Abstract
Porphyromonas gingivalis is a bacterial pathogen associated with chronic periodontitis that results in destruction of the tooth's supporting tissues. The major virulence determinants of P. gingivalis are its cell surface Arg- and Lys-specific cysteine proteinases, RgpA/B and Kgp. Lactoferrin (LF), an 80-kDa iron-binding glycoprotein found in saliva and gingival crevicular fluid, is believed to play an important role in innate immunity. In this study, bovine milk LF displayed proteinase inhibitory activity against P. gingivalis whole cells, significantly inhibiting both Arg- and Lys-specific proteolytic activities. LF inhibited the Arg-specific activity of purified RgpB, which lacks adhesin domains, and also inhibited the same activity of the RgpA/Kgp proteinase-adhesin complexes in a time-dependent manner, with a first-order inactivation rate constant (kinact) of 0.023 min−1 and an inhibitor affinity constant (KI) of 5.02 μM. LF inhibited P. gingivalis biofilm formation by >80% at concentrations above 0.625 μM. LF was relatively resistant to hydrolysis by P. gingivalis cells but was cleaved into two major polypeptides (53 and 33 kDa) at R284 to S285, as determined by in-source decay mass spectrometry; however, these polypeptides remained associated with each other and retained inhibitory activity. The biofilm inhibitory activity of LF against P. gingivalis was not attributed to direct antibacterial activity, as LF displayed little growth inhibitory activity against planktonic cells. As the known RgpA/B and Kgp inhibitor N-α-p-tosyl-l-lysine chloromethylketone also inhibited P. gingivalis biofilm formation, the antibiofilm effect of LF may at least in part be attributable to its antiproteinase activity.
INTRODUCTION
Chronic periodontitis is an inflammatory disease that results in the destruction of the supporting tissues of the teeth. It is associated with specific bacteria in subgingival dental plaque. The disease is a major public health problem in all societies and is estimated to affect around 30% of the adult population, with severe forms affecting 5 to 10% (1, 3, 41). Chronic periodontitis poses a significant public health challenge as it is not only a major cause of tooth loss in adults but also a risk factor for cardiovascular diseases (12, 47). In addition, chronic periodontitis has recently been associated with an increased risk of pancreatic cancer (20, 29, 48) and squamous cell carcinoma of the head, neck, and esophagus (18).
The periodontal pathogen Porphyromonas gingivalis is a black-pigmented, Gram-negative, anaerobic coccobacillus. This bacterium has been implicated as a major opportunistic pathogen in the progression of chronic periodontitis, and its invasion into the host may be associated with the increased risk of systemic disease (35). It is usually found only in small numbers or not at all in the subgingival plaque of periodontally healthy individuals, but its numbers substantially increase in subjects with chronic periodontitis, especially at sites where bleeding occurs (46). P. gingivalis is normally found as part of a polymicrobial biofilm, called subgingival plaque, accreted to the nonshedding surface of the tooth root below the gum line. Biofilms are communities of bacteria that live associated with a surface and embedded in a polymeric matrix, which complicates treatment of chronic infections such as periodontitis by protecting bacteria from the immune system, decreasing antibiotic/antimicrobial efficacy, and allowing dispersal of planktonic cells to invade the host and colonize at distant sites (10, 11). P. gingivalis releases antigens, toxic metabolites, and hydrolytic enzymes such as proteinases that enter host tissues and subvert the host's immune response (35). The proteinases have been shown to be essential for virulence and tissue invasion by P. gingivalis (35, 38). The major proteinases of P. gingivalis are its cell surface-located complexes of Arg- and Lys-specific cysteine proteinases (RgpA/B and Kgp) (35).
Lactoferrin (LF), an 80-kDa iron-binding glycoprotein of the transferrin family, is present in the milk of various mammals, other exocrine secretions such as tears, saliva, gingival crevicular fluid, and synovial fluid, and the secondary granules of neutrophils and blood. It is believed to play an important role in innate immunity, exhibiting antibacterial, antiviral, antifungal, antitumor, parasiticidal, immunomodulatory, and anti-inflammatory activities (for reviews, see references 16, 22, 26, and 37). The iron-binding capacity of LF sequesters iron from the microbial environment, contributing to its ability to inhibit the growth of bacteria and yeasts (39, 40). In addition, LF can directly interact with microbial membranes to alter their permeability through dispersion of membrane components, such as lipopolysaccharide (LPS), thereby causing cell death (4, 15, 52).
