Abstract
DNA gyrase is an essential enzyme in bacteria, and its inhibition results in the disruption of DNA synthesis and, subsequently, cell death. The pyrrolamides are a novel class of antibacterial agents targeting DNA gyrase. These compounds were identified by a fragment-based lead generation (FBLG) approach using nuclear magnetic resonance (NMR) screening to identify low-molecular-weight compounds that bind to the ATP pocket of DNA gyrase. A pyrrole hit with a binding constant of 1 mM formed the basis of the design and synthesis of a focused library of compounds that resulted in the rapid identification of a lead compound that inhibited DNA gyrase with a 50% inhibitory concentration (IC50) of 3 μM. The potency of the lead compound was further optimized by utilizing iterative X-ray crystallography to yield DNA gyrase inhibitors that also displayed antibacterial activity. Spontaneous mutants were isolated in Staphylococcus aureus by plating on agar plates containing pyrrolamide 4 at the MIC. The resistant variants displayed 4- to 8-fold-increased MIC values relative to the parent strain. DNA sequencing revealed two independent point mutations in the pyrrolamide binding region of the gyrB genes from these variants, supporting the hypothesis that the mode of action of these compounds was inhibition of DNA gyrase. Efficacy of a representative pyrrolamide was demonstrated against Streptococcus pneumoniae in a mouse lung infection model. These data demonstrate that the pyrrolamides are a novel class of DNA gyrase inhibitors with the potential to deliver future antibacterial agents targeting multiple clinical indications.
INTRODUCTION
The emergence of drug resistance in both community- and hospital-acquired infections has outpaced the development and delivery of new antibacterial drugs to the clinic. As a result, there is a serious threat to global health that currently available therapies will no longer be effective in treating infections due to increasing resistance (1, 5, 24). One approach to combating the emergence of resistance to current antibacterial drugs is to discover novel agents that inhibit known drug targets through a unique binding site, chemistry, or mechanism of inhibition, thereby evading existing resistance mechanisms.
DNA gyrase, consisting of the subunits GyrA and GyrB, is a member of the type II family of topoisomerases that control the topological state of DNA in cells (27). DNA gyrase couples ATP hydrolysis by the GyrB subunit to supercoiling of DNA, which is required for maintenance of DNA topology during the replication process. It is an essential enzyme across bacterial species, and inhibition results in disruption of DNA synthesis and, subsequently, cell death. DNA gyrase has long been known as an attractive target for antibacterial drugs (15). Two classes of antibiotics have clinically validated DNA gyrase as a viable target—quinolones and aminocoumarins. Fluoroquinolones inhibit DNA gyrase by interfering with the DNA cleavage/resealing function of the enzyme. The inhibition of the enzyme function may be only partially responsible for the bactericidal effects of fluoroquinolones. DNA damage resulting from the cellular response to fluoroquinolone stabilization of the covalent gyrase-DNA complex and the potential formation of reactive oxygen species have been postulated as likely key contributors to cell death induced by this class of antibacterials (8, 9). Aminocoumarin antibiotics (e.g., novobiocin) inhibit DNA gyrase by competing with ATP for binding within the GyrB subunit and blocking the ATP hydrolysis function of the enzyme. Although novobiocin is no longer used in the clinic, several generations of the fluoroquinolone class of antibacterial drugs continue to be used extensively. However, increasing prevalence of fluoroquinolone-resistant bacterial strains is eroding the utility of even these successful drugs (4, 6, 21). The aim of the program described here was to discover novel compounds that target the ATP-binding site of GyrB, since antibacterial agents with this mechanism of action should avoid cross-resistance with existing target-based fluoroquinolone-resistant pathogens.
Breakthroughs in genome sequence analysis and gene knockout capabilities in the 1990s jump started a frenzy of interest in identifying novel, essential targets for antibacterial drug discovery. However, the lack of progression of new drug classes into the clinic from these efforts has proven disappointing (20). While novel target identification and validation are important components of drug discovery, success in the antibacterial therapy area is also critically dependent on the attributes of the chemical series. Successful antibacterial compounds must have appropriate properties to penetrate bacterial cells while also exhibiting in vivo and physicochemical properties suitable for achieving relatively high doses with acceptable safety margins. One possible reason for the lack of success in identifying novel antibacterial drugs is the reliance on internal compound collections as the source of chemical starting points for the novel bacterial targets. Such corporate collections are typically populated with compounds from previous projects accumulated over the years and are therefore biased toward inhibitors of human targets according to the legacy interests/expertise of the company. Therefore, most corporate collections do not offer compounds optimally suited to antibacterial discovery programs.
Fragment-based lead generation (FBLG) offers an alternative screening approach to novel hit identification that avoids these common issues associated with typical high-throughput screening (HTS) of corporate compound collections (2, 11). By utilizing low-molecular-mass (generally <350-Da) compound “fragments” as chemical starting points rather than fully elaborated compounds from an HTS library, the resulting novel scaffolds can be specifically tailored to bind the intended target, resulting in compounds with greater specificity and ligand binding efficiency for the target. In addition, physicochemical properties necessary to support penetration into bacterial cells can be built into the scaffold at an earlier stage rather than modifying existing properties of a more elaborated HTS hit.
