Abstract
A total of 149 porcine Staphylococcus isolates with florfenicol MICs of ≥16 μg/ml were screened for the presence of the multiresistance gene cfr, its location on plasmids, and its genetic environment. In total, 125 isolates carried either cfr (16 isolates), fexA (92 isolates), or both genes (17 isolates). The 33 cfr-carrying staphylococci, which included isolates of the species Staphylococcus cohnii, S. arlettae, and S. saprophyticus in which the cfr gene has not been described before, exhibited a wide variety of SmaI pulsed-field gel electrophoresis patterns. In 18 cases, the cfr gene was located on plasmids. Four different types of cfr-carrying plasmids—pSS-01 (n = 2; 40 kb), pSS-02 (n = 3; 35.4 kb), pSS-03 (n = 10; 7.1 kb), and pBS-01 (n = 3; 16.4 kb)—were differentiated on the basis of their sizes, restriction patterns, and additional resistance genes. Sequence analysis revealed that in plasmid pSS-01, the cfr gene was flanked in the upstream part by a complete aacA-aphD-carrying Tn4001-like transposon and in the downstream part by a complete fexA-carrying transposon Tn558. In plasmid pSS-02, an insertion sequence IS21-558 and the cfr gene were integrated into transposon Tn558 and thereby truncated the tnpA and tnpB genes. The smallest cfr-carrying plasmid pSS-03 carried the macrolide-lincosamide-streptogramin B resistance gene erm(C). Plasmid pBS-01, previously described in Bacillus spp., harbored a Tn917-like transposon, including the macrolide-lincosamide-streptogramin B resistance gene erm(B) in the cfr downstream region. Plasmids, which in part carry additional resistance genes, seem to play an important role in the dissemination of the gene cfr among porcine staphylococci.
INTRODUCTION
Florfenicol is a fluorinated derivative of chloramphenicol that was licensed in China in 1999 for the control of bacterial infections in cattle, swine, and chickens. It acts by reversible binding to the peptidyltransferase center at the 50S ribosomal subunit of 70S ribosomes, thus inhibiting protein synthesis in bacteria (28). The chloramphenicol-associated adverse side effects, in particular the dose-independent irreversible aplastic anemia, have not been observed in animals treated with florfenicol (29). Florfenicol has been approved exclusively for use in veterinary medicine. In veterinary practice in China, florfenicol is used extensively in swine farms to prevent and cure diseases caused by a variety of bacterial pathogens including staphylococci.
In staphylococci, two different florfenicol resistance genes have been identified thus far. The florfenicol-chloramphenicol exporter gene fexA encodes a protein of 475 amino acids (aa) with 14 transmembrane domains which represents a novel type of efflux protein within the major facilitator superfamily (17). The gene fexA, which has been detected mainly in staphylococci of animal origin (1, 8, 11, 16), was first identified on the plasmid pSCFS2 from Staphylococcus lentus and shown to be part of the Tn554-like transposon Tn558 (18). In contrast, the multiresistance gene cfr has been found in staphylococci of both human and veterinary origins (2, 25, 31). The cfr gene codes for a 23S rRNA methyltransferase which modifies the position A2503 in 23S rRNA and thereby confers resistance not only to phenicols but also to lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics (PhLOPSA of phenotype) (19). The expression of this gene renders five important classes of antibiotics ineffective in the treatment of infections in either human or veterinary medicine (10, 19). In this regard, resistance to oxazolidinones is of particular relevance since these antibiotics may represent the last option in the treatment of infections caused by methicillin-resistant Staphylococcus aureus (MRSA) or vancomycin-resistant enterococci in humans (21, 34).
While the cfr gene was found in the chromosomal DNA in some staphylococcal isolates (13, 34), most of the previous reports identified this gene on plasmids in staphylococci. To date, four different cfr-carrying staphylococcal plasmids—pSCFS1 (17.1 kb), pSCFS3 (35.7 kb), pSCFS6 (ca. 43 kb), and pSCFS7 (ca. 45 kb)—have been sequenced completely or in part (16, 31, 32). Among them, plasmid pSCFS7 was detected in a Panton-Valentine leukocidin-positive ST8-MRSA-IVa (USA300) strain of human origin (32). This observation underlines the increasing threat of dissemination of this resistance gene.