In this study, we show that LF inhibited the P. gingivalis proteinases in a time-dependent manner and that LF was relatively resistant to cleavage by the proteinases. LF inhibited P. gingivalis biofilm formation while having only limited direct antibacterial activity against the bacterium. The initial cleavage site of LF was identified using in-source decay (ISD) mass spectrometry (MS) and was located on an external, hydrophilic loop of the protein. The results suggest that this cleavage did not cause a major disruption of the tertiary structure of the molecule. Even after incubation with P. gingivalis for extended time periods, LF still retained antiproteinase and biofilm inhibitory activity.
MATERIALS AND METHODS
Lactoferrin preparation and iron analysis.
Native bovine LF was provided by MG Nutritionals, Murray Goulburn Co-operative Ltd., Victoria, Australia. The purity of the LF was more than 95%, as determined by SDS-PAGE and in-gel digestion of protein bands with trypsin and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometric analysis (see below).
Apo-LF, the iron-free form of bovine LF, and holo-LF, the iron-loaded form of bovine LF, were produced according to the methods of Shimazaki (43) and Brisson et al. (8), respectively. Apo-LF had an iron saturation of 0.8% and native LF had an iron saturation of 7.4%, while holo-LF was 98.7% iron saturated, as determined by atomic absorption spectrometry (AA240 spectrometer; Varian, Inc., CA) at a wavelength of 248.3 nm.
Proteinase activity of P. gingivalis whole cells.
The Arg- and Lys-specific proteinase activities of P. gingivalis whole cells were measured in a 96-well plate using the chromogenic substrates N-α-benzoyl-Arg-p-nitroanilide (l-BApNA) and N-(p-tosyl)-Gly-Pro-Lys 4-nitroanilide acetate salt (GPK-NA) (Sigma), respectively, essentially as described previously (33). P. gingivalis ATCC 33277 was anaerobically grown in brain heart infusion (BHI) broth (37 g/liter) (Oxoid Australia Pty Ltd., South Australia, Australia), supplemented with hemin (5 μg/ml), vitamin K3 (5 μg/ml), and cysteine (0.5 mg/ml), at 37°C to the late exponential phase. P. gingivalis cells were harvested by centrifugation (8,000 × g, 4°C, 20 min), washed with 20 ml, and suspended in 2 ml ice-cold TC150 buffer (pH 8.0) comprising 2.8 mM cysteine, 50 mM Tris-HCl, 150 mM NaCl, and 5 mM CaCl2 to a cell density of 4.5 × 107 CFU/ml.
P. gingivalis cell suspensions (2.5 μl) were preincubated with LF preparations at 37°C for between 0 and 90 min at concentrations between 0.01 and 10 mg/ml in TC150 buffer in 96-well plates with a total volume of 100 μl in each well. To these a substrate preparation (100 μl) that contained 2 mM BApNA or GPK-NA, 30% (vol/vol) isopropanol, 400 mM Tris-HCl (pH 8), 100 mM NaCl, and 2 mM cysteine was added, the absorbance at a wavelength of 405 nm was monitored for 10 min, and initial rates were calculated.
Purification of P. gingivalis RgpA/Kgp proteinase-adhesin complexes and RgpB.