This paper describes initial prototypes of a novel class of DNA gyrase inhibitors, the pyrrolamides, with potent activity against a broad spectrum of primarily Gram-positive bacterial pathogens, including those resistant to current drugs. The pyrrolamide antibacterials originated from an FBLG approach using nuclear magnetic resonance (NMR) to identify and characterize low-molecular-weight compounds that bind to the ATP pocket of GyrB. A library of compounds was designed based on a selected pyrrole hit that had a low-millimolar dissociation constant for GyrB. This library led to an initial pyrrolamide lead compound that inhibited the ATPase function of DNA gyrase with micromolar potency. Further elaboration of the series was guided by computational design and X-ray crystallography, resulting in potent enzyme inhibitors with promising antibacterial activity and cellular mode of action through GyrB. A representative compound from the pyrrolamide series demonstrated efficacy in a mouse lung infection model, indicating that the chemical series has further potential to yield novel antibacterial drugs.
(Data from these studies were presented in part at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy in Washington, DC, in 2008 [10, 13, 14, 22, 26]).
MATERIALS AND METHODS
Bacterial strains and compounds.
Staphylococcus aureus ARC516, Streptococcus pneumoniae ARC548, Enterococcus faecium ARC521, Haemophilus influenzae KW20 (ATCC 51907), Escherichia coli ARC523, and E. coli ARC524 were from the AstraZeneca bacterial culture collection. The pyrrolamide compounds were synthesized as previously described (A. Breeze, O. Green, K. Hull, H. Ni, S. Hauck, G. Mullen, N. Hales, and D. Timms, 24 March 2005, international patent application WO/2005/026149). Other antibiotics (levofloxacin and novobiocin) were obtained from Sigma-Aldrich Inc. (St. Louis, MO).
Production of recombinant DNA gyrase proteins.
The methods for expression and purification of DNA gyrase subunits used in enzyme assays and biophysical studies are described in detail in the supplemental material.
NMR screening and hit characterization.
Samples of uniformly 15N-labeled E. coli GyrB 24-kDa N-terminal domain were prepared for NMR experiments in a buffer comprising 50 mM sodium phosphate, pH 6.8, 1 mM dithiothreitol (DTT), 0.1 mM EDTA, 0.02% (wt/vol) sodium azide, and 5% (vol/vol) D2O. The final protein concentration was 300 μM. NMR screening was carried out on mixtures of 10 compounds derived from targeted or generic (2) fragment libraries by addition of deuterated-dimethyl sulfoxide (DMSO) stocks to a final concentration of 1 mM per compound. 1H-15N heteronuclear single quantum coherence (HSQC) experiments were acquired on a Varian 600-MHz NMR spectrometer at a probe temperature of 298 K. Mixtures containing active binders were identified by their residue-specific amide chemical shift perturbations (CSPs); the individual hit compounds were confirmed by deconvolution of the mixtures through serial addition of the mixture components to a fresh 15N-labeled protein sample until the HSQC spectrum of the original mixture was recapitulated. The KD values for certain weakly binding compounds were determined by NMR using nonlinear fitting of CSP data as a function of the total ligand concentration to a tight-binding saturation isotherm model using the program GraFit (Erithacus Software, London, United Kingdom).
Gyrase ATPase assay.
Details of the assay used to monitor compound inhibitory effects on the ATP hydrolysis activity of E. coli DNA gyrase are described in the supplemental material. Briefly, an assay using malachite green detection of inorganic phosphate was performed to measure the ATP hydrolysis activity of E. coli DNA gyrase reconstituted in vitro from purified, recombinant GyrA and GyrB subunits. Pyrrolamide compounds were tested at various concentrations in this assay to yield the 50% inhibitory concentrations (IC50s) (see Table 2).
Table 2.
Enzyme and antibacterial activities of selected pyrrolamide analogs
| Compound | E. coli ATPase IC50 (nM) | MIC (μg/ml) |
|||||
|---|---|---|---|---|---|---|---|
| E. coli ARC523 | E. coli ARC524a | H. influenzae KW20 | S. aureus ARC516 | S. pneumoniae ARC548 | E. faecium ARC521 | ||
| 1 | 3,000 | >64 | >64 | >64 | >64 | >64 | >64 |
| 2 | 14 | >64 | 2 | 64 | 2 | 1 | 1 |
| 3 | 0.9 | 8 | <0.06 | 0.25 | 0.5 | 0.5 | 2 |
| 4 | 25 | >64 | 0.25 | 2 | 8 | 0.5 | 2 |
Strain ARC524 is equivalent to ARC523 with a Tn10 insertion in tolC; efflux mutant.
GyrB crystallography and structure refinement.
Details of crystallization of the S. aureus GyrB 24-kDa N-terminal loop deletion domain with pyrrolamide 1 and pyrrolamide 2 and structure refinement to resolutions of 1.85 Å and 1.64 Å, respectively, are described in the supplemental material.
Susceptibility assays.
MIC values for bacterial species and for Candida albicans were determined according to Clinical and Laboratory Standards Institute guidelines (17, 19). Minimum bactericidal concentrations for all species were determined according to Clinical and Laboratory Standards Institute guidelines (18).