In China, the cfr gene has been detected on plasmids pBS-01 and pBS-02 in Bacillus strains of porcine origin (6, 36). Currently, no data about the presence of the genes cfr and fexA in staphylococci of animal origin are available, although florfenicol has been used in animals in China for more than 10 years. We sought here to gain insight into the presence of the multiresistance gene cfr among florfenicol-resistant porcine staphylococci, its location on plasmids, and its genetic environment.
MATERIALS AND METHODS
Bacterial isolates and antimicrobial susceptibility testing.
In 2010, a total of 149 Staphylococcus isolates were identified from nasal swabs taken from 557 swine by growth on brain heart infusion (BHI) agar containing 10 μg of florfenicol/ml. Isolates growing on these selective media have an MIC of florfenicol of at least 16 μg/ml. Although no clinical breakpoints applicable to staphylococci are currently available, isolates with an MIC of ≥16 μg/ml were tentatively considered as florfenicol resistant (16). The nasal swabs were collected from three geographically distinct and unrelated swine farms in the Shandong province, China. All 149 Staphylococcus isolates were subjected to 16S rRNA gene sequencing. For this, a 1,466-bp amplicon obtained with the universal prokaryotic primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGCTACCTTGTTACGACTT-3′) (20) was analyzed. In addition, the Staphylococcus isolates were further confirmed by the ID32 STAPH system (bioMérieux, Craponne, France). The MICs of all cfr-positive original Staphylococcus isolates, the recipient strain S. aureus RN4220, and transformants were determined by broth microdilution according to the recommendations given in documents M100-S21 (4) and M31-A3 (5) of the Clinical and Laboratory Standards Institute (CLSI). The reference strain S. aureus ATCC 29213 served as a quality control.
DNA isolation and florfenicol resistance gene detection.
Whole-cell DNA of the Staphylococcus isolates was isolated using a commercial kit (TianGen, Beijing, China) and according to the manufacturer's instruction. Plasmid DNA was extracted using a Qiagen plasmid extraction Midi kit (Qiagen, Germany) with one modification: after the pelleted bacteria were suspended in buffer P1, lysostaphin was added to a final concentration of 50 μg/ml, and the mixture was incubated for 30 min at 37°C. The presence of the florfenicol resistance genes cfr and fexA was investigated by previously described PCR assays (6).
Molecular typing.
Genetic diversity of the cfr-positive staphylococcal isolates was determined by SmaI pulsed-field gel electrophoresis (PFGE). The Harmony PFGE protocol (26) was used with some modifications as follows: For each isolate, 200 μl of a staphylococcal suspension (optical density at 600 nm of 1.3) in 1× TE buffer (10 mM Tris-HCl [pH 7.5], 10 mM EDTA) containing 4 μl of lysostaphin (1000 μg/ml; Sigma) was mixed with 1.2% low-melting-point agarose (Seakem Gold; Bio-Rad, Hercules, CA) and embedded in a plug mold (Bio-Rad). The solidified plugs were placed in 1× TE buffer containing 10 μg of lysostaphin/ml and incubated for 4 h at 37°C. Subsequently, the plugs were transferred to a buffer consisting of 1 M Tris-HCl (pH 8.0), 0.5 M EDTA (pH 8.0), 10% sodium sarcosyl, and 100 μg of proteinase K (Merck, Darmstadt, Germany) and then incubated for 2 h at 55°C with agitation at 130 rpm. After PFGE, digital images were analyzed using InfoQuestFP software, version 4.5. The similarities between the profiles were calculated using the Dice coefficient, with 0.5% optimization and a maximum position tolerance of 1.0%. The patterns were clustered by using the unweighted pair group method with arithmetic averages (UPGMA). The definition of a PFGE cluster was based on a similarity cutoff value of 80% (23). Different PFGE clusters were indicated by capital letters in alphabetical order.
Analysis of cfr-carrying plasmids.
Purified plasmids extracted from cfr-carrying staphylococcal strains were transformed into the S. aureus recipient strain RN4220 by electrotransformation. The transformants were selected by incubation for 24 h on BHI agar supplemented with 10 μg of florfenicol/ml. Transformants were screened for their plasmid content and their resistance phenotype. Southern blot analysis performed with DNA probes specific for the cfr and fexA genes were used to confirm the location of these genes. The corresponding gene probes, which consisted of PCR-amplified internal segments of the genes cfr and fexA, were nonradioactively labeled by using a DIG High Prime DNA labeling and detection kit (Roche Diagnostics, Mannheim, Germany). If not sequenced completely, the sizes of the cfr-carrying plasmids extracted from transformants were estimated by calculation of the sums of the different fragment sizes obtained after BglII digestion.