RgpA/Kgp proteinase-adhesin complexes were prepared using a modification of previously described methods (38). P. gingivalis W50 cells were harvested from 1 liter planktonic culture grown to the late exponential phase, washed, and resuspended in 30 ml TC50 buffer (50 mM Tris-HCl, pH 7.4, 5 mM CaCl2, 50 mM NaCl). Following the addition of 60 U/ml Benzonase (Sigma) and 2 mM MgCl2, the cell suspension was ultrasonicated using a Branson Sonifier 250 as described previously (5). The resulting cell sonicate was clarified by centrifugation (40,000 × g, 30 min, 4°C), made up to 50 ml with TC50 buffer containing 2 mM MgCl2, then incubated with freshly added Benzonase (140 U/ml) for 1 h on ice. After filtration (0.22 μm), the Benzonase-treated cell sonicate was loaded onto a 50-ml arginine-Sepharose 4B column at 1 ml/min and monitored at 280 nm using an ÄKTA explorer 100 automatic liquid chromatography system (GE Healthcare Bio-Science, Uppsala, Sweden). After being step washed with 150, 200, and 250 mM NaCl in 50 mM Tris-HCl (pH 7.4) buffer containing 5 mM CaCl2, the column was reequilibrated in TC50 buffer and then eluted with 500 mM arginine (Sigma) in the same buffer at 1.6 ml/min to recover the bound RgpA/Kgp proteinase-adhesin complexes. The eluted protein peak fraction of 25 ml was concentrated using a Centriprep YM-10 centrifugal filter device (Millipore) and buffer exchanged into TC50 buffer using a PD-10 desalting column (GE Healthcare). RgpB was purified from P. gingivalis strain HG66 as described previously (9).
LF hydrolysis by P. gingivalis.
P. gingivalis ATCC 33277 was cultured and harvested as described above. The cells were washed and resuspended to the initial volume with Pga buffer (pH 7.5), which was modified from that described by Milner et al. (30). It contained 2.8 mM cysteine, 10 mM NaH2PO4, 10 mM KCl, 2 mM citric acid, 1.25 mM MgCl2, 20 μM CaCl2, 25 μM ZnCl2, 50 μM MnCl2, 5 μM CuCl2, 10 μM CoCl2, 5 μM H3BO3, and 1 μM Na2MoO4, with the pH adjusted to 7.5 using 5 M NaOH at 37°C.
LF was dissolved in Pga buffer to a concentration of 2 mg/ml. Equal volumes of LF solution and cell suspension were mixed thoroughly and incubated at 37°C for between 10 min and 3 days. To terminate P. gingivalis proteolytic activity, 0.46 M acetic acid was added to lower the pH to 4.4. The preparations were filtered (0.22 μm) to remove the cells, and the degree of hydrolysis and biofilm inhibitory activity were determined by SDS-PAGE and a biofilm assay, respectively.
SDS-PAGE.
To denature PAGE in the presence of ionic detergent, all LF and LF treated with P. gingivalis (LF-Pg) samples were analyzed on NuPAGE (Invitrogen, Carlsbad, CA) 4 to 12% Bis-Tris precast gels used with 3-(N-morpholino) propane sulfonic acid (MOPS) SDS running buffer at pH 7.7. Protein samples (1 mg/ml) were mixed with NuPAGE lithium dodecyl sulfate (LDS) sample buffer and reducing agent, heated at 90°C for 5 min, and pulse centrifuged at 6,000 × g prior to being loaded on gels. The gels were stained overnight with 0.1% (wt/vol) Coomassie blue G-250 in 17% (wt/vol) ammonium sulfate, 34% (vol/vol) methanol, and 3% (vol/vol) o-phosphoric acid (31).
SEC.
Size-exclusive chromatography (SEC) was applied to separate LF-Pg by using a Superdex 75 10/300 GL column (GE Healthcare) connected to an ÄKTA explorer 100 (GE Healthcare). The elution buffer was 50 mM phosphate buffer (pH 6.0) with 750 mM NaCl. The flow rate was 0.5 ml/min with an injection volume of 250 μl of filtered (0.22-μm) sample.
Fractionation of LF-Pg by RP-HPLC.
LF treated with P. gingivalis, prepared as described above and referred to as Lf-Pg, was incubated for 6 h and subjected to fractionation using an Aquapore OD-300 reversed-phase (RP) column (7 μm, 4.6 × 250 mm; PerkinElmer Brownlee Columns, Shelton, CT) connected to an Agilent 1200 series high-pressure liquid chromatography (HPLC) system (Agilent Technologies, Santa Clara, CA). The eluting solvents consisted of (i) 0.01% (vol/vol) trifluoroacetic acid (TFA) in deionized water and (ii) 0.01% (vol/vol) TFA in 80% acetonitrile and 20% deionized water (HPLC grade). The injection volume was 100 μl per sample, and the system was operated at a flow rate of 1 ml/min. The protein content was detected by a diode array detector at wavelengths of 214 and 280 nm. The column was initially equilibrated with 100% mobile phase A for 10 min, followed by elution with a linear gradient from 0% to 40% of mobile phase B for 40 min, from 40% to 50% for 30 min, from 50% to 60% for 10 min, and from 60% to 100% for 5 min. The elution peaks were collected, freeze-dried, and reconstituted with deionized water. The protein concentrations were determined using the Bradford microassay from Bio-Rad (6).