Mammalian MICs were determined against A549 human lung carcinoma cells (ATCC) in RPMI medium (Sigma-Aldrich Inc., St. Louis, MO) containing 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA) and 1% l-glutamine at a density of 1,000 cells/well. After incubation of the cells with compound in a CO2 atmosphere at 37°C for 72 h, cell viability was determined by absorbance at 490 nM with Cell Titer 96 AQ One Solution Cell Proliferation Assay Reagent (Promega Corporation, Madison, WI). The MIC was the concentration that produced ≥50% transmission.
Isolation and characterization of pyrrolamide-resistant bacteria.
Suspensions of cultures of methicillin-sensitive S. aureus (ARC516) were transferred onto plates of Mueller-Hinton agar with 2.5% blood containing pyrrolamide 4 at a concentration equivalent to the MIC (8 μg/ml), as well as similar plates without compound. The frequency of spontaneous resistance was determined in two independent studies performed in triplicate on each occasion and was calculated from the number of colonies on the compound-containing plates divided by the number of colonies on compound-free plates after incubation for 7 days. Values from across the triplicate plates in the two studies were averaged to yield the reported frequency of spontaneous resistance. Similar methods were used to determine the spontaneous-resistance frequency for levofloxacin, with the exception that bacteria were plated on agar plates containing 0.25 μg/ml levofloxacin (a concentration equivalent to the MIC). When bacteria were plated on agar plates containing a 4-fold multiple of the MIC value for pyrrolamide 4 (32 μg/ml) or levofloxacin (1 μg/ml), no resistant colonies were recovered.
Twenty colonies were selected from the pyrrolamide-containing plates, frozen in glycerol stocks, and recovered on drug-free plates. Genomic DNA was prepared from 8 of these isolated resistant variants, as well as from the original susceptible parent strain, and the genes for the topoisomerase subunits (gyrA, gyrB, parC, and parE) were amplified by PCR and sequenced using standard protocols described previously (7). Gene sequences from the variant strains were compared to those from the parent strain and found to be identical, except for two different point mutations found in the gyrB gene. One point mutation was observed to be common to 5 of the 8 strains sequenced (resulting in Arg144Leu), and the 3 remaining strains were observed to share a different mutation (resulting in Thr73Ala). A more detailed description of these methods, including a list of primers and sequence comparison for the gyrB genes from the parent and resistant strains, can be found in the supplemental material.
In vivo efficacy.
In vivo activity against S. pneumoniae ARC548 was determined in an immunocompetent-mouse model of pneumonia (3, 16). The animals used in the infection model were maintained in accordance with the criteria of the American Association for Accreditation of Laboratory Animal Care. All animal studies were carried out according to protocols approved by the Institutional Animal Care and Use Committee at AstraZeneca R&D Boston. Groups of 7 C57BL6 mice (Charles River Laboratories, MA) were infected intratracheally with 1 × 105 CFU/lung while under anesthesia with ketamine/xylazine. Treatment with pyrrolamide 4, by oral gavage, commenced 18 h after infection with a twice daily (b.i.d.) every 12 hours (q12 h) regimen. Activity was quantified by viable counts on serial dilutions of lung homogenates 24 h after the start of treatment and reported as mean CFU/lung (±standard error).
Protein structure accession numbers.
The coordinates for the complexes with pyrrolamides 2 and 3 have been deposited in the Protein Data Bank with accession numbers 3U2D and 3U2K, respectively.
RESULTS
Identification of NMR hits that bind GyrB.
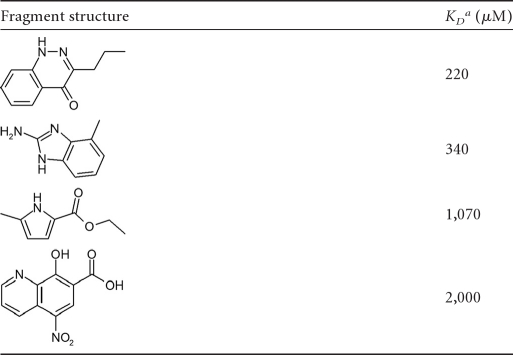
NMR screening (23) was used to identify low-molecular-weight hits that bind to the 24-kDa N-terminal ATP-binding domain of E. coli GyrB (25). The screening library consisted of ∼1,000 diverse low-molecular-mass (100- to 370-Da) compounds, together with a number of fragments derived from “deconstruction” of known GyrB inhibitors, such as the pyrrole and noviose sugar from clorobiocin. NMR CSP-mapping experiments were performed with 15N-labeled protein to identify hits based on shifts observed in the 1H-15N HSQC spectrum. Binding constants for hits were determined by titrating labeled protein with ligand and plotting shift change versus concentration. The resulting KD values for selected hits are shown in Table 1.
Table 1.
Fragment hits from NMR screen
KD, dissociation constant against the E. coli GyrB N terminal 24-kDa ATP-binding domain measured using NMR.