Cloning and sequencing experiments.
All PCR amplicons were cloned into the vector pEASY-T1 Simple and transformed into competent Escherichia coli Trans1-T1 cells by CaCl2 transformation according to the manufacturer's instructions (TransGen Biotech, Beijing, China). The partial or complete nucleotide sequences of the cfr-carrying plasmids extracted from the respective transformants were determined by primer walking (Invitrogen, Beijing, China) or a modified random primer sequencing walking strategy (35). The obtained sequences were annotated by using the VectorNTI program (Invitrogen, Carlsbad, CA), and the predicted coding sequences (CDSs) were identified by using the GLIMMER software. The DNA sequences and deduced amino acid sequences were compared to those deposited in GenBank using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST).
Nucleotide sequence accession numbers.
The nucleotide sequences of a 15,702-bp segment of plasmid pSS-01, an 8,580-bp fragment of plasmid pSS-02, and the complete sequence of cfr-carrying plasmid pSS-03 (7,057 bp) have been deposited in GenBank under accession numbers JQ041372 (pSS-01), JF834910 (pSS-02), and JQ219851 (pSS-03), respectively.
RESULTS
Identification of the florfenicol resistance genes cfr and fexA in Staphylococcus isolates.
Of the 149 staphylococci with florfenicol MICs of ≥16 μg/ml that were studied, 125 isolates carried either cfr (n = 16) or fexA (n = 92) or both genes (n = 17). The most prevalent species of cfr-harboring staphylococci were Staphylococcus sciuri (n = 11) and Staphylococcus cohnii (n = 11), followed by Staphylococcus arlettae (n = 6), and Staphylococcus saprophyticus (n = 4). Detailed information of the 149 Staphylococcus isolates from the three swine farms is given in Table 1. The cfr amplicons of all 33 cfr-positive isolates were sequenced and proved to be indistinguishable. Moreover, they exhibited 100% identity to the corresponding cfr sequences of plasmids pSCFS1 (accession no. AJ579365), pSCFS6 (AM408573), pSCFS7 (FR675942), and 99.9% identity to cfr of pSCFS3 (AM086211). The fexA amplicons of 15 of the 17 fexA-positive isolates displayed 100% identity to the corresponding fexA sequence of plasmid pSCFS3 (AM086211), while one nucleotide exchange change (A→G) at position 913 was detected in the amplicons of the remaining two isolates.
Table 1.
Distribution of cfr and fexA among Staphylococcus isolates from three pig farms
| Farm | Staphylococcus species | No. of isolates |
||||
|---|---|---|---|---|---|---|
| Total | fexA | cfr | Both fexA and cfr | Neither fexA nor cfr | ||
| 1 | S. cohnii | 16 | 7 | 4 | 3 | 2 |
| S. sciuri | 4 | 0 | 2 | 1 | 1 | |
| S. saprophyticus | 9 | 4 | 1 | 1 | 3 | |
| S. arlettae | 2 | 2 | 0 | 0 | 0 | |
| S. pseudintermedius | 3 | 3 | 0 | 0 | 0 | |
| S. aureus | 4 | 4 | 0 | 0 | 0 | |
| Subtotal | 38 | 20 | 7 | 5 | 6 | |
| 2 | S. cohnii | 15 | 10 | 1 | 2 | 2 |
| S. sciuri | 3 | 1 | 1 | 0 | 1 | |
| S. saprophyticus | 10 | 7 | 1 | 1 | 1 | |
| S. arlettae | 7 | 3 | 0 | 1 | 3 | |
| S. haemolyticus | 4 | 3 | 0 | 0 | 1 | |
| S. hominis | 1 | 1 | 0 | 0 | 0 | |
| S. aureus | 6 | 5 | 0 | 1 | 0 | |
| Subtotal | 46 | 30 | 3 | 5 | 8 | |
| 3 | S. cohnii | 14 | 10 | 0 | 1 | 3 |
| S. sciuri | 14 | 4 | 4 | 3 | 3 | |
| S. saprophyticus | 4 | 4 | 0 | 0 | 0 | |
| S. arlettae | 18 | 10 | 2 | 3 | 3 | |
| S. epidermidis | 1 | 1 | 0 | 0 | 0 | |
| S. equorum | 1 | 1 | 0 | 0 | 0 | |
| S. chromogenes | 1 | 1 | 0 | 0 | 0 | |
| S. hyicus | 8 | 8 | 0 | 0 | 0 | |
| S. xylocus | 4 | 3 | 0 | 0 | 1 | |
| Subtotal | 65 | 42 | 6 | 7 | 10 | |
| Total | 149 | 92 | 16 | 17 | 24 | |
Antimicrobial resistance and plasmid profiles of cfr-positive Staphylococcus isolates.