In-gel digestion of LF and mass spectrometry.
Protein bands from SDS-PAGE were excised and subjected to in-gel tryptic digestion and mass spectrometry analysis as published previously (34). Briefly, gel pieces were washed in 50 mM NH4HCO3/ethanol 1:1 (vol/vol), reduced and alkylated with dithiothreitol (DTT) and iodoacetamide, respectively, and digested with sequencing-grade modified trypsin (10 ng/μl) (Promega, NSW, Australia) overnight at 37°C. Peptides were analyzed on an Ultraflex TOF-TOF III mass spectrometer (Bruker Daltonics, Bremen, Germany) using 4-hydroxy-α-cyanocinnamic acid as the matrix (34). Peaks were assigned to LF by using an in-house Mascot search engine (Matrix Science, London, United Kingdom) with bovine LF as the sequence database. Sample preparation in sinapinic acid and spectra acquisition for in-source decay (ISD) were performed according to the FlexControl 3.0 user manual (Bruker Daltonics). Briefly, 1 μl matrix A (sinapinic acid saturated in ethanol) was applied to a steel target to create a thin layer. LF fractions were mixed 1:1 with matrix B (sinapinic acid saturated in 0.1% TFA, 30% acetonitrile), and 1 μl was deposited onto the thin layer and allowed to dry. ISD spectra were acquired using a standard reflectron method optimized for peptides except for an increased pulsed ion extraction (PIE) delay of 200 ns and enhanced sensitivity settings on the digitizer and reflector detector. Laser power was increased until ISD peaks appeared, and then spectra from 1,600 laser shots were acquired.
P. gingivalis biofilm assay.
The biofilm assay was modified from the method of Wen and Burne (56). P. gingivalis ATCC 33277 was grown in a batch culture as described above. The culture was then diluted 10 times with supplemented BHI to give a cell density of 2.8 × 109 CFU/ml and kept on ice until used. LF and other antimicrobials were prepared 10 times more concentrated than the final concentration, and they were sterilized by being passed through a 0.22-μm filter. All antimicrobial solutions (20 μl) were added to 96-well plates with 6 replicates, while control samples contained 20 μl distilled water. The bacterial culture (180 μl) was added to each well to make the total volume 200 μl. The plate was incubated at 37°C for 24 h in an anaerobic chamber (MK3 anaerobic work station; Don Whitley Scientific Ltd., Sydney, NSW, Australia) with an atmosphere of 5% hydrogen, 10% carbon dioxide, and 85% nitrogen. To determine the effect of hemin on biofilm formation, hemin was not added to the growth medium.
Following incubation, the plate was shaken at 100 rpm at 37°C for 15 min, and all media were removed. Each well was washed with deionized water and blow dried by air for at least 3 h. Crystal violet (0.1%) was used to stain the biofilm on each well surface at room temperature for 15 min. Unbound crystal violet was then removed by washing twice with deionized water. The plate was blow dried by air briefly, and 100 μl ethanol containing 20% (vol/vol) acetone was used to dissolve the bound color from the well surface. The mass of the biofilm on the well surface was expressed as the absorbance at 600 nm for each sample in the well, using a UV spectrometer (Victor3 1420 multilabel counter; PerkinElmer, MA).
P. gingivalis planktonic growth assay.
The effect of LF on P. gingivalis ATCC 33277 was determined in a 96-well plate assay using supplemented BHI growth medium under anaerobic conditions at 37°C as described previously (27).
Statistical analyses.
A one-way analysis of variance (ANOVA) was used to analyze the data, and significantly different values (P < 0.05) were identified using a post hoc Tukey test. A kinetic analysis of the time-dependent inactivation of the proteinases was determined using the methodology of Maurer and Fung (28), where KI represents the inhibitor binding affinity and kinact is the first-order inactivation rate constant.
