciferase assay and 1.8 nM by RT-PCR on day 13. Therefore, the drug concentrations examined in this study were such that the smallest concentration was approximately 3.33 to 5.45 times and the largest concentration was approximately 16.67 to 27.27 times the EC50 at day 13. These large exposures were unable to suppress the amplification of replicon mutant populations, indicating that combination chemotherapy is the most promising option to prevent emergence of resistance for HCV. The BelloCell system can easily be adapted to investigate optimal dosage regimens with multiple compounds, illustrating the versatility of this system.
The BelloCell system holds many advantages over traditional tissue culture methods used to measure antiviral activity against HCV replicons. However, the ability to simulate any drug concentration-time profile for a compound is arguably the most important property of the system. We employed this aspect of the BelloCell system to design optimal dosing regimens for MK-4519 when used against a genotype 1b HCV replicon. We showed that the dynamically linked PD parameter for the inhibition of replicon replication is the time that the concentration remains greater than the EC50. Additionally, we were able to detect replicons containing known genetic mutations that confer resistance to serine protease inhibitors as a consequence of MK-4519 therapy. The mutations selected under drug pressure in this study are the same mutations described in patients receiving monotherapy with other serine protease inhibitors, including boceprevir and telaprevir (18, 24). These data indicate that the BelloCell system is an important and clinically relevant in vitro tool that is likely to have significant implications for optimizing dosing strategies of anti-HCV compounds. Future studies will focus on testing in the BelloCell system anti-HCV compounds which have already been used in clinical trials. These studies are paramount for further validation of the clinical utility of the BelloCell system.
Footnotes
Published ahead of print 12 December 2011
REFERENCES
- 1. Adiwijaya BS, et al. 2010. A multi-variant, viral dynamic model of genotype 1 HCV to assess the in vivo evolution of protease-inhibitor resistant variants. PLoS Comput. Biol. 6:e1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown AN, et al. 2011. Effect of half-life on the pharmacodynamic index of zanamivir against influenza virus delineated by a mathematical model. Antimicrob. Agents Chemother. 55:1747–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown AN, et al. 2011. Zanamivir, at 600 milligrams twice daily, inhibits oseltamivir-resistant 2009 pandemic H1N1 influenza virus in an in vitro hollow-fiber infection model system. Antimicrob. Agents Chemother. 55:1740–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown AN, et al. 2010. In vitro system for modeling influenza A virus resistance under drug pressure. Antimicrob. Agents Chemother. 54:3442–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Drusano GL, et al. 2001. Hollow-fiber unit evaluation of a new human immunodeficiency virus type 1 protease inhibitor, BMS-232632, for determination of the linked pharmacodynamic variable. J. Infect. Dis. 183:1126–1129 [DOI] [PubMed] [Google Scholar]
- 6. Drusano GL, et al. 2002. Pharmacodynamics of abacavir in an in vitro hollow-fiber model system. Antimicrob. Agents Chemother. 46:464–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghany MG, Strader DB, Thomas DL, Seeff LB. 2009. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49:1335–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ho L, Greene CL, Schmidt AW, Huang LH. 2004. Cultivation of HEK 293 cell line and production of a member of the superfamily of G-protein coupled receptors for drug discovery applications using a highly efficient novel bioreactor. Cytotechnology 45:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klumpp K, et al. 2006. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 281:3793–3799 [DOI] [PubMed] [Google Scholar]
- 10. Kuntzen T, et al. 2008. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology 48:1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin C, et al. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508–17514 [DOI] [PubMed] [Google Scholar]
- 12. Lin K, Perni RB, Kwong AD, Lin C. 2006. VX-950, a novel hepatitis C virus (HCV) NS3-4A protease inhibitor, exhibits potent antiviral activities in HCV replicon cells. Antimicrob. Agents Chemother. 50:1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liverton NJ, et al. 2008. Molecular modeling based approach to potent P2-P4 macrocyclic inhibitors of hepatitis C NS3/4A protease. J. Am. Chem. Soc. 130:4607–4609 [DOI] [PubMed] [Google Scholar]
- 14. Lopez-Labrador FX, Moya A, Gonzalez-Candelas F. 2008. Mapping natural polymorphisms of hepatitis C virus NS3/4A protease and antiviral resistance to inhibitors in worldwide isolates. Antivir. Ther. 13:481–494 [PubMed] [Google Scholar]
- 15. McSharry JJ, Deziel MR, Zager K, Weng Q, Drusano GL. 2009. Pharmacodynamics of cidofovir for vaccinia virus infection in an in vitro hollow-fiber infection model system. Antimicrob. Agents Chemother. 53:129–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McSharry JJ, Weng Q, Brown A, Kulawy R, Drusano GL. 2009. Prediction of the pharmacodynamically linked variable of oseltamivir carboxylate for influenza A virus using an in vitro hollow-fiber infection model system. Antimicrob. Agents Chemother. 53:2375–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Merck & Co. Inc 13 May 2011. FDA approves Merck's VICTRELIS™ (boceprevir), first-in-class oral hepatitis C virus (HCV) protease inhibitor. Merck & Co. Inc., Whitehouse Station, NJ [Google Scholar]
- 18. Merck & Co Inc 2011. VICTRELIS (boceprevir) prescribing information. Merck & Co. Inc., Whitehouse Station, NJ [Google Scholar]
- 19. Reesink HW, et al. 2010. Rapid HCV-RNA decline with once daily TMC435: a phase I study in healthy volunteers and hepatitis C patients. Gastroenterology 138:913–921 [DOI] [PubMed] [Google Scholar]
- 20. Sarrazin C, et al. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777 [DOI] [PubMed] [Google Scholar]
- 21. Thompson AJ, McHutchison JG. 2009. Antiviral resistance and specifically targeted therapy for HCV (STAT-C). J. Viral Hepat. 16:377–387 [DOI] [PubMed] [Google Scholar]
- 22. Toriniwa H, Komiya T. 2007. Japanese encephalitis virus production in Vero cells with serum-free medium using a novel oscillating bioreactor. Biologicals 35:221–226 [DOI] [PubMed] [Google Scholar]
- 23. Vertex Pharmaceuticals Inc 27 May 2011 FDA advisory committee unanimously recommends approval of telaprevir for people with hepatitis C. Vertex Pharmaceuticals Inc., Cambridge, MA [Google Scholar]
- 24. Vertex Pharmaceuticals Inc 2011. INCIVEK (telaprevir) prescribing information. Vertex Pharmaceuticals Inc., Cambridge, MA [Google Scholar]
- 25. Welsch C, et al. 2008. Molecular basis of telaprevir resistance due to V36 and T54 mutations in the NS3-4A protease of the hepatitis C virus. Genome Biol. 9:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. White PW, et al. 2010. Preclinical characterization of BI 201335, a C-terminal carboxylic acid inhibitor of the hepatitis C virus NS3-NS4A protease. Antimicrob. Agents Chemother. 54:4611–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 103:2310–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Y, et al. 2008. Phenotypic characterization of resistant Val36 variants of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 52:110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Y, et al. 2007. Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha. J. Biol. Chem. 282:22619–22628 [DOI] [PubMed] [Google Scholar]