All 33 cfr-carrying isolates exhibited resistance to chloramphenicol, clindamycin, and erythromycin and showed elevated MICs to florfenicol and tiamulin. The MIC values for florfenicol of these isolates varied from 64 to ≥128 μg/ml, and those for chloramphenicol varied from 16 to ≥128 μg/ml. Nine (27.3%) and eleven (33.3%) of these isolates exhibited resistance to ciprofloxacin and gentamicin, respectively. The MICs for linezolid were 4 μg/ml in 23 isolates and 8 μg/ml in the remaining 10 isolates.
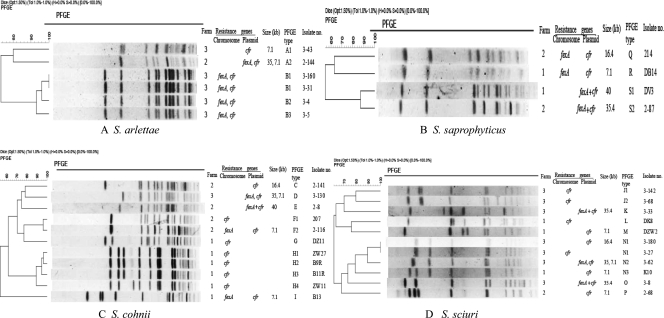
Southern blot hybridization indicated that 15 isolates harbored the cfr gene in their chromosomal DNA, while plasmid-borne cfr genes were present in 18 isolates. The cfr-carrying plasmids were transformed into S. aureus RN4220 and subjected to restriction analysis. Moreover, the transformants were investigated for their resistance phenotypes. On the basis of plasmid sizes, BglII restriction patterns and antimicrobial resistance patterns, four different types of cfr-carrying plasmids were differentiated. These included the ∼40-kb plasmid pSS-01 present in S. cohnii (n = 1) and S. saprophyticus (n = 1), the ∼35.4-kb plasmid pSS-02 present in S. saprophyticus (n = 1) and S. sciuri (n = 2), the 16.4-kb plasmid pBS-01 present in single isolates of S. cohnii, S. saprophyticus, and S. sciuri, and the 7.1-kb plasmid pSS-03 present in S. arlettae (n = 2), S. cohnii (n = 3), S. saprophyticus (n = 1), and S. sciuri (n = 4). The three 16.4-kb plasmids identified in the present study (Fig. 1B to D) revealed the same size, indistinguishable restriction patterns, and resistance phenotype as the previously described plasmid pBS-01 of Bacillus origin (6). Thus, the designation pBS-01 was also used for this plasmid type in the present study.
Fig 1.
Dendrogram and associated SmaI PFGE patterns of 32 cfr-carrying Staphylococcus isolates and the localization of cfr and fexA gene. (A) S. arlettae; (B) S. saprophyticus; (C) S. cohnii; (D) S. sciuri. The single S. aureus strain with chromosomally located genes cfr and fexA was not included.
Hybridization experiments confirmed that the cfr-carrying plasmids pSS-01 and pSS-02 also harbored the fexA gene. In addition, the fexA gene was detected on a ∼35.0-kb plasmid in three isolates, which also had the 7.1-kb cfr-carrying plasmid pSS-03. MIC testing revealed that besides the cfr-mediated PhLOPSA phenotype, transformants carrying plasmid pSS-01 exhibited high MIC values for gentamicin (128 μg/ml), whereas transformants carrying plasmids pSS-03 showed high MIC values for erythromycin (>64 μg/ml). These observations suggested that the corresponding plasmid types carried genes for resistance to gentamicin and macrolides, respectively. To gain insight into the genetic environment of the cfr gene in the three newly identified plasmid types, the 7.1-kb plasmid pSS-03 from S. arlettae was sequenced completely, whereas 15.7- and 8.5-kb segments encompassing the cfr gene of the ∼40-kb plasmid pSS-01 from S. cohnii and the ∼35.4-kb plasmid pSS-02 from S. saprophyticus, respectively, were sequenced. The corresponding maps are shown in Fig. 2.