Molecular dynamics simulation of the LF-RgpB interaction.
Crystallographic models of RgpB (14) and the C-lobe of lactoferrin (57) (PDB ID 1cvr and 3taj, respectively) were downloaded from the Protein Data Bank. Structures were prepared for further modeling using the AMBER-99 force field and the program Sybyl (51). Crystallographic water molecules were removed, and the resulting structures initially energy minimized to a maximum energy derivative of 0.5 kcal mol−1 Å−1. The 3taj (C-lobe lactoferrin) structure was then manually docked against the 1cvr (RgpB) structure so that Glu659 of the C-lobe of lactoferrin that binds a zinc ion in the crystal structure could be constrained to the zinc ion bound to the catalytic histidine of RgpB, His244. The atoms of the RgpB inhibitor d-Phe-Phe-Arg-chloromethylketone (DFFR-chloromethylketone) were removed for the dynamics simulation. The docked structures were then solvated with “TIP3P” waters using a “droplet” solvation model and energy minimized to a maximum energy derivative of 0.5 kcal mol−1 Å−1. A 10-ns molecular dynamics simulation was then performed for all atoms within a 20-Å sphere centered on the Zn ion, with atoms more than 15 Å from the Zn ion and all atoms in the RgpB structure being frozen at their initial coordinates. The constant number (of particles), temperature, and volume (NTV) dynamics simulation used the AMBER-99 force field, a constant dielectric of 4.0, a van der Waal's scale factor of 0.7827, a nonbonded cutoff of 12 Å, and a set temperature of 300 K.
RESULTS
Effect of LF on P. gingivalis proteinase activity.
LF inhibited both the Arg- and Lys-specific proteinase activities of P. gingivalis whole cells by approximately 40% at 1.0 mg/ml (12.5 μM) and over 70% at 10 mg/ml (125 μM) (Table 1). Bovine serum albumin (BSA) had no effect on P. gingivalis Arg- and Lys-specific proteinase activities at concentrations up to 1 mg/ml. At 10 mg/ml, BSA displayed a small effect on the hydrolysis of the chromogenic substrate, which was attributed to substrate competition (Table 1).
Table 1.
Arg- and Lys-specific proteinase activities of P. gingivalis whole cells in the presence of LF and BSA as determined using the chromogenic substrates l-BApNA and GPK-NA, respectively
| Protein concn (mg/ml) | Lys-specific proteinase activity of P. gingivalis whole cellsa |
Arg-specific proteinase activity of P. gingivalis whole cellsa |
||
|---|---|---|---|---|
| LF | BSA | LF | BSA | |
| 0 | 27.6 ± 0.6 A | 27.6 ± 0.6 A | 55.4 ± 0.5 A | 55.4 ± 0.5 A |
| 0.001 | 27.2 ± 0.9 A | 27.8 ± 1.1 A | —b | — |
| 0.01 | 27.4 ± 0.7 A | 27.5 ± 1.0 A | 56.0 ± 2.3 A | 47.9 ± 7.7 A |
| 0.1 | 24.8 ± 0.7 A | 26.1 ± 0.7 AB | 47.8 ± 2.5 B | — |
| 1.0 | 15.9 ± 1.0 B | 24.5 ± 1.8 B | 34.7 ± 2.3 C | 61.4 ± 7.0 A |
| 10 | 8.5 ± 0.2 C | 14.3 ± 0.7 C | 6.4 ± 1.9 D | 33.7 ± 3.6 B |
Units of proteinase activity/1011 cells. Values are significantly different (P < 0.05) to all other values not similarly marked in the same column, as determined by ANOVA and a post hoc Tukey test.
—, not determined.
LF inhibited both the Arg-specific and Lys-specific activities of purified P. gingivalis RgpA/Kgp proteinase-adhesin complexes by >96% at a concentration of 5 mg/ml. As the levels of inhibition were similar and there is a very high degree of similarity between RgpA and Kgp, we then focused on the characterization of the Arg-specific inhibitory activity.