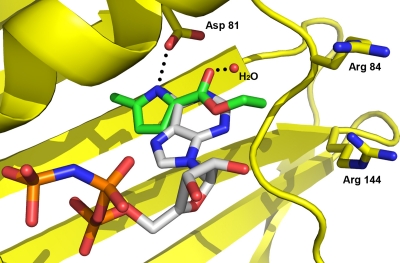
The most potent hits shared a common hydrogen bond donor/acceptor motif also seen in the adenine ring of the natural substrate, ATP, and induced amide CSPs consistent with binding in the vicinity of the known adenine binding pocket. These data supported computational-modeling efforts to predict the binding mode for selected hits using a crystal structure of the S. aureus GyrB 24-kDa N-terminal domain. The predicted binding mode for the pyrrole hit (Table 1, third line) is shown in Fig. 1 overlaid with ATP. This hit was selected as the starting point for inhibitor design based on potency and ligand efficiency (12), as well as an attractive binding mode for future elaboration. By repeating the screen with the GyrB adenine pocket fully occupied, a second-site binder (Table 1, fourth line) was identified that appeared by NMR CSPs to be binding in a more distal region of the binding pocket (close to Arg84 and Arg144) (Fig. 1) known to be exploited by the aminocoumarin antibiotics. Despite its weak binding (KD = 2 mM), the identification of a chemotype that bound to a second site suggested the possibility of elaborating the primary pyrrole hit to access additional interactions in the GyrB binding pocket and thereby gain potency.
Fig 1.
Computational docking of a pyrrole NMR fragment hit overlaid with ATP in the crystal structure of the S. aureus GyrB 24-kDa N-terminal domain. Hydrogen bonds between the adenine of ATP, Asp81, and a water molecule (red sphere) are shown with dotted lines. Carbons of the pyrrole NMR hit are colored green, while those of ATP are colored gray. Other atoms are shown in standard coloring. The GyrB protein is colored yellow and displayed as a ribbon structure, with the exception of key amino acid side chains labeled by sequence number.
Pyrrole library design and testing.
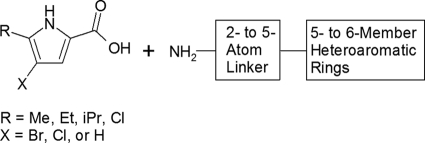
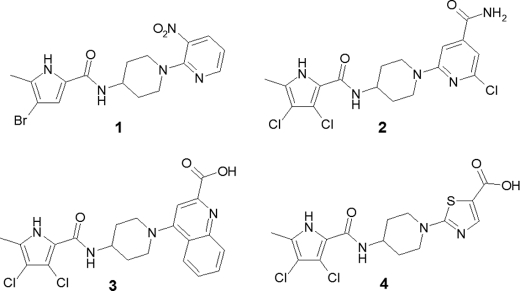
A library of pyrrole-containing compounds was designed and prepared using multiple parallel synthesis with commercially available starting materials. Selected pyrrole-carboxylate groups were combined using standard peptide chemistry with a variety of heteroaryl rings with terminal amino-substituted alkyl linkers (Fig. 2). The resulting library of compounds was screened for inhibition of the ATPase activity of E. coli DNA gyrase. Pyrrolamide 1 (Fig. 3) was the most potent hit, with an IC50 of 3 μM against E. coli DNA gyrase (Table 2). IC50s for the pyrrolamide compounds against E. coli topoisomerase IV correlated well with DNA gyrase inhibition but were consistently >200-fold less potent against this related bacterial topoisomerase (data not shown).
Fig 2.
Design of a compound library based on a pyrrole NMR hit. Me, methyl-; Et, ethyl-; iPr, isoproyl.
Fig 3.
Chemical structures of representative pyrrolamides with identification numbers used in the text.
Progression of pyrrolamide series from ATPase inhibition to antibacterial activity.
Pyrrolamide 1 did not display antibacterial activity but formed the basis of a medicinal chemistry program aimed at improving enzyme and cellular potencies in the series. This effort was supported by iterative crystallography using the S. aureus GyrB 24-kDa N-terminal ATP-binding domain to guide improvements in the pyrrolamide series. Figure 4A shows the crystal structure of pyrrolamide 1 bound to the S. aureus GyrB ATP-binding domain. The pyrrole group occupies the same pocket as the adenine group of ATP and forms hydrogen bond donor/acceptor interactions similar to those seen for the natural substrate. Specifically, the pyrrole nitrogen is within hydrogen bond distance of a conserved aspartic acid 81 (Asp81), and the carbonyl substituent on the pyrrole is within hydrogen bond distance of a conserved water molecule that is positioned by Asp81. This crystal structure closely mimics the binding pose predicted from computational docking of the pyrrole NMR hit (Fig. 1). The pyridine heterocycle extends outside the ATP pocket and forms stacking interactions with protein residues, while the nitro substituent is oriented toward solvent. This crystal structure guided the design of additional features on the pyrrolamide scaffold to exploit new binding interactions with GyrB and improve the potency of subsequent compounds.
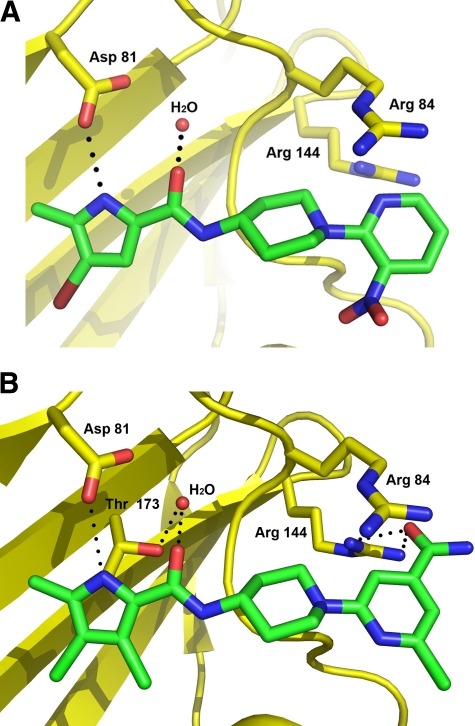
Fig 4.