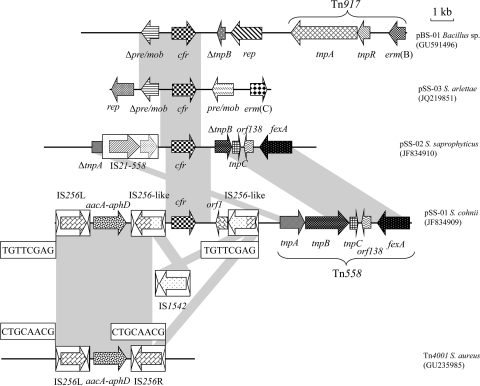
Fig 2.
Schematic presentation of the genetic environment of the cfr gene in plasmids pSS-01, pSS-02, pSS-03, and pBS-01. The arrows indicate the positions and directions of the transcription of the genes. The regions of >96% homology are marked using gray shading. The direct target site duplication is boxed. Δ indicates a truncated gene. A distance scale in kilobases is displayed in the upper right corner.
Analysis of the genetic environment of cfr in plasmid pSS-01.
In the 15.7-kb cfr-containing fragment of plasmid pSS-01, the cfr gene was bracketed by two transposons: in the upstream part by a complete aacA-aphD-carrying Tn4001-like transposon and in the downstream part by a complete fexA-carrying Tn558 transposon. The composite transposon Tn4001, originally detected in S. aureus (22), has a central region carrying the bifunctional aminoglycoside resistance gene aacA-aphD. Two copies of 1,324-bp IS256 elements located in opposite orientations represent the termini of Tn4001. Each IS256 element had 26-bp terminal imperfect inverted repeats (accession no. GU235985, Fig. 2). In addition, IS256 generates 8-bp direct repeats at the integration site (33). In the present study, the novel Tn4001-like transposon found on plasmid pSS-01 showed an identical sequence compared to the Tn4001 from S. aureus (accession no. GU235985), except for the right-hand terminal IS256R-like element. This IS256R-like element harbored a 1,173-bp transposase gene that showed only 88.3% (1,036/1,173 bp) nucleotide sequence identity to the transposase gene of IS256R of Tn4001 (3, 22). It was most closely related to IS1542 from Enterococcus faecium (7) with an overall nucleotide sequence identity of 91.5% (1,212/1,324 bp) and 93.6% aa sequence identity (365/390 aa) of the transposase protein. Downstream of the cfr gene, an orf1 coding for a protein of 170 aa was detected. The ORF1 protein showed 46.6% aa identity to an internal segment of a transcriptional regulator of the mercury resistance (mer) operon of Paenibacillus vortex V453 (accession no. EFU39289). The orf1 was followed by a second IS256R-like element in which the noncoding sequence downstream of the transposase gene, including the terminal inverted repeat, was deleted. The transposase protein also showed 93.6% aa sequence identity (365/390 aa) to the transposase protein of IS1542. The observation that the typical 8-bp repeats (5′-TGTTCGAG-3′) were found immediately upstream of the intact left-hand IS256 and downstream of the second copy of the IS256R-like element (Fig. 2) suggested that the cfr gene region including an IS1542-like insertion sequence were integrated into the original IS256R element of Tn4001, most likely by homologous recombination. Downstream of this second IS256-like element, a complete Tn558 transposon including the fexA gene was present (Fig. 2).
Analysis of the genetic environment of cfr in plasmid pSS-02.
The 8.5-kb cfr-carrying segment of the ca. 35.4-kb plasmid pSS-02 resembled closely (99.8% identity; 8565/8580 bp) the corresponding region of the 35.7-kb plasmid pSCFS3 (16). In both cases, a segment containing an IS21-558 insertion sequence and the cfr gene was integrated into a Tn558 element and thereby truncated the transposase genes tnpA and tnpB. The observation that plasmid pSS-02 had a similar size, exhibited a closely related BglII fragment pattern (data not shown), carried the same in part truncated Tn558 element with an IS21-558/cfr integrate, and did not harbor additional resistance genes suggested that plasmid pSS-02 might be similar to the plasmid pSCFS3.
Analysis of the genetic environment of cfr in plasmid pSS-03.
