Abstract
Genotypic tropism testing methods are emerging as the first step before prescription of the CCR5 antagonist maraviroc (MVC) to HIV-infected patients in Europe. Studies validating genotypic tests have included other active drugs that could have potentially convoluted the effects of MVC. The maraviroc clinical test (MCT) is an in vivo drug sensitivity test based on the virological response to a short-term exposure to MVC monotherapy. Thus, our aim was to compare the results of genotypic tropism testing methods with the short-term virological response to MVC monotherapy. A virological response in the MCT was defined as a ≥1-log10 decrease in HIV RNA or undetectability after 8 days of drug exposure. Seventy-three patients undergoing the MCT were included in this study. We used both standard genotypic methods (n = 73) and deep sequencing (n = 27) on MCT samples at baseline. For the standard methods, the most widely used genotypic algorithms for analyzing the V3 loop sequence, geno2pheno and PSSM, were used. For deep sequencing, the geno2pheno algorithm was used with a false-positive rate cutoff of 3.5. The discordance rates between the standard genotypic methods and the virological response were approximately 20% (including mostly patients without a virological response). Interestingly, these discordance rates were similar to that obtained from deep sequencing (18.5%). The discordance rates between the genotypic methods (tropism assays predictive of the use of the CCR5 coreceptor) and the MCT (in vivo MVC sensitivity assay) indicate that the algorithms used by genotypic methods are still not sufficiently optimized.
INTRODUCTION
The first coreceptor antagonist approved for the treatment of HIV-1 infection by inhibiting the entry of CCR5 (R5) viruses is maraviroc (MVC) (1, 5). Determining HIV coreceptor usage is essential before prescribing R5 antagonists (17). Currently, the most widely used coreceptor tropism test is the recombinant phenotypic Trofile assay (Monogram Biosciences) (39) or its newer version, the enhanced-sensitivity Trofile assay (ESTA) (26). However, this phenotypic assay has some limitations, including an approximately 20% rate of nonreportable results, and specimens must be shipped to the unique reference laboratory in the United States. For these reasons, other clinical (8), phenotypic (10, 11, 23), or genotypic (12) alternatives for determining viral tropism have been examined.
Genotypic methods based on analysis of the third variable region (V3) of the HIV envelope are emerging in Europe as widely available alternatives for determining HIV tropism (13). The reliability of genotypic tools to determine HIV tropism in clinical samples has been compared with that of phenotypic studies and reveals the low sensitivity of genotypic assays for detection of CXCR4-using (X4) variants (15, 25, 30). Improving this low sensitivity has been attempted through simple modifications in the interpretation of the algorithms or by combining the results given by different algorithms (2, 29). Another genotypic approach is ultradeep sequencing (UDS), which, although currently not common in clinical practice, provides encouraging results by allowing minor variant detection (4, 35). However, validation of genotypic tropism prediction methods ultimately requires not just evidence of concordance with phenotypic methods but also concordance with the virological response to drug exposure. Recent retrospective analyses from MVC clinical trials (MOTIVATE/A4001029 and MERIT) have shown that specific genotypic tools have an ability to predict the virological response to MVC that is similar to or better than that of Trofile (18, 33). However, although these results were score corrected, a potential confounding effect of the other active drugs accompanying MVC is still possible.
Recently, our group developed a clinical test based on the virological response to short-term exposure to MVC monotherapy, the maraviroc clinical test (MCT), to use prior to recommendation of R5 antagonist therapy (8). Using this model, we have already found a discordance rate of 15% between ESTA results and this clinical approach (7). Genotypic analysis of samples from the MCT would be an attractive technique for comparing these results with short-term MVC virological effects because the MCT is not based on a tropism prediction but rather on real-time sensitivity to drug exposure.
Thus, the aim of this study was to compare the results from genotypic tropism testing methods with the virological response after short-term MVC monotherapy (MCT).
MATERIALS AND METHODS
Patients.
The study was performed at the Infectious Diseases Service of the Virgen del Rocio University Hospital (Seville, Spain). From 1 July 2008 until 1 February 2011, 73 patients were included in the MCT. Briefly, the MCT consists of an 8-day exposure to MVC as monotherapy. A “virological response” was defined as a 1-log10 reduction of HIV RNA copies/ml or an undetectable viral load (<50 HIV RNA copies/ml) on the eighth day after the addition of MVC during the MCT (8). Forty-eight patients on structured treatment interruption (66%) underwent real monotherapy (only MVC) during the MCT, and the other 25 patients were on a previously failing regimen with a persistently detectable viral load; therefore, they underwent functional monotherapy (MVC add-on) during the MCT. The inclusion criteria for participation in the study were a persistently detectable viral load (>50 HIV RNA copies/ml) during the previous 6 months and no previous treatment with R5 antagonists.
