Abstract
The antiviral profile of BMS-790052, a potent hepatitis C virus (HCV) replication complex inhibitor targeting nonstructural protein NS5A, is well characterized for HCV genotype-1. Here, we report that BMS-790052 inhibits hybrid replicons containing HCV genotype-4 NS5A genes with 50% effective concentrations (EC50s) ranging from 7 to 13 pM. NS5A residue 30 was an important site for BMS-790052-selected resistance in the hybrid replicons. Our results support the potential of BMS-790052 as a valuable component of combination therapy for HCV genotype-4 chronic infection.
TEXT
Hepatitis C virus (HCV) is classified into at least six different genotypes and multiple subtypes based on phylogeny (10). HCV genotype-4 (HCV4) is the predominant genotype in the Middle East and Africa, and HCV4 accounts for ∼20% of all HCV chronic infections worldwide (6). Only 43% to 70% of patients with HCV4 chronic infections achieve a sustained viral response when treated with the combination of pegylated alpha interferon and ribavirin (6). BMS-790052 is a potent inhibitor of HCV RNA replication that targets the essential replication factor NS5A (5). Once-daily treatment with BMS-790052 generated robust and rapid viral load declines in subjects with chronic HCV genotype-1 (HCV1) infections, although viral breakthrough associated with amino acid substitutions in the N-terminal region of NS5A was also observed (4, 5, 8). In HCV1a, NS5A residues 28 (M28T), 30 (Q30E/R/H), 31 (L31M/V), and 93 (Y93H) were the major sites of drug-induced amino acid substitutions both in vivo and in vitro, while residues 31 (L31V) and 93 (Y93H) were the major residues associated with resistance in HCV1b (3–5).
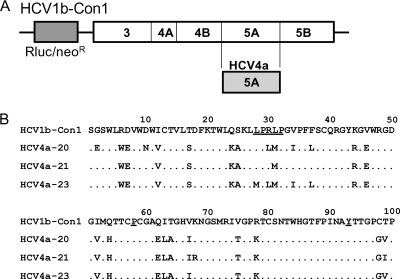
To examine the effectiveness of BMS-790052 treatment with respect to the most prevalent HCV4 subtype, HCV4a, NS5A sequences in a Con1 subgenomic replicon were replaced by standard cloning techniques with NS5A cDNA sequences that had been isolated by reverse transcription-PCR (RT-PCR) from three HCV4a-positive sera (Cliniqa Corporation, San Marcos, CA; Fig. 1). In transient replicon assays (3), the hybrid Con1/4a replicons, labeled HCV4a-20, HCV4a-21, and HCV4a-23, replicated at only 7% to 10% of the level of the parental Con1 replicon (data not shown). The activity of BMS-790052 against each of the hybrid replicons was therefore assessed with neomycin-resistant replicon cell lines that were established and tested as previously described (7, 9). BMS-790052 was highly potent against all three hybrid replicons (50% effective concentrations [EC50s] = 7 to 13 pM; Table 1). To identify BMS-790052 resistance mutations, the HCV4a replicon cell lines were treated with BMS-790052 at concentrations ranging from 0.1 to 10 nM (Table 1). After approximately 4 weeks, replicon cells with resistance to BMS-790052 were obtained. NS5A cDNA was isolated by RT-PCR from resistant and dimethyl sulfoxide (DMSO)-treated control cells as previously described (3). Amino acid substitutions predicted from sequence analysis of the population of cDNA from the pooled cells under each set of selection conditions are summarized in Table 1. In HCV4a-20 cells treated with either 0.2 or 1 nM BMS-790052, a consensus NS5A L30H amino acid substitution was the only noticeable change. L30H was also the only substitution identified in HCV4a-23 replicon cells that were selected with 10 nM BMS-790052. Selection of HCV4a-23 replicon cells with 0.1 or 1 nM BMS-790052 yielded a mixture of L30H and Y93H amino acid substitutions (Table 1). The parental HCV4a-21 replicon encodes an arginine at NS5A position 30 rather than a leucine (Fig. 1). A mixture of some combination of R30G, R30H, and R30S was observed in cDNA isolated from the HCV4a-21 replicon cells that were selected with 0.1 or 1.0 nM BMS-790052. In cDNA from cells selected at 10 nM, R30G was the only evident substitution, suggesting that the R30H and R30S variants were suppressed at this higher inhibitor concentration. To more clearly define the mixtures of replicon variants present in bulk cDNA from the BMS-790052-treated HCV4a-21 and HCV4a-23 cells selected at 0.1 nM and 1 nM, sequence was obtained from NS5A cDNA clones as previously described (3) (Table 2). This analysis largely confirmed the population sequencing results and also identified additional, less prevalent mutations at codons 30 (L30R/V), 31 (M31I/V), 32, (P32L), 58 (P58H), and 93 (Y93C/R).
Fig 1.