The smallest plasmid pSS-03 was found to be present in 10 isolates (four S. sciuri, three S. cohnii, two S. arlettae, and one S. saprophyticus) from the three farms included in the present study. Analysis of the pSS-03 sequence revealed the presence of five reading frames for proteins of >100 aa (Fig. 2). The rep gene encoded a 327-aa plasmid replication protein which showed 98.5% identity to the 326-aa RepU replication protein of the S. saprophyticus plasmid pSES22 (9) (accession no. CAJ43791). Moreover, a macrolide-lincosamide-streptogramin B (MLSB) resistance gene was detected, which encoded a 244-aa Erm(C) rRNA methylase that differed only by three aa exchanges (N74D, G130E, and S210N) from the Erm(C) protein from plasmid pSES22. It should be noted that the 22-bp duplication detected in the erm(C) translational attenuator of plasmid pSES22 (9) was absent in plasmid pSS-03. Nucleotide sequence comparisons suggested that a 3,096-bp segment from a pSCFS1-related plasmid, which comprised the cfr gene and a pre/mob gene, was integrated into a pSES22-like plasmid. This integration process resulted in the deletion of a 508-bp segment of plasmid pSES22 and the truncation of the original pre/mob gene of pSES22. As such, the cfr gene of plasmid pSS-03 was flanked in the upstream part by a truncated copy of a pre/mob gene whose deduced amino acid sequences showed 99.6% identity to the C-terminal 271 aa of the Pre/Mob protein from plasmid pSES22 (accession no. CAJ43794). In the cfr downstream part, a reading frame for a 376-aa Pre/Mob protein was detected which exhibited 95.2% identity to the corresponding part of the 392-aa Pre/Mob protein of plasmid pSCFS1 (accession no. CAE18143).
Clonal analysis of cfr-positive staphylococci.
PFGE analysis of the 32 cfr-positive staphylococci (11 S. sciuri, 11 S. cohnii, 6 S. arlettae, and 4 S. saprophyticus) revealed 16 major SmaI patterns A to P (Fig. 1). A similarity cutoff ≥80% was used to assign isolates to the same clonal group. The 40-kb plasmid pSS-01 was present in two isolates (S. saprophyticus and S. cohnii) obtained from farm 1. The 35.4-kb plasmid pSS-02 was present in three isolates (one S. saprophyticus from farm 2 and two S. sciuri from farm 3) with the two S. sciuri strains exhibiting different PFGE patterns (K and O; Fig. 1). The cfr- and erm(B)-carrying 16.4-kb plasmid pBS-01 was present in three different isolates, one S. saprophyticus and one S. cohnii from farm 2 and one S. sciuri from farm 3. The 7.1-kb plasmid pSS-03, which also carried the MLSB resistance gene erm(C), was present in 10 isolates, including one S. saprophyticus, two S. arlettae (with related PFGE patterns A1 and A2) separately obtained from farms 2 and 3, three S. cohnii with different PFGE patterns (D, F2 and I) obtained from all three farms, and four S. sciuri with three different major PFGE patterns M, P, and N. The two strains showing patterns N2 and N3 originated from farms 3 and 1, respectively.
DISCUSSION
Since no information has been available about the distribution of the multiresistance gene cfr among staphylococci in China, we screened 149 porcine Staphylococcus isolates for the presence of this gene. The observation that we detected the gene cfr in 33/149 (22.1%) florfenicol-resistant staphylococcal isolates from pigs suggested that this multiresistance gene might be widely disseminated among staphylococci of porcine origin in the Shangdong Province, China. To the best of our knowledge, this is also the first time that the cfr was found in isolates of S. cohnii, S. arlettae, and S. saprophyticus. However, among the isolates included in the present study, the predominant florfenicol resistance gene was fexA, which was detected among 61.7% (92/149) of the florfenicol-resistant staphylococcal isolates. Interestingly, a significant fraction of florfenicol-resistant isolates (24/149 isolates) did not harbor either cfr or fexA gene (Table 1). This observation indicated that other resistance genes might be involved in the florfenicol resistance of staphylococci, which needs further investigation.
In the present study, four different types of cfr-carrying plasmids (pSS-01, pSS-02, pSS-03, and pBS-01) could be differentiated on the basis of their sizes, restriction patterns, and additional resistance genes. The occurrence of cfr-carrying plasmids of type pBS-01 in both Staphylococcus and Bacillus isolates from swine (6) suggests that this plasmid has the ability to spread among different genera of Gram-positive bacteria. Similar observations have been made with other small antimicrobial resistance plasmids, such as the aadD-carrying kanamycin/neomycin resistance plasmid pUB110 (24) or the tet(L)-carrying tetracycline plasmid pBC16 (12, 27) which proved to be able to replicate and express their resistance genes in Staphylococcus and Bacillus hosts.