Patients or their representatives (for those under 18 years of age) gave written informed consent, and the ethics committee of the hospital approved the study.
Quantification of viral RNA.
HIV-1 RNA was measured in fresh plasma samples by quantitative PCR (Cobas Ampliprep/Cobas TaqMan HIV-1 test; Roche Molecular Systems, Basel, Switzerland) according to the manufacturer's instructions. This assay has a detection limit of 50 HIV RNA copies/ml. Plasma hepatitis C virus (HCV) RNA was detected by a commercially available PCR procedure (Cobas Amplicor; Roche Diagnostics, Barcelona, Spain) with a detection limit of 15 IU/ml.
Determination of HIV-1 coreceptor usage by genotypic methods. (i) Standard methods.
V3 loop amplification and the sequencing of PCR products were performed on plasma samples at baseline, immediately prior to MVC treatment, as previously described (36). Briefly, HIV RNA was purified from 500 μl of plasma by use of a viral RNA purification kit (Qiagen, Valencia, CA), and cDNA was synthesized by reverse transcription (RT). PCR was performed with 35 cycles of 15 s at 94°C, 15 s at 50°C, and 45 s at 72°C in a reaction volume of 50 μl. For amplification of the V3 loop, the following primers were used: outer primers V1 (5′-GCACAGTACAATGTACACATGG) and V2 (5′-ACAGTTGTGTTGAATTACAGTAG) and inner primers V3 (5′-CTGTTAAATGGCAGTCTAGCAG) and V4 (5′-TTTCTGGGTCCCCTCCTGAGG). Sequencing reactions were performed with BigDye terminators and an ABI 310 sequencer (Applied Biosystems, Foster City, CA). The V3 sequences were interpreted using the most widely used bioinformatic genotypic tropism predictors, PSSM and geno2pheno (G2P), which are freely available online (http://coreceptor.bioinf.mpi-inf.mpg.de/index.php and http://fortinbras.us/cgi-bin/fssm/fssm.pl). Clonal versions of G2P with false-positive rates (FPR) of 5% (G2P5%clo) and 10% (G2P10%clo) were used. In addition, clinical versions of G2P that include clinical parameters to improve viral tropism predictions (G2P5%cli and G2P10%cli) were used. Furthermore, PSSM using Poveda's threshold values (PSSMp), optimized to enhance the sensitivity for detection of dual/mixed-tropism (D/M) or X4 variants by changing the original threshold for R5 categorization (21), was also employed. In all cases, HIV-1 variants were classified as R5 or D/M, with the latter including pure X4 and dual-tropic viruses.
(ii) Ultradeep sequencing.
For a subgroup of 27 consecutive patients, samples were deep sequenced at baseline, immediately prior to MVC treatment, on a 454 GS-FLX sequencer using titanium chemistry (Roche/454 Life Sciences). Viral RNA was extracted from frozen plasma by use of a NucliSENS easyMAG system (bioMérieux), and from this RNA, viral cDNA was generated using RT-PCR. To prepare samples for deep sequencing, the V3 loop of the gp120 protein was amplified using nested PCR in triplicate with tagged second-round primers. PCR triplicates were quantified and combined in equal proportions. Emulsion PCR with magnetic beads was used to clonally amplify the DNA libraries; 5 molecules of viral DNA were present for every DNA capture bead in the reaction mix. DNA-covered beads were purified and prepared for sequencing according to the 454 GS-FLX manual. The picotiter plate was loaded such that approximately 790,000 beads were loaded onto each of the plate regions. A more detailed description of the methodology has been published previously (2). The sequence data were interpreted using G2P with an FPR of 3.5%; this cutoff was optimized based on retrospective analyses of samples from the MOTIVATE and A4001029 clinical trials (33, 34).
Determination of plasma MVC concentrations.