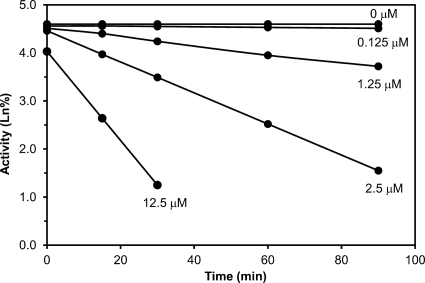
A kinetic analysis of the inhibition of Arg-specific proteolytic activity of purified P. gingivalis RgpA/Kgp proteinase-adhesin complexes by LF demonstrated time-dependent inhibition with a first-order inactivation rate constant (kinact) of 0.023 min−1 and an inhibitor affinity constant (KI) of 5.02 μM (Fig. 1). To confirm that LF was interacting with RgpA and Kgp by binding to the catalytic domain of the proteinases, LF was incubated with purified RgpB, which lacks the adhesin domains of RgA and Kgp. LF inhibited RgpB activity by 77% at a concentration of 1.0 mg/ml and by 95% at 10 mg/ml, confirming that the inhibition was independent of adhesins.
Fig 1.
Time-dependent inhibition of RgpA activity by lactoferrin. The figure shows the effect of lactoferrin at different concentrations (μM) on the activity (Ln%) of RgpA over time. The slope of these lines provided the apparent inactivation rate constants (kapp). Plotting 1/kapp versus 1/[I], where I is the lactoferrin concentration, yielded a straight line (r = 0.9995) with an intercept of 1/kinact and a slope of KI/kinact (28). This produced a first-order inactivation rate constant (kinact) of 0.023 min−1 and a lactoferrin binding affinity (KI) of 5.02 μM.
Molecular dynamics simulation of the LF-RgpB interaction.
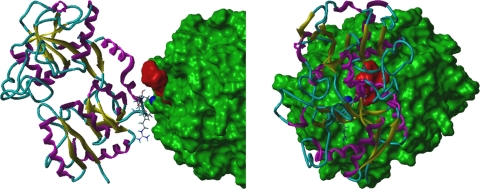
As LF inhibited both the Arg- and Lys-specific proteinases (RgpA/B and Kgp) of P. gingivalis, this suggested a common inhibitory mechanism. The crystal structure of RgpB (14) shows a zinc ion-binding site involving residues Glu152 and the catalytic histidine, His211. Modeling of Kgp based on the known structure of RgpB indicates that residues Asp388 and His444 of Kgp would similarly bind a zinc ion in the active site of the proteinase. As lactoferrin is known to bind zinc ions (21), then the interaction between LF and the zinc ion in the active site of RgpA/B and Kgp would explain the inactivation of the proteinases. The molecular modeling of the interaction between LF and RgpB (Fig. 2) demonstrates that these two proteins could readily be cross-linked by sharing the zinc ion bound by Glu152 and His211 of the RgpB active site. Energy minimization followed by molecular dynamics indicated that the unfavorable van der Waal's contacts observed between the proteins were readily relaxed and that additional favorable interactions between the two proteins developed on a relatively short time scale, consistent with the time-dependent manner of the observed inactivation. As modeled, there would be significant unfavorable van der Waal's interactions between Glu658 of LF and the terminal d-phenylalanyl residue of DFFR-chloromethylketone, the substrate analogue inhibitor of RgpB crystalized with the proteinase. This indicates that the interaction of LF with the zinc ion in the RgpB active site would sterically hinder substrate access to the active site (Fig. 2).
Fig 2.
An orthographic view of the modeled lactoferrin-RgpB complex. The solvent-accessible surface of RgpB is shown in green, and the dark blue, space-filling atom is the zinc ion bound to the catalytic histidine (His244) of RgpB. Lactoferrin is shown as a ribbon structure (β-strand in yellow, α-helix in magenta, and coil in cyan), and the side-chains of residues that moved to within 3 Å of RgpB during the dynamics simulation are shown as “capped sticks.” The location of the atoms of the RgpB inhibitor, DFFR-chloromethylketone, are shown by red space-filling atoms.
Effect of P. gingivalis proteinase activity on LF.