(A) Schematic diagram of the bicistronic Con1 replicon with Renilla luciferase or neomycin resistance genes. Hybrid replicons were constructed by replacing the entire NS5A coding region with sequences derived from the HCV4a-20, -21, and -23 clinical isolates. Each of the replicons contained an engineered S2204I adaptive mutation in NS5A. (B) Alignment of the N-terminal 100 amino acids of NS5A deduced from cDNA isolated from neomycin-resistant replicon cell lines. Amino acid identities are indicated with dots. Residues identified in this study as potentially important sites of resistance development in HCV4a are underlined.
Table 1.
BMS-790052 potency on HCV4a replicon cells
| Replicon | EC50 (nM) for parental cellsa | BMS-790052 selection concn (nM) | EC50 (nM) for selected cellsa | NS5A amino acid substitution(s)c |
|---|---|---|---|---|
| HCV4a-20 | 0.010 ± 0.002 | 0b | 0.01 ± 0.004 | None |
| 0.2 | 0.64 ± 0.35 | L30H (100) | ||
| 1.0 | 1.32 ± 0.78 | L30H (100) | ||
| HCV4a-21 | 0.007 ± 0.0002 | 0b | 0.01 ± 0.002 | None |
| 0.1 | 2.55 ± 0.07 | R30G/H/S (20/30/50) | ||
| 1.0 | 1.00 ± 0.06 | R30G/H/S (5/40/50) | ||
| 10 | 21.1 ± 0.25 | R30G (100) | ||
| HCV4a-23 | 0.013 ± 0.0001 | 0b | 0.02 ± 0.001 | None |
| 0.1 | 4.4 ± 1.5 | L30H (40), Y93H (20) | ||
| 1.0 | 14.2 ± 0.4 | L30H (80), Y93H (10) | ||
| 10 | 16.0 ± 3.0 | L30H (100) |
Mean ± standard deviation (n ≥ 3).
DMSO-treated control cells.
Substitutions were deduced from bulk NS5A sequence traces; numbers in parentheses represent the estimated percentages of the indicated mutation.
Table 2.
Number of cDNA clones encoding the indicated amino acids in NS5Aa
| Replicon | Amino acid substitution | No. of cDNA clones at indicated BMS-790052 selection concn (nM) |
|
|---|---|---|---|
| 0.1 | 1 | ||
| HCV4a-21 | R30G | 4 | 4 |
| R30H | 5 | 7 | |
| R30S | 10 | 12 | |
| R30H/Y93C | 1 | ||
| HCV4a-23 | L30H | 7 | 16 |
| L30R | 2 | ||
| L30V | 1 | ||
| M31I | 1 | ||
| M31V | 1 | ||
| P32L | 1 | ||
| P58H | 1 | ||
| Y93H | 2 | 3 | |
| Y93R | 1 | ||
| L30I/Y93R | 1 | ||
cDNA clones were isolated from HCV4a replicon cells selected for resistance to BMS-790052 at the indicated concentrations.
In order to assess the impact on BMS-790052 potency of the R30G, R30H, and R30S amino acid substitutions in the HCV4a-21 background and the L30H, Y93H and Y93R substitutions in the HCV4a-23 replicon background, mutant replicons encoding these individual amino acid replacements were constructed (3) and used to select G418-resistant replicon cell lines. While each of these substitutions reduced the potency of BMS-790052, the greatest effects were seen with R30G in the HCV4a-21 replicon and L30H in the HCV4a-23 replicon (Table 3). These results are consistent with R30G and L30H being the predominant variants selected with 10 nM BMS-790052 (Table 1).
Table 3.
Effect of NS5A amino acid substitutions on BMS-790052 potency
| Replicon | Variant | BMS-790052 EC50 (nM)a | Fold resistance |
|---|---|---|---|
| HCV4a-21 | Wild type | 0.006 ± 0.0002 | 1 |
| R30G | 12.1 ± 2.0 | 2,017 | |
| R30H | 0.93 ± 0.10 | 155 | |
| R30S | 1.0 ± 0.053 | 167 | |
| R30H+Y93C | No replicationb | ||
| HCV4a-23 | Wild type | 0.013 ± 0.0001 | 1 |
| L30H | 15.8 ± 7.2 | 1,215 | |
| Y93H | 2.2 ± 0.1 | 169 | |
| Y93R | 5.9 ± 0.4 | 454 | |
| M28L+L30H | 0.83 ± 0.17 | 64 | |
| L30I+Y93R | 118.3 ± 42.8 | 9,100 |
Means ± standard deviations of the results for replicon cells (n ≥ 3).
No replicon cell line was established due to poor replication of mutant replicon.
In HCV1 replicons, linked resistance mutations generally confer high levels of resistance to BMS-790052 (3). Mutations that yielded linked amino acid substitutions were observed in only two cDNA clones from the BMS-790052-treated HCV4a replicon cells (R30H plus Y93C in HCV4a-21 and L30I plus Y93R in HCV4a-23; Table 2). An HCV4a-23 replicon with linked L30I and Y93R substitutions was highly resistant to BMS-790052 (EC50 = 118 nM; Table 3), but an HCV4a-21 replicon with linked R30H and Y93C substitutions did not replicate at levels sufficient to select a replicon cell line (Table 3). The less prevalent mutations identified in NS5A cDNA clones (Table 2) have not yet been characterized.