To determine adherence to treatment, but not for pharmacokinetic purposes, plasma MVC was detected by high-performance liquid chromatography–tandem mass spectrometry (HPLC-MS/MS). Plasma samples (100 μl) were spiked with an internal standard (10 μl of 1-mg/ml metergoline) or DMSO (dimethyl sulfoxide; 10 μl to mimic the standard solutions) and acetonitrile (400 μl) for protein denaturation. Subsequently, the samples were subjected to centrifuge filtration using a 30,000-molecular-weight spin column (Millipore, Bedford, MA), diluted 1 to 1 with 5 mM ammonium acetate buffer, and injected into an HPLC-MS/MS system (HP 1100 chromatograph [Agilent Technologies, Palo Alto, CA] with an Applied Biosystems API 2000 mass spectrometer [AB Sciex, Concord, Ontario, Canada]). An Agilent XDB-C8 guard column and an Agilent Eclipse XDB-C18 column (50 mm) were used. MVC was separated using a gradient mobile phase (0.5 ml/min) from 80% phase A (acetate/acetic acid-buffered 10% methanol, pH 3.6 ± 0.2) to 100% phase B (acetate/acetic acid-buffered 90% methanol, pH 6.6 ± 0.2) and then analyzed using an API 2000 MS/MS instrument. The analytical range for MVC was 25 ng/ml to 5,000 ng/ml.
Statistical analysis.
Statistical analyses were performed using SPSS software (SPSS 16.0, Inc., Chicago, IL). Differences between groups were analyzed using the Mann-Whitney U test for continuous variables and the chi-square test for categorical variables. All continuous variables were expressed as medians (interquartile ranges [IQR]), and the categorical variables were expressed as percentages. All differences with P values of <0.05 were considered statistically significant.
RESULTS
Baseline characteristics of patients.
Seventy-three consecutive patients were included in this study (Table 1). Virological responses were observed in 53 patients, whereas 20 had no virological response. Patients with a virological response experienced a median viral load change of −1.32 (IQR, −1.03 to −1.59) log HIV RNA copies/ml. In contrast, the viral loads from patients with no virological response remained unchanged (0.03 [−0.21 to 0.23] log copies/ml). No differences in baseline parameters were found between patients undergoing real (MVC only) versus functional (MVC add-on) monotherapy (Table 2). Notably, adherence to treatment was confirmed for all patients by self-reported information and pharmacy refill records. In addition, we tested the plasma MVC levels in a subgroup of consecutive patients (n = 24), and all had MVC levels above the minimal therapeutic concentration required to suppress HIV (2.3 ng/ml) on day 8 (Table 3).
Table 1.
Baseline characteristics of the patients in this study (n = 73), separated by virological response
| Parameter | Valuea |
P valuee | |
|---|---|---|---|
| VR (n = 53) | No VR (n = 20) | ||
| Age (yr) | 43 (36–48) | 41 (31–45) | NS |
| No. (%) of males | 39 (79) | 17 (85) | NS |
| No. (%) of patients with HCV coinfectionb | 18 (34) | 8 (40) | NS |
| CD4+ count (cells/mm3) | 355 (243–475) | 63 (16–175) | <0.001 |
| No. (%) of patients in risk group | |||
| IDUc | 19 (35.8) | 11 (55) | NS |
| Sexual risk | 31 (58.5) | 4 (20) | 0.004 |
| Other | 3 (5.7) | 5 (25) | NS |
| No. (%) of patients with stage C disease (according to CDC) | 7 (13.2) | 9 (45) | 0.009 |
| Plasma HIV-1 RNA load (log10 copies/ml) | 4.45 (3.66–4.88) | 4.87 (3.89–5.21) | NS |
| No. (%) of patients with plasma HIV-1 RNA load of <1,000 copies/ml | 9 (17) | 3 (15) | NS |
| ΔVL after MCT (log10 copies/ml)d | −1.32 (−1.03–−1.59) | 0.03 (−0.21–0.23) | <0.001 |
| No. (%) of patients with real MVC monotherapy | 38 (71.7) | 10 (50) | NS |
Values other than numbers with percentages are expressed as medians (IQR). VR, virological response.
PCR positive for hepatitis C virus.
Intravenous drug users.
ΔVL, viral load change.
NS, P > 0.05.
Table 2.