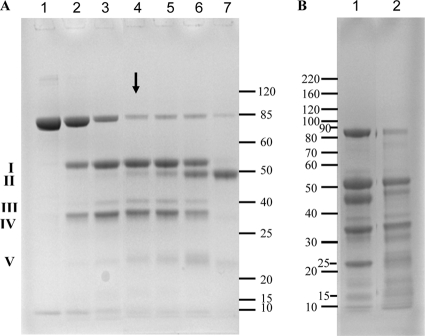
LF was incubated anaerobically with P. gingivalis whole cells for 3 days in Pga buffer, which was formulated to ensure the survival of the bacterium and activity of the cell surface proteinases. Samples taken at specified time points were subjected to SDS-PAGE analysis and used in the P. gingivalis biofilm assay. There was a limited initial hydrolysis of LF by P. gingivalis, resulting in two major products, fragment I (53 kDa) and fragment IV (33 kDa), and a minor product, fragment III (40 kDa) (Fig. 3A, lanes 2 to 4). This was followed by a slower, more extensive hydrolysis (Fig. 3A, lanes 5 to 7) that after 3 days resulted in the almost complete disappearance of the intact LF band (85 kDa) and the production of fragment II (48 kDa). When LF was digested by trypsin, a more extensive hydrolysis was seen with two major bands (23 and 46 kDa) detected in addition to the 33 and 53 kDa bands seen in LF digested by P. gingivalis (Fig. 3B).
Fig 3.
(A) SDS-PAGE of LF incubated with P. gingivalis. Lane 1, LF (1 μg); lanes 2 to 7, LF and P. gingivalis; lane 2, 1-min incubation; lane 3, 10-min incubation; lane 4, 3-h incubation; lane 5, 6-h incubation; lane 6, 18-h incubation; lane 7, 3-day incubation. The arrow indicates the sample fractionated by gel filtration (6-h incubation). Major fragments of LF are labeled with Roman numerals, and the gel bands have been analyzed by MS. (B) LF was treated with different concentrations of trypsin at 37°C for 18 h. Lane 1, LF/trypsin = 2,000/1; lane 2, LF/trypsin = 5,000/1.
Identification of LF cleavage site.
In order to identify the protein fragments that resulted from incubation of LF with P. gingivalis cells, the gel bands shown in Fig. 3A were subjected to in-gel digestion and analysis by mass spectrometry (MS). The protein bands were grouped as fragments I, II, III, IV, and V with relative molecular masses of 53, 48, 40, 33, and 22 kDa, respectively, while intact LF had a relative molecular mass of 85 kDa (Fig. 3A). Fragments I and IV were the main products during the first 6 h of incubation, and their predicted masses suggested that they resulted from cleavage at a single site in LF. Peptide mass fingerprint analysis indicated that fragment I was a C-terminal fragment of LF and that fragment IV was an N-terminal fragment (Fig. 4). It appears that fragment IV was further hydrolyzed by loss of the N-terminal portion to yield fragment V. Fragment II, which was only generated in significant quantities after 6 h of incubation with P. gingivalis, corresponds to a C-terminal fragment of fragment I (Fig. 4).
Fig 4.
The primary sequence of LF. Underlined sequences denote the peptides identified by peptide mass fingerprint (PMF) analysis of fragments I to V. The cleavage site (‖) between fragments I and IV (the major polypeptides in the LF-Pg sample) was determined by in-source decay (ISD) MALDI-TOF MS. All arginine and lysine residues are shown in bold.