X-ray crystal structure of the S. aureus GyrB 24-kDa N-terminal domain in complex with the initial lead, pyrrolamide 1 (A), and a later series representative, pyrrolamide 2 (B). Carbons of the pyrrolamide inhibitors are colored green, while the other atoms are shown in standard coloring. The GyrB protein is colored yellow and displayed as a ribbon structure, with the exception of key amino acid side chains labeled by sequence number. Hydrogen bonds are shown with dotted lines, and a conserved water molecule is shown as a red sphere.
The crystal structure of GyrB in complex with pyrrolamide 2 (Fig. 4B) provides evidence that new, specific interactions with the protein contributed to the potency improvement of >150-fold relative to the initial lead (pyrrolamide 1). The new 3,4-dichloro substituents on the pyrrole provided increased hydrophobic interactions in the adenine pocket. Furthermore, these electron-withdrawing groups reduce the pKa of the pyrrole NH group, which is expected to make it a better hydrogen bond donor to Asp81. In addition, new hydrogen bond interactions between a conserved GyrB arginine (Arg144) and the carbonyl of the amide substitution on the pyridine heterocycle likely contributed to the potency improvement of pyrrolamide 2. The improvement in enzyme potency also resulted in an improvement in antibacterial activity. While pyrrolamide 1 had MIC values of >64 μg/ml across all bacteria tested, pyrrolamide 2 displayed a significant improvement in in vitro activity against the Gram-positive bacteria S. aureus, S. pneumoniae, and E. faecium, with MIC values of ≤2 μg/ml (Table 2). In contrast, the antibacterial potency of pyrrolamide 2 against the Gram-negative species E. coli and H. influenzae was significantly weaker (≥64 μg/ml). However, more potent activity was observed (MIC = 2 μg/ml) when tested in efflux mutants such as a tolC deletion strain (ARC524) of E. coli, suggesting that activity was likely limited by cell outer membrane permeability and/or efflux mechanisms in Gram-negative pathogens rather than poor target potency.
Further modifications to the scaffold are exemplified by pyrrolamide 3 and pyrrolamide 4, which maintain potency through key interactions with GyrB while accommodating structural and physical chemical diversity through changes in the heterocycle distal to the pyrrole. These changes primarily impacted antibacterial potency, as shown in Table 2, with pyrrolamide 3 displaying the greatest potency against both Gram-positive and Gram-negative bacteria.
Due to a combination of suitable potency and desirable physical properties (e.g., solubility), pyrrolamide 4 was selected as the optimal series representative for further profiling. The minimal bactericidal concentration (MBC) values for pyrrolamide 4 against S. aureus, S. pneumoniae, and H. influenzae were determined to be 8, 0.5, and 4 μg/ml, respectively. These values are similar to the MIC values against these pathogens (Table 2), indicating that the compound is bactericidal against these species. In addition, time-kill studies were performed with pyrrolamide 4 and found to vary across different species, with observed bacterial killing ranging from 100- to 10,000-fold relative to untreated cells after 24 h. The general cytotoxicity of pyrrolamide 4 appears to be low, as MIC values for inhibition of the growth of mammalian and fungal cell lines were >64 μg/ml, resulting in >100-fold selectivity for killing of S. pneumoniae cells.
Spontaneous resistance and mutation mapping.
The frequency of spontaneous resistance was examined in two independent studies by plating S. aureus ARC516 bacteria onto agar plates containing 8 μg/ml of pyrrolamide 4, a concentration equivalent to the MIC of the compound. In these studies, spontaneous resistance was observed, with an average frequency of approximately 2.5 × 10−9. When suspensions of 1 × 1010 CFU were plated in triplicate, 27, 32, and 34 colonies were recovered in the first determination, and in the second determination, when 1.3 × 1010 CFU were plated, 19, 27, and 30 colonies were recovered from drug-containing plates. In comparison, the average resistance frequency was approximately 1 × 10−9 for the same strain of S. aureus plated on agar plates containing levofloxacin at a concentration equivalent to the MIC (0.25 μg/ml).
Strains isolated in this study demonstrated a 4- to 8-fold increase in the MIC of pyrrolamide 4 relative to the parent strain (Table 3). Elevated MIC values were also observed with some of these strains for novobiocin, a known DNA gyrase inhibitor that competes with ATP for binding the enzyme. In all cases, however, no change in MIC was observed for these variant strains against levofloxacin, an inhibitor that binds DNA gyrase at a site remote from the ATP pocket. The growth rates and morphologies of these variant strains were similar to those of the parent strain, suggesting that the overall fitness of the variants was not greatly compromised by the resistance mutations.
Table 3.