Baseline characteristics of the patients in this study (n = 73), separated by type of MVC therapy
| Parameter | Valuea |
|
|---|---|---|
| MVC monotherapy (n = 48) | MVC add-on therapy (n = 25) | |
| Age (yr) | 43 (36–47) | 41 (25.5–46) |
| No. (%) of males | 39 (75) | 20 (80) |
| No. (%) of patients with HCV coinfectionb | 19 (39.6) | 7 (28) |
| CD4+ count (cells/mm3) | 274 (119–424) | 302 (171–521) |
| No. (%) of patients in risk group | ||
| IDUc | 19 (39.6) | 11 (44) |
| Sexual risk | 26 (54.2) | 9 (36) |
| Other | 3 (6.3) | 4 (16) |
| No. (%) of patients with stage C disease (according to CDC) | 8 (16.6) | 8 (32) |
| Plasma HIV-1 RNA load (log10 copies/ml) | 4.59 (4.15–4.98) | 3.86 (2.79–4.98) |
| ΔVL after MCT (log10 copies/ml)d | −1.16 (−0.66–−1.54) | −0.71 (−1.32–0.11) |
Values other than numbers with percentages are expressed as medians (IQR). VR, virological response. Differences between groups were analyzed using the Mann-Whitney U test, and P was >0.05 (not significant) in all cases.
PCR positive for hepatitis C virus.
Intravenous drug use.
ΔVL, viral load change.
Table 3.
Plasma MCV concentrations on day 8 of MCTa
| Patient | Plasma MVC concn on day 8 (ng/ml) |
|---|---|
| P1 | 202.0 |
| P3 | 55.7 |
| P4 | 37.2 |
| P6 | 66.1 |
| P7 | 138.0 |
| P8 | 92.1 |
| P9 | 498.0 |
| P10 | 243.0 |
| P11 | 143.0 |
| P12 | 89.0 |
| P13 | 198.0 |
| P15 | 2.6 |
| P16 | 174.0 |
| P17 | 752.0 |
| P18 | 52.6 |
| P19 | 582.0 |
| P20 | 637.0 |
| P21 | 435.0 |
| P22 | 75.8 |
| P23 | 1,090.0 |
| P24 | 120.0 |
| P25 | 557.0 |
| P26 | 54.8 |
| P27 | 121.0 |
Plasma MVC concentrations were measured to check adherence to treatment but not for pharmacokinetic purposes.
Discordance rates of genotypic tropism test results for patients with a virological response. (i) Standard methods.
Samples from 7/53 patients (13.21%) could not be amplified (see Table S1 in the supplemental material). Therefore, comparison analysis was performed for 46 patients with a virological response, and a D/M result was considered discordant. G2P5%cli gave D/M results for 2/46 (4.30%) patients, while G2P5%clo gave D/M results for 5/46 (10.87%) patients, G2P10%cli gave D/M results for 3/46 (6.52%) patients, and G2P10%clo gave D/M results for 8/46 (17.39%) patients. In contrast, PSSM gave D/M results for 9/46 (19.56%) patients, and PSSMp gave D/M results for 12/46 (26.09%) patients, resulting in unexpected score values (−6.63 to 9.64) associated with D/M viruses.
(ii) Ultradeep sequencing.
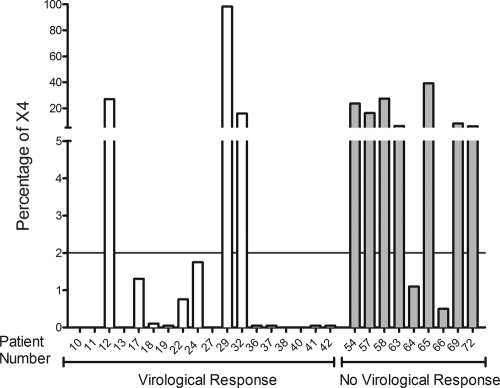
UDS was performed for a subgroup of 27 consecutive patients, 18 of whom had a virological response (see Table S1 in the supplemental material). In a previous study, UDS indicated a CCR5-positive virus if the X4 percentage was <2% (34). UDS revealed an X4-tropic strain percentage of >2% for 3/18 (16.66%) patients with a virological response (15.95%, 27%, and 98.52%); these results were considered to be discordant. For the remaining patients, X4 percentages were lower than 2% (Fig. 1).
Fig 1.
X4 prediction by deep sequencing. %X4, percentage of X4-tropic viruses detected by deep sequencing. Vertical white bars represent patients with a virological response, and vertical gray bars represent patients with no virological response. The line represents the established cutoff of 2%.