Of note, BMS-790052 inhibited an HCV4a-21 R30H mutant replicon with an EC50 of 0.93 nM (Table 3). In contrast, the EC50 of BMS-790052 on the comparable HCV4a-23 L30H mutant replicon was 15.8 nM (Table 3). These results suggested that one or more differences in the genetic background between the HCV4a-21 and HCV4a-23 replicons modulated the impact of a histidine at NS5A position 30. Among the differences within the NS5A N-terminal region, HCV4a-21 has a leucine, and HCV4a-23 has a methionine, at position 28 (Fig. 1B). Residue 28 is an important site for resistance development in the HCV1a replicon (3). To determine if position 28 contributed to the differential effects of histidine at position 30 between the HCV4a-21 and HCV4a-23 replicons, a mutant HCV4a-23 replicon with linked M28L and L30H substitutions was constructed. The EC50 of BMS-790052 on this replicon was 0.83 nM, very similar to the EC50 of BMS-790052 on the HCV4a-21 R30H replicon (0.93 nM; Table 3). These results indicate that methionine at NS5A position 28, while conferring little resistance on its own, acts in concert with histidine at position 30 to increase resistance to BMS-790052. NS5A position 28 is therefore another potential site for resistance development in HCV4a strains.
In this study, we identified NS5A positions 28, 30, 32, 58, and 93 as possible sites for BMS-790052 resistance development in HCV4a strains. Overall, these sites are very similar to those previously identified in studies with HCV1 and HCV2 replicons (2, 3), suggesting that the location of the BMS-790052 binding site is conserved among diverse HCV strains. The naturally occurring variability at these positions in HCV4 NS5A sequences in the European HCV database (1) is summarized in Table 4. Importantly, none of the database sequences contained a glycine or histidine at NS5A position 30, the predominant amino acid replacements associated with BMS-790052 resistance in this study. However, 10% of the sequences had a serine at position 30, and 5% had a histidine at position 93. BMS-790052 inhibited hybrid replicons with these amino acid substitutions (R30S and Y93H) with EC50s of 1.0 and 2.2 nM, respectively (Table 3). Based on a mean trough concentration of ∼260 nM, as achieved with HCV1-infected subjects who received 60 mg of BMS-790052 once daily for 14 days (8), HCV4a variants with these preexisting mutations would likely be effectively suppressed at this inhibitor dose. Since HCV4a variants with greater resistance to BMS-790052 may be encountered in individuals with chronic HCV4a infections, BMS-790052 would probably be most effective in combination therapy with other antivirals.
Table 4.
Variability at NS5A positions important for BMS-790052 resistancea
| Amino acid (no. of corresponding sequences in database) at NS5A position: | ||||||
|---|---|---|---|---|---|---|
| 28 | 29 | 30 | 31 | 32 | 58 | 93 |
| L (33) | P (40) | R (20) | M (37) | P (40) | P (36) | Y (35) |
| M (5) | L (12) | L (3) | T (4) | H (2) | ||
| I (1) | S (4) | S (2) | ||||
| V (1) | Q (2) | T (1) | ||||
| A (1) | ||||||
| T (1) | ||||||
Numbers of HCV4 sequences in the European HCV database with the indicated amino acid are shown in parentheses (n = 40).
In summary, we have demonstrated that BMS-790052 is a highly potent inhibitor of hybrid replicons with NS5A sequences derived from three HCV4a strains. Resistance selection studies with these hybrid replicons suggest that NS5A residue 30 would be a major site for BMS-790052 resistance development in HCV4a strains in vivo. Our results support the continued exploration of BMS-790052 as a component of combination therapy for treatment of chronic infection with HCV4.
Nucleotide sequence accession numbers.
The HCV4a NS5A nucleotide sequences are available in GenBank under accession numbers JQ347513 to JQ347515.
Footnotes
Published ahead of print 27 December 2011
REFERENCES
- 1. Combet C, et al. 2007. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res. 35:D363–D366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fridell RA, et al. 2011. Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052. J. Virol. 85:7312–7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fridell RA, Qiu D, Wang C, Valera L, Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob. Agents Chemother. 54:3641–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fridell RA, et al. 2011. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology 54:1924–1935 [DOI] [PubMed] [Google Scholar]
- 5. Gao M, et al. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khattab MA, et al. 2011. Management of hepatitis C virus genotype 4: recommendations of an international expert panel. J. Hepatol. 54:1250–1262 [DOI] [PubMed] [Google Scholar]
- 7. Lemm JA, et al. 2010. Identification of hepatitis C virus NS5A inhibitors. J. Virol. 84:482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nettles RE, et al. 2011. Multiple ascending dose study of BMS-790052, a nonstructural protein 5A replication complex inhibitor, in patients infected with hepatitis C virus genotype 1. Hepatology 54:1956–1965 [DOI] [PubMed] [Google Scholar]
- 9. O'Boyle DR, et al. 2005. Development of a cell-based high-throughput specificity screen using a hepatitis C virus-bovine viral diarrhea virus dual replicon assay. Antimicrob. Agents Chemother. 49:1346–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173–3188 [DOI] [PubMed] [Google Scholar]