Activities of pyrrolamide 4 and comparators against GyrB mutant strainsa of S. aureus
| Compound |
S. aureus MIC (μg/ml) |
||
|---|---|---|---|
| Parent | GyrB R144I | GyrB T173A | |
| Pyrrolamide 4 | 8 | 32 | 64 |
| Novobiocin | 0.25 | 8 | 0.25 |
| Levofloxacin | 0.25 | 0.25 | 0.25 |
Resistant strains of S. aureus were selected on media containing pyrrolamide 4 at 8 μg/ml. Point mutations in the gyrB genes were identified and mapped to the amino acids shown by sequencing genomic DNA isolated from different resistant strains.
Genomic DNA from 8 of the S. aureus strains for which pyrrolamide 4 displayed elevated MIC values (as described above) was purified, and DNA sequencing of the genes encoding the topoisomerase subunits (gyrA, gyrB, parC, and parE) revealed point mutations only in the gyrB genes compared to sequences determined from the pyrrolamide-sensitive parent strain. In 5 of the strains sequenced, a single nucleotide difference that is predicted to result in an amino acid change from arginine (in the parent strain) to isoleucine (in the resistant strains) was observed at position 144 of GyrB (Arg144Ile). In the other 3 strains, a distinct nucleotide difference that is predicted to result in an amino acid change from threonine (in the parent strain) to alanine (in the resistant strains) was observed at position 173 in GyrB (Thr173Ala). The locations of these residues in the ATP-binding domain of GyrB are shown in Fig. 4B.
Both of the amino acids predicted to be altered in the resistant strains relative to those in the parent strain make key interactions with the pyrrolamide 2 inhibitor, as observed in the GyrB crystal structure (Fig. 4B). Arg144 forms hydrogen bond interactions with the carboxamide substitution on the right-side heterocycle, and Thr173 forms a hydrogen bond with the water molecule that interacts with the amide group on the pyrrole. Alteration of these amino acids to isoleucine and alanine, respectively, would likely result in disruption of those interactions with the pyrrolamide inhibitors and thus would be expected to reduce potency. It should be noted, however, that the variant GyrB proteins were not produced and tested to confirm the prediction that decreased potency would be observed for the inhibitors. Therefore, while these data support the hypothesis that GyrB may be a primary target of the pyrrolamides, it is possible that the observed mutations are compensatory rather than direct target mutations. Also, because whole-genome sequencing was not performed with these strains, the presence of additional mutations outside the genes sequenced cannot be ruled out.
As expected, the MIC values of levofloxacin were unaffected in the resistance strains with the observed GyrB mutations, since the fluoroquinolone binds to the DNA cleavage/resealing region of the enzyme, remote from the ATP pocket. Interestingly, while both variants resulted in elevated MIC values of pyrrolamide 4, the novobiocin MIC was increased only for the strain with the Arg144Ile change, while the MIC for the strain with the Thr173Ala change remained the same as that for the parent strain (Table 3). This observation suggests that although the binding sites between novobiocin and the pyrrolamides overlap, key interactions with GyrB differ between these distinct scaffolds, and as such, not all potential resistance mutations will be cross-resistant to the same extent.
In vivo efficacy.
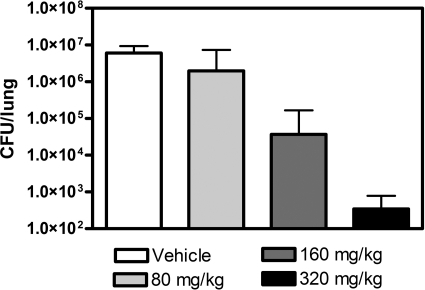
The potential for in vivo efficacy of the pyrrolamide series was investigated with pyrrolamide 4 in an immunocompetent-mouse pneumonia model with S. pneumonae ARC548 (3, 16). Infection with S. pneumoniae was established in the lungs of mice at 1 × 105 CFU/lung. Treatment was initiated with pyrrolamide 4 or vehicle control by oral gavage, commencing 18 h postinfection. Total daily doses of 80, 160, and 320 mg/kg of body weight, administered b.i.d. q12 h, were tested in the model. In the vehicle-treated mice, the bacterial burden in the lungs increased to 6.0 × 106 ± 3.3 × 106 CFU/lung over the 24 h after treatment was initiated. Treatment with escalating doses of pyrrolamide 4 resulted in a concentration-dependent decrease in viable bacteria recovered from the lungs after 24 h of oral treatment (Fig. 5). Relative to the vehicle-treated control, 160 and 320 mg/kg/day resulted in reductions in viable bacterial counts of >100-fold and >10,000-fold, respectively. Full assessment of the toxicology risk for the series was not performed as part of this study; however, based on this efficacy study, mice tolerated pyrrolamide 4 at doses up to 320 mg/kg/day for the duration of the study.
Fig 5.
Efficacy of pyrrolamide 4 in an S. pneumoniae lung infection model in mice. Shown are the numbers of S. pneumoniae CFU recovered from the lungs of mice treated with 0, 80, 160, and 320 mg/kg of pyrrolamide 4 administered orally twice per day, every 12 h. The doses shown are the total daily dose. The error bars represent standard errors in the CFU measurements.