Discordance rates of genotypic tropism test results for patients with no virological response. (i) Standard methods.
One sample (1/20 samples [5%]) could not be amplified (see Table S2 in the supplemental material). Therefore, comparison analysis was performed for 19 patients, and an R5 result was considered discordant. G2P5%cli gave R5 results for 10/19 (52.63%) patients, while G2P5%clo gave R5 results for 8/19 (42.11%) patients, G2P10%cli gave R5 results for 10/19 (52.63%) patients, and G2P10%clo gave R5 results for 6/19 (31.58%) patients. In addition, PSSM gave R5 results for 8/19 (42.11%) patients, and PSSMp gave R5 results for 3/19 (15.79%) patients, resulting in unexpected score values (−9.59 to −11.57) associated with R5-tropic viruses.
(ii) Ultradeep sequencing.
Of the 27 patients for whom UDS was performed, 9 lacked a virological response (see Table S2 in the supplemental material). UDS detected an X4-tropic strain percentage of <2% for 2/9 (22.22%) patients (0.5% and 1.1%), and these results were considered discordant. For the remaining patients, X4 percentages were higher than 2% (Fig. 1).
Sensitivities, specificities, and discordance rates of genotypic tropism testing methods for patients with a virological response.
The percentage of unreported results obtained by standard genotypic methods was 10.96% (8/73 patients). Values for sensitivity/specificity for patients with a virological response by standard methods and deep sequencing methods were highest for PSSMp (84.2%/73.9%), followed by UDS (83.3%/77.8%), G2P10%clo (68.4%/82.6%), G2P5%clo (57.9%/89.1%), PSSM (57.9%/80.4%), G2P5%cli (47.4%/95.7%), and G2P10%cli (47.4%/93.5%) (Table 4).
Table 4.
Global tropism assay resultsa
| Parameter | Value (%) for test |
||||||
|---|---|---|---|---|---|---|---|
| G2P5%cli | G2P5%clo | G2P10%cli | G2P10%clo | PSSM | PSSMp | UDS | |
| Concordance | 81.5 | 80.0 | 80.0 | 78.5 | 73.9 | 76.9 | 81.5 |
| Sensitivity | 47.4 | 57.9 | 47.4 | 68.4 | 57.9 | 84.2 | 83.3 |
| Specificity | 95.7 | 89.1 | 93.5 | 82.6 | 80.4 | 73.9 | 77.8 |
| PPV | 81.8 | 68.8 | 75.0 | 61.9 | 55.0 | 57.1 | 88.2 |
| NPV | 81.5 | 83.7 | 81.1 | 86.4 | 82.2 | 91.9 | 70.0 |
PPV, positive predictive value; NPV, negative predictive value; G2P5%cli, clinical G2P test at 5%; G2P5%clo, clonal G2P test at 5%; G2P10%cli, clinical G2P test at 10%; G2P5%clo, clonal G2P test at 10%; PSSMp, PSSM test using Poveda's score; UDS, ultradeep sequencing with a cutoff of 2%.
Concordance values for standard methods were also analyzed and were found to be highest for G2P5%cli (53/65 samples [81.54%]), followed by G2P5%clo, G2P10%cli (both with 52/65 samples [80.0%]), G2P10%clo (51/65 samples [78.46%]), PSSMp (50/65 samples [76.92%]), and PSSM (48/65 samples [73.85%]). For UDS, the concordance value was similar to those for the standard tests (22/27 samples [81.48%]) (Table 4).
DISCUSSION
In this study, we compared the genotyping methods used most widely to predict successful CCR5 antagonist treatment with an in vivo drug sensitivity assay (MCT).
Using this model, discordance rates of >15% between ESTA results and the MCT have been reported (7). To date, most genotypic studies have attempted to correlate genotypic tests with Trofile or ESTA results (22, 37). In these studies, a strong correlation between this phenotypic assay and genotypic results was obtained. However, the same error rates achieved by this phenotypic method could be extrapolated to genotypic methods due to the virological response observed after the MCT. In contrast, recent studies have validated results from genotypic methods in relation to a virological response (18, 25, 33). These studies have shown that genotypic tests have a similar ability to that of Trofile to predict a virological response to MVC. Nevertheless, it is important that in these studies, MVC was combined with other active drugs that, although score corrected, may have masked the real effect of MVC.