Plasmid pSS-02 was identified in the present study in isolates of S. sciuri and S. saprophyticus. This plasmid was found to be similar to plasmid pSCFS3 in its size, BglII restriction pattern and the absence of other resistance genes. In addition, plasmids indistinguishable from pSCFS3 have been isolated previously from bovine and porcine Staphylococcus lentus and from porcine S. aureus (including one MRSA ST398) in Germany (14, 16). The observation that similar plasmids were found in S. sciuri and S. saprophyticus from swine in China suggests a wide dissemination of these closely related plasmids.
With a size of 7,057 bp, plasmid pSS-03 is the smallest cfr-carrying plasmid known to date. In addition, the macrolide-lincosamide-streptogramin B (MLSB) resistance gene erm(C) was also present on this plasmid. The coexistence of cfr and erm genes on the same plasmid have been previously reported in both animal and human isolates. The MLSB resistance gene erm (33), which is an in vivo-derived “in-frame” recombination product of the MLSB resistance genes erm(A) and erm(C), was located together with the cfr gene on plasmid pSCFS1 (15, 30). The 16.5-kb plasmid pBS-01, which was identified in three staphylococcal isolates of the present study but had also been described before in one Bacillus strain, carried the cfr gene along with transposon Tn917 harboring the MLSB resistance gene erm(B) (6). Moreover, the erm(B) gene was found in close proximity of the cfr gene in the chromosomal DNA of MRSA strain CM05 (34).
The cfr-positive staphylococci identified in the present study were compared for their macrorestriction patterns and their cfr plasmid carriage to gain insight into the dissemination of the different cfr-carrying plasmids among unrelated strains of the same or different staphylococcal species obtained from the different swine farms. A wide variety of SmaI PFGE patterns was seen among the cfr-positive staphylococci. The same plasmid type was detected not only in members of different staphylococcal species but also in strains of the same species, which differed in their PFGE patterns. This observation strongly suggested that plasmids played an important role in the dissemination of the gene cfr among staphylococci from the same and different farms. The smallest cfr-carrying plasmid pSS-03 of 7.1 kb was most widespread and was found in four different staphylococcal species obtained from all three farms. The PFGE results indicated that horizontal transmission of cfr-carrying plasmids as well as the clonal spread of cfr-positive isolates were the two main ways leading to the dissemination of this multiresistance gene among the same and different species of porcine staphylococci.
In conclusion, the results of the present study confirmed not only the wide dissemination of the multidrug resistance gene cfr in five different species of staphylococci from three unrelated swine farms but also showed that the cfr gene is present on novel plasmid types which carry additional resistance genes, such as aacA-aphD or erm(C), and thus allow for persistence and coselection of the cfr gene also under selective pressure imposed by the use of aminoglycosides, such as gentamicin, tobramycin, or kanamycin, or macrolides. The findings presented here underline the role of plasmids in the dissemination of the gene cfr across species and genus boundaries and the risk of acquisition of expanded multidrug resistance by the uptake of a single cfr-carrying plasmid.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (grants 31001087 and U1031004), the Program for Chang Jiang Scholars, and the Innovative Research Team at the University of China (IRT0866).