DISCUSSION
The pyrrolamide series described in this paper is a novel class of DNA gyrase inhibitors with potent activity against a broad spectrum of primarily Gram-positive bacterial pathogens. The series was designed based on a pyrrole hit identified by NMR screening of a library of low-molecular-weight fragments for those that bound to E. coli GyrB. The fragment library consisted of generic compounds with masses of less than 370 Da and desirable physical properties. Also, the library included fragments of known DNA gyrase inhibitors, such as aminocoumarin antibiotics, which are known to bind the ATP pocket of DNA gyrase (15). The pyrrole fragment that formed the basis of the pyrrolamide series is, in fact, a feature of the natural-product antibiotic clorobiocin.
Although a structure of a fully intact DNA gyrase tetramer has not been determined, truncated domains of the protein subunits retain partial function and are amenable to biophysical techniques, such as NMR and iterative X-ray crystallography (15, 25, 28). One such domain is the 24-kDa N-terminal fragment of GyrB, which retains ATP binding and offers a robust and reliable protein for NMR and crystallography (15, 25). This form of the GyrB protein provided the receptor for hit identification by NMR fragment screening and the follow-up crystal structures of the GyrB ATP domain in complex with pyrrolamide leads. Functional assessment of the ATPase activity of the full DNA gyrase tetramer (GyrA2/GyrB2 with DNA) was used to monitor enzyme inhibition properties during the course of lead identification and progression of the pyrrolamide series. This structure-guided approach enabled rapid progression from a fragment hit with millimolar binding affinity for the GyrB target to pyrrolamide lead compounds with submicromolar enzyme inhibition potency.
Compounds in the pyrrolamide series not only display potent enzyme inhibition of DNA gyrase, but also have in vitro antibacterial activity against Gram-positive and selected Gram-negative pathogens (Table 2). Outer-membrane permeability and/or efflux mechanisms limit the potency of current pyrrolamides against a broader range of Gram-negative pathogens, including E. coli, as demonstrated by a >200-fold improvement in potency against a tolC deletion strain of E. coli for pyrrolamide 4 relative to wild-type E. coli. Improvement in activity against wild-type E. coli can be achieved by driving enzyme potency, as demonstrated by pyrrolamide 3; however efflux remains an issue that must be addressed with future analogs if Gram-negative potency is to be reliably achieved for the series.
Data for a series representative (pyrrolamide 4) indicate that the pyrrolamides are bactericidal, with equivalent MBC and MIC values against S. aureus, S. pneumoniae, and H. influenzae. While the mechanism of action for the pyrrolamides is not likely to induce downstream lethal effects similar to those described for fluoroquinolones, gyrase inhibition through competition with ATP apparently does result in bactericidal activity for these compounds against the pathogens tested thus far.
The frequency of spontaneous resistance for a representative pyrrolamide was determined to be approximately 2 × 10−9 in S. aureus, suggesting that rapid development of pyrrolamide resistance is not likely. MIC values for these variant strains against the pyrrolamide compound are elevated 4- to 8-fold relative to the wild-type parent strain, and characterized resistance mutations map to amino acids in the pyrrolamide binding site of GyrB that make key contacts with the inhibitor. While not conclusive, these data suggest that the mode of action for the pyrrolamides is through inhibition of DNA gyrase. However, full genome sequencing was not performed to rule out the presence of other genetic changes in these variants, and mutant GyrB proteins were not isolated in the present study to confirm the expected reduced affinity for the inhibitor.
The absence of resistance mutations mapping to topoisomerase IV (ParE) suggests that inhibition of this bacterial enzyme does not contribute significantly to the cellular activity of the pyrrolamides. That observation is supported by the >200-fold difference in potency observed at the enzyme level for these compounds. The apparent selectivity of the current compounds for GyrB is surprising given the similarity of sequence and structure between GyrB and ParE, particularly in key amino acids making contact with the pyrrolamide inhibitors. However, subtle conformational or dynamic differences between the DNA gyrase and topoisomerase IV enzymes could contribute to the observed differences in potency. A more complete understanding of this apparent selectivity offers a potential area of improvement for future analogs in the series, as dual targeting of both gyrase and topoisomerase IV would likely contribute to improved potency, as well as reduced frequencies of resistance.
A representative of the series (pyrrolamide 4) demonstrated efficacy in an immunocompetent-mouse pneumonia model with S. pneumoniae. Although relatively high oral doses of 160 mg/kg and 320 mg/kg were required to yield >100-fold and >10,000-fold reductions, respectively, in bacteria relative to vehicle-treated controls in this model, these data demonstrate the potential of the pyrrolamides for in vivo efficacy. Further improvements to potency and in vivo properties are the current aim for progression of this promising lead series to candidate drugs. In summary, the pyrrolamide series of gyrase inhibitors demonstrates promise as a novel class of antibacterials with the potential to deliver compounds that target multiple clinical indications, including those caused by pathogens resistant to current drugs.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following individuals for valuable discussion and/or technical contributions to this project: Barbara Arsenault, Michael Block, Claire Brassington, Jason Breed, Jeremy Colls, Amanda Doucette, David Ehmann, Richard Ernill, Lena Grosser, Ian Haigh, Sussie Hopkins, Jeanette Jones, Heather Kamp, Irene Karantzeni, Peter Kenny, Kathy MacCormack, Scott Mills, Linda Otterson, Phil Poyser, Jon Read, and John Stawpert. We also thank Tom Dougherty and Boudewijn deJonge for critical reading of the manuscript prior to submission.