Standard genotypic tropism testing methods have overcome some of the cost- and time-related limitations of phenotypic tropism tests; however, in our study, the standard genotypic method results had discordance rates of approximately 20% with the virological response after MVC monotherapy. These discordance rates between tropism prediction and an in vivo drug sensitivity test indicate the necessity to improve the current prediction algorithms for CCR5 antagonist prescription.
The concordance rates between UDS and the virological response were similar to those found for standard methods. This result was unexpected due to the ability of UDS to detect and amplify minor viral populations. However, to predict the viral tropism of each individual viral variant in UDS, the same algorithms as those used in standard sequencing, for instance, G2P, are used. For this reason, UDS has the same advantages but also the same disadvantages as the standard approaches in terms of the algorithm used. The X4 strains found in patients 12, 29, 32, 64, and 66 are difficult to explain based on the virological responses (Fig. 1). The absence of a virological response in patients 64 and 66 could be explained by possible resistance mutations to MVC (31), despite the fact that the V3 sequences in these samples were analyzed and known mutations were not detected (see Table S3 in the supplemental material). However, the virological response observed in other patients, especially patient 29, points to a failure of the genotypic algorithm predictions as the more plausible explanation. The discordance rates between the virological response and genotypic algorithm prediction could be explained by the use of only the V3 loop sequence in the modeling of these algorithms, because other variable regions of the HIV envelope, and even gp41, can be involved in HIV tropism (3, 14, 16, 27). Furthermore, although the discordance rates were similar for comparing UDS and standard genotypic methods, they did not occur in the same set of patients.
According to previous results (21), a UDS cutoff for X4-tropic strains of 2% fits better with our model. Nevertheless, this observation should be verified with a larger number of patients, covering values from 2 to 6%, to consolidate the threshold of the 2% X4-tropic strains in this scenario (20).
We recently reported that the only variable independently associated with the virological response after MCT was the X4 virus level (28). To increase the sensitivity to X4 variant detection, some studies recommend performing two RT-PCR assays (24). Performing a single PCR for each sample for standard analysis could be a limitation of the present study. Nonetheless, recent studies in which V3 amplicons were obtained by unique RT-PCRs gave concordance rates with the virological response of up to 85% (19, 32, 38). In addition, our results should be evaluated according to the inherent limitations of the MCT: it is a short-term test and has not been validated in a randomized clinical trial. However, the MCT is not a coreceptor-based method but instead gives real-time evidence of drug sensitivity; therefore, the use of the MCT as a reference could be an attractive way to compare genotypic methods. For the purposes of this study, it was not necessary to perform a prospective analysis of patient immunovirological evolution following the MCT, but it is important that after MVC treatment based on the MCT results, these patients had a good immunovirological response with an undetectable viral load and significant CD4 T-cell increases after 48 weeks of combined antiretroviral therapy, as previously reported (6, 9).
In summary, standard V3-based genotypic tropism assays give considerable discordance rates with the virological response after MVC monotherapy. In addition, UDS gives discordance rates similar to those of standard methods and does not improve tropism predictions, although this study included a small number of patients. These results suggest that the algorithms used by both genotypic methods need to be improved.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all of the patients who participated in this study and to Marien Gutiérrez, Francisca Cano, and Magdalena Rodriguez for their clinical support. Patient samples were kindly provided by the HIV BioBank integrated into the Spanish AIDS Research Network (RIS).
A. Gonzalez-Serna performed phenotypic testing and data analysis and wrote the manuscript. R. A. McGovern, P. R. Harrigan, and A. F. Y. Poon performed deep sequencing and data analysis. F. Vidal and S. Ferrando-Martinez participated in the study design. M. A. Abad and M. Genebat performed the standard genotypic tests. M. Leal and E. Ruiz-Mateos were responsible for the study conception and design.
We have no conflicts of interest to declare.
This study was supported by Redes Temáticas de Investigación en SIDA (ISCIII RETIC RD06/0006/0021, RD06/0006/0035, and RD06/0006/1004), by Fondo de Investigación Sanitaria grants PI07/0976 and PS09/01595, and by the Fondos Europeos para el Desarrollo Regional (FEDER). A.G.-S. received a grant from Fundación FISEVI. S.F.-M. and E.R.-M. received grants from the Fondo de Investigaciones Sanitarias (CD10/00382 and CP08/00172, respectively).