Footnotes
Published ahead of print 19 December 2011
REFERENCES
- 1. Argudin MA, et al. 2011. Virulence and resistance determinants of German Staphylococcus aureus ST398 isolates from nonhuman sources. Appl. Environ. Microbiol. 77:3052–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bonilla H, et al. 2010. Multicity outbreak of linezolid-resistant Staphylococcus epidermidis associated with clonal spread of a cfr-containing strain. Clin. Infect. Dis. 51:796–800 [DOI] [PubMed] [Google Scholar]
- 3. Chen L, et al. 2010. Identification of a novel transposon (Tn6072) and a truncated staphylococcal cassette chromosome mec element in methicillin-resistant Staphylococcus aureus ST239. Antimicrob. Agents Chemother. 54:3347–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clinical Laboratory Standards Institute 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard M31-A3, 3rd ed CLSI, Wayne, PA [Google Scholar]
- 5. Clinical Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement M100–S21. CLSI, Wayne, PA [Google Scholar]
- 6. Dai L, et al. 2010. First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob. Agents Chemother. 54:3953–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Darini AL, Palepou MF, Woodford N. 1999. Nucleotide sequence of IS1542, an insertion sequence identified within VanA glycopeptide resistance elements of enterococci. FEMS Microbiol. Lett. 173:341–346 [DOI] [PubMed] [Google Scholar]
- 8. Feßler A, et al. 2010. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65:619–625 [DOI] [PubMed] [Google Scholar]
- 9. Hauschild T, Lüthje P, Schwarz S. 2006. Characterization of a novel type of MLSB resistance plasmid from Staphylococcus saprophyticus carrying a constitutively expressed erm(C) gene. Vet. Microbiol. 115:258–263 [DOI] [PubMed] [Google Scholar]
- 10. Jensen SO, Lyon BR. 2009. Genetics of antimicrobial resistance in Staphylococcus aureus. Future Microbiol. 4:565–582 [DOI] [PubMed] [Google Scholar]
- 11. Kadlec K, et al. 2009. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J. Antimicrob. Chemother. 64:1156–1164 [DOI] [PubMed] [Google Scholar]
- 12. Kadlec K, Schwarz S. 2009. Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet(L). Antimicrob. Agents Chemother. 53:776–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kehrenberg C, Aarestrup FM, Schwarz S. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51:483–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kehrenberg C, Cuny C, Strommenger B, Schwarz S, Witte W. 2009. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 53:779–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kehrenberg C, Ojo KK, Schwarz S. 2004. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J. Antimicrob. Chemother. 54:936–939 [DOI] [PubMed] [Google Scholar]
- 16. Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kehrenberg C, Schwarz S. 2004. fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicol. Antimicrob. Agents Chemother. 48:615–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kehrenberg C, Schwarz S. 2005. Florfenicol-chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob. Agents Chemother. 49:813–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol, and clindamycin resistance: methylation of 23S rRNA at A2503. Mol. Microbiol. 57:1064–1073 [DOI] [PubMed] [Google Scholar]
- 20. Kim TW, et al. 2010. Identification and distribution of Bacillus species in Doenjang by whole-cell protein patterns and 16S rRNA gene sequence analysis. J. Microbiol. Biotechnol. 20:1210–1214 [DOI] [PubMed] [Google Scholar]
- 21. Livermore DM. 2003. Linezolid in vitro: mechanism and antibacterial spectrum. J. Antimicrob. Chemother. 51(Suppl. 2):ii9–ii16 [DOI] [PubMed] [Google Scholar]
- 22. Lyon BR, May JW, Skurray RA. 1984. Tn4001: a gentamicin and kanamycin resistance transposon in Staphylococcus aureus. Mol. Gen. Genet. 193:554–556 [DOI] [PubMed] [Google Scholar]
- 23. McDougal LK, et al. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McKenzie T, Hoshino T, Tanaka T, Sueoka N. 1986. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid 15:93–103 [DOI] [PubMed] [Google Scholar]
- 25. Morales G, et al. 2010. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 50:821–825 [DOI] [PubMed] [Google Scholar]
- 26. Murchan S, et al. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palva A, Vigren G, Simonen M, Rintala H, Laamanen P. 1990. Nucleotide sequence of the tetracycline resistance gene of pBC16 from Bacillus cereus. Nucleic. Acids Res. 18:1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schlunzen F, et al. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814–821 [DOI] [PubMed] [Google Scholar]
- 29. Schwarz S, Chaslus-Dancla E. 2001. Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet. Res. 32:201–225 [DOI] [PubMed] [Google Scholar]
- 30. Schwarz S, Kehrenberg C, Ojo KK. 2002. Staphylococcus sciuri gene erm(33), encoding inducible resistance to macrolides, lincosamides, and streptogramin B antibiotics, is a product of recombination between erm(C) and erm(A). Antimicrob. Agents Chemother. 46:3621–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shore AC, et al. 2010. Identification and characterization of the multidrug resistance gene cfr in a Panton-Valentine leukocidin-positive sequence type 8 methicillin-resistant Staphylococcus aureus IVa (USA300) isolate. Antimicrob. Agents Chemother. 54:4978–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic. Acids Res. 34:D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Toh SM, et al. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang K, McClure JA, Elsayed S, Conly JM. 2009. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang WJ, et al. 2011. The new genetic environment of cfr on plasmid pBS-02 in a Bacillus strain. J. Antimicrob. Chemother. 66:1174–1175 [DOI] [PubMed] [Google Scholar]