Footnotes
Published ahead of print 19 December 2011
Supplemental material for this article may be found at http://aac.asm.org.
REFERENCES
- 1. Alanis AJ. 2005. Resistance to antibiotics: Are we in the post-antibiotic era? Arch. Med. Res. 36:697–707 [DOI] [PubMed] [Google Scholar]
- 2. Albert JS, et al. 2007. An integrated approach to fragment-based lead generation: philosophy, strategy and case studies from AstraZeneca's drug discovery programmes. Curr. Top. Med. Chem. 7:1600–1629 [DOI] [PubMed] [Google Scholar]
- 3. Azoulay-Dupuis E, Vallee E, Hardy DJ, Swanson RN, Pocidalo JJ. 1991. Antipneumococcal activity of ciprofloxacin, ofloxacin, and temafloxacin in an experimental mouse pneumonia model at various stages of disease. J. Infect. Dis. 163:319–324 [DOI] [PubMed] [Google Scholar]
- 4. Biedenbach DJ, Farrell DJ, Mendes RE, Ross JE, Jones RN. 2010. Stability of linezolid activity in the era of mobile oxazolidinone resistance determinants: results from the 2009 Zyvox Annuaj Appraisal of Potency and Spectrum program. Diagn. Microbiol. Infect. Dis. 68:459–467 [DOI] [PubMed] [Google Scholar]
- 5. Boucher HW, et al. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 6. Boyd LB, et al. 2008. Increased fluoroquinolone resistance with time in Escherichia coli from >17,000 patients at a large county hospital as a function of culture site, age, sex and location. BMC Infect. Dis. 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Jonge BLM, Kutschke A, Uria-Nickelsen M, Kamp HD, Mills SD. 2009. Pyrazolopyrimidinediones are selective agents for Helicobacter pylori that suppress growth through inhibition of glutamate racemase (MurI). Antimicrob. Agents Chemother. 53:3331–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Drlica K, Malik M, Kerns RJ, Zhao X. 2008. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52:385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. 2007. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3:91–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Green O, et al. 2008. Novel DNA gyrase inhibitors: structure guided discovery and optimization of pyrrolamides, abstr F1-2025. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 11. Hajduk PJ. 2006. Fragment-based drug discovery: how big is too big? J. Med. Chem. 49:6972–6976 [DOI] [PubMed] [Google Scholar]
- 12. Hopkins AL, Groom CR, Alex A. 2004. Ligand efficiency: a useful metric for lead selection. Drug Discov. Today 9:430–431 [DOI] [PubMed] [Google Scholar]
- 13. Hull K, et al. 2008. Novel DNA gyrase inhibitors: the effect of pyrrolamide variations at site 1 and site 2 upon potency, abstr F1-2027. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 14. Illingworth RN, Uria-Nickelsen M, Bryant J, Eakin AE. 2008. Novel DNA gyrase inhibitors: microbiological characterization and in vivo efficacy of pyrrolamides, abstr F1-2028. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 15. Maxwell A, Lawson DM. 2003. The ATP binding site of type II topoisomerases as a target for antibacterial drugs. Curr. Top. Med. Chem. 3:283–303 [DOI] [PubMed] [Google Scholar]
- 16. Moine P, et al. 1994. In vivo efficacy of a broad-spectrum cephalosporin, ceftriaxone, against penicillin-susceptible and -resistant strains of Streptococcus pneumoniae in a mouse model. Antimicrob. Agents Chemother. 380:1953–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. National Committee for Clinical Laboratory Standards 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A. National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 18. National Committee for Clinical Laboratory Standards 2005. Methods for determining bacteriocidal activity of antimicrobial agents, vol 12, no 9. Approved Standard M26. National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 19. National Committee for Clinical Laboratory Standards 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 8th ed, vol 29, no 2. Approved standard M07-A8. National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 20. Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29–40 [DOI] [PubMed] [Google Scholar]
- 21. Rhomberg PR, Jones RN. 2009. Summary trends for the meropenem yearly susceptibility test information collection program: a 10-year experience in the United States (1999–2009). Diagn. Microbiol. Infect. Dis. 65:414–426 [DOI] [PubMed] [Google Scholar]
- 22. Sherer B, et al. 2008. Novel DNA gyrase inhibitors: the effect of pyrrolamide linker variations upon potency, abstr F1-2026. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 23. Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. 1996. Discovering high-affinity ligands for proteins: SAR by NMR. Science 274:1531–1534 [DOI] [PubMed] [Google Scholar]
- 24. Talbot GH, et al. 2006. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 42:657–668 [DOI] [PubMed] [Google Scholar]
- 25. Tsai, et al. 1997. The high-resolution crystal structure of a 24-kDa gyrase B fragment from Escherichia coli complexed with one of the most potent coumarin inhibitors, clorobiocin. Proteins 28:41–52 [PubMed] [Google Scholar]
- 26. Uria-Nickelsen M, Blodgett A, Eakin AE. 2008. Novel DNA gyrase inhibitors: investigation of the mechanism of action of pyrrolamides, abstr F1-2029. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 27. Wang JC. 2009. Untangling the double helix: DNA entanglement and the action of the DNA topoisomerases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Wigley DB, Davies GJ, Dodson EJ, Maxwell A, Dodson G. 1991. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351:624–629 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.