Footnotes
Published ahead of print 5 December 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Berger EA, Murphy PM, Farber JM. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657–700 [DOI] [PubMed] [Google Scholar]
- 2. Chueca N, et al. 2009. Improvement in the determination of HIV-1 tropism using the V3 gene sequence and a combination of bioinformatic tools. J. Med. Virol. 81:763–767 [DOI] [PubMed] [Google Scholar]
- 3. Del Prete GQ, et al. 2010. Distinct molecular pathways to X4 tropism for a V3-truncated human immunodeficiency virus type 1 lead to differential coreceptor interactions and sensitivity to a CXCR4 antagonist. J. Virol. 84:8777–8789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Mendoza C, et al. 2008. Performance of a population-based HIV-1 tropism phenotypic assay and correlation with V3 genotypic prediction tools in recent HIV-1 seroconverters. J. Acquir. Immune Defic. Syndr. 48:241–244 [DOI] [PubMed] [Google Scholar]
- 5. Dorr P, et al. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49:4721–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Genebat M, et al. 2011. Maraviroc clinical test (MCT) effectiveness for CCR5 prescription in clinical practice, abstr PE7.9/8. Abstr. 13th Eur. AIDS Conf., Belgrade, Serbia [Google Scholar]
- 7. Genebat M, et al. 2011. Discordance rates between Trofile(®) test and short-term virological response to maraviroc. Antiviral Res. 89:182–185 [DOI] [PubMed] [Google Scholar]
- 8. Genebat M, et al. 2009. Correlation between the Trofile test and virological response to a short-term maraviroc exposure in HIV-infected patients. J. Antimicrob. Chemother. 64:845–859 [DOI] [PubMed] [Google Scholar]
- 9. Genebat M, et al. 2010. Long-term immunovirological effect and tolerability of a maraviroc-containing regimen in routine clinical practice. Curr. HIV Res. 8:482–486 [DOI] [PubMed] [Google Scholar]
- 10. González N, et al. 2010. A sensitive phenotypic assay for the determination of human immunodeficiency virus type 1 tropism. J. Antimicrob. Chemother. 65:2493–2501 [DOI] [PubMed] [Google Scholar]
- 11. Gonzalez-Serna A, et al. 2010. TROCAI (tropism coreceptor assay information): a new phenotypic tropism test and its correlation with Trofile enhanced sensitivity and genotypic approaches. J. Clin. Microbiol. 48:4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harrigan PR, Geretti AM. 2011. Genotypic tropism testing: evidence-based or leap of faith? AIDS 25:257–264 [DOI] [PubMed] [Google Scholar]
- 13. Jensen MA, van't Wout AB. 2003. Predicting HIV-1 coreceptor usage with sequence analysis. AIDS Rev. 5:104–112 [PubMed] [Google Scholar]
- 14. Lin G, et al. 2007. Replication-competent variants of human immunodeficiency virus type 2 lacking the V3 loop exhibit resistance to chemokine receptor antagonists. J. Virol. 81:9956–9966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Low AJ, et al. 2007. Current V3 genotyping algorithms are inadequate for predicting X4 receptor usage in clinical isolates. AIDS 21:F17–F24 [DOI] [PubMed] [Google Scholar]
- 16. Marozsan AJ, et al. 2005. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D). Virology 338:182–199 [DOI] [PubMed] [Google Scholar]
- 17. McGovern RA, Harrigan PR, Swenson LC. 2010. Genotypic inference of HIV-1 tropism using population-based sequencing of V3. J. Vis. Exp. doi:10.3791/2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McGovern RA, et al. 2010. Population-based V3 genotypic tropism assay: a retrospective analysis using screening samples from the A4001029 and MOTIVATE studies. AIDS 24:2517–2525 [DOI] [PubMed] [Google Scholar]
- 19. Obermeier M, et al. 2010. Tropism testing from proviral DNA—analysis of a subgroup from the Berlin maraviroc cohort, abstr 23. Abstr. 8th Eur. HIV Drug Resist. Workshop, Sorrento, Italy [Google Scholar]
- 20. Poveda E, et al. 2010. Genotypic determination of HIV tropism—clinical and methodological recommendations to guide the therapeutic use of CCR5 antagonists. AIDS Rev. 12:135–148 [PubMed] [Google Scholar]
- 21. Poveda E, et al. 2009. Design and validation of new genotypic tools for easy and reliable estimation of HIV tropism before using CCR5 antagonists. J. Antimicrob. Chemother. 63:1006–1010 [DOI] [PubMed] [Google Scholar]
- 22. Prosperi MC, et al. 2010. Comparative determination of HIV-1 co-receptor tropism by enhanced sensitivity Trofile, gp120 V3-loop RNA and DNA genotyping. Retrovirology 7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raymond S, et al. 2010. Development and performance of a new recombinant virus phenotypic entry assay to determine HIV-1 coreceptor usage. J. Clin. Virol. 47:126–130 [DOI] [PubMed] [Google Scholar]
- 24. Raymond S, et al. 2011. Improved V3 genotyping with duplicate PCR amplification for determining HIV-1 tropism. J. Antimicrob. Chemother. 66:1972–1975 [DOI] [PubMed] [Google Scholar]
- 25. Recordon-Pinson P, et al. 2010. Evaluation of the genotypic prediction of HIV-1 coreceptor use versus a phenotypic assay and correlation with the virological response to maraviroc: the ANRS GenoTropism study. Antimicrob. Agents Chemother. 54:3335–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reeves JD, Coakley E, Petropoulos CJ, Whitcomb JM. 2009. An enhanced sensitivity Trofile HIV coreceptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5: a review of analytical and clinical studies. J. Viral Entry 3:94–102 [Google Scholar]
- 27. Rossi F, et al. 2008. The V1–V3 region of a brain-derived HIV-1 envelope glycoprotein determines macrophage tropism, low CD4 dependence, increased fusogenicity and altered sensitivity to entry inhibitors. Retrovirology 5:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ruiz-Mateos E, et al. 2011. Virological response after a short-term CCR5 antagonist exposure in HIV-infected patients: frequency of subjects with virological response and associated factors. Antimicrob. Agents Chemother. 55:4664–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sánchez V, et al. 2011. A highly sensitive and specific model for predicting HIV-1 tropism in treatment-experienced patients combining V3 loop sequences interpretation and clinical parameters. J. Acquir. Immune Defic. Syndr. 56:51–58 [DOI] [PubMed] [Google Scholar]
- 30. Sánchez V, et al. 2010. Performance of genotypic algorithms for predicting HIV-1 tropism measured against the enhanced-sensitivity Trofile coreceptor tropism assay. J. Clin. Microbiol. 4:4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seclén E, et al. 2010. Primary resistance to maraviroc in a large set of R5-V3 viral sequences from HIV-1-infected patients. Antimicrob. Chemother. 65:2502–2504 [DOI] [PubMed] [Google Scholar]
- 32. Sierra S, et al. 2010. Tropism determination and clinical outcome of 61 patients under maraviroc treatment, abstr 20. Abstr. 8th Eur. HIV Drug Resist. Workshop, Sorrento, Italy [Google Scholar]
- 33. Swenson LC, et al. 2011. Deep sequencing to infer HIV-1 co-receptor usage: application to three clinical trials of maraviroc in treatment-experienced patients. J. Infect. Dis. 203:237–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Swenson LC, et al. 2010. Improved detection of CXCR4-using HIV by V3 genotyping: application of population-based and “deep” sequencing to plasma RNA and proviral DNA. J. Acquir. Immune Defic. Syndr. 54:506–510 [DOI] [PubMed] [Google Scholar]
- 35. Tsibris AM, et al. 2009. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One 4:e5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vallejo A, et al. 2006. Immunovirologic characteristics of human immunodeficiency virus-infected patients consisting mainly of injecting drug users on highly active antiretroviral treatment with prolonged virologic failure. Viral Immunol. 19:759–767 [DOI] [PubMed] [Google Scholar]
- 37. Vandekerckhove L, et al. 2010. Comparison of phenotypic and genotypic tropism determination in triple-class-experienced HIV patients eligible for maraviroc treatment. J. Antimicrob. Chemother. 66:265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van Lelyveld S, et al. 2010. Correlation of clinical outcome of maraviroc treatment with different methods to determine HIV tropism: genotypic assays, MT-2 assay and Trofile, abstr. 41. Abstr. 8th Eur. HIV Drug Resist. Workshop, Sorrento, Italy [Google Scholar]
- 39. Whitcomb JM, et al. 2007. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob. Agents Chemother. 51:566–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.