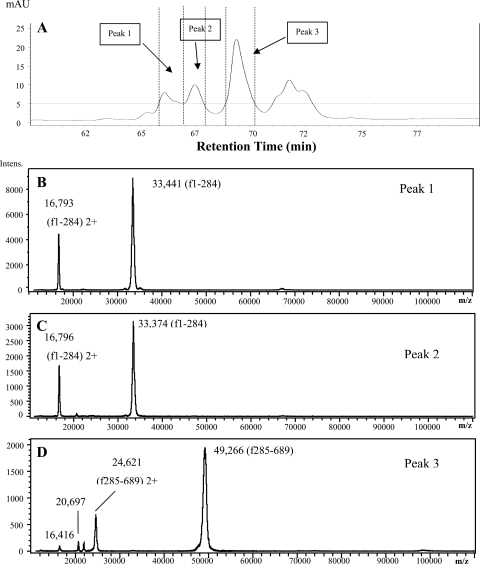
RP-HPLC was used to obtain further information about these LF fragments, and the fractions of LF-Pg (6 h of incubation) were analyzed by MS (Fig. 5). Peaks 1 and 2 consisted of a polypeptide with a mass of 33 kDa, whereas peak 3 consisted mainly of a polypeptide with a mass of 49 kDa (Fig. 5). These peaks correspond to fragment IV (33 kDa) and fragment I (53 kDa) in SDS-PAGE analysis (Fig. 3), respectively. The masses of these fragments as measured by MS total between 82.66 and 82.71 kDa, which is close to the measured mass of 82.8 kDa for whole LF. This confirms that these two fragments are the products of cleavage at a single site in LF. However, due to potential glycosylation of both fragments I and IV, it was not possible to use these measured masses to identify the cleavage site.
Fig 5.
(A) RP-HPLC analysis of LF after 6 h of hydrolysis by P. gingivalis whole cells. (B to D) MS spectra of RP-HPLC peaks 1 to 3, respectively.
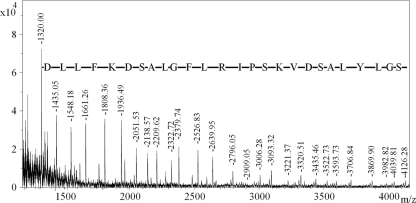
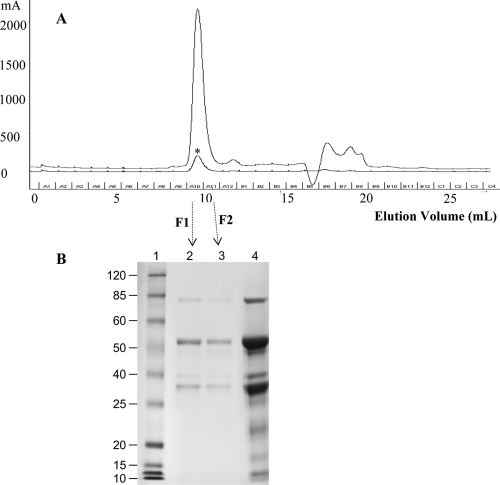
To determine the exact cleavage site, in-source decay mass spectrometry (ISD MS), which enables extensive N-terminal (and sometimes C-terminal) protein sequence to be elucidated, was employed on the fractions. ISD MS of fragment IV was unsuccessful; however, fragment I was successfully fragmented, yielding a strong series of peaks that corresponded to N-terminal fragment ions (Fig. 6). The sequence DLLFKDSALGFLRI PSKVDSALYLGSRY was directly determined from the series of peaks, while the mass of each peak indicated that the N terminus of fragment I was S285. The primary cleavage site of LF when exposed to P. gingivalis was therefore R284 to S285 (Fig. 4). Analysis of LF-Pg samples with size-exclusion chromatography (SEC) revealed a single peak (Fig. 7A), and SDS-PAGE analysis of the material in this peak showed that it consisted of the two fragments (33 and 53 kDa) as well as a much smaller amount of native LF (Fig. 7B).
Fig 6.
ISD MS spectrum of fragment I of LF-Pg. The fragment ions labeled correspond to N-terminal (C-type) fragments of lactoferrin, starting from C-12 and extending to C-38. The mass difference between each peak corresponds to the amino acid residue shown. Extrapolation of these data indicated that the N-terminal sequence of this polypeptide was 285SFQLFGSPPGQRDLLFKDSALGFLRIPSKVDSALYLGS, with the underlined portion evident in the spectrum.
Fig 7.
(A) LF-Pg was fractionated by size-exclusion chromatography (SEC), while native LF was eluted as a control (indicated by an asterisk). The fractions were collected at an interval of 1 ml, and two fractions (marked as F1 and F2) were collected from the peak at the elution volume between 9 and 11 ml. (B) LF-Pg and the SEC fractions from LF-Pg were analyzed by using SDS-PAGE. Lane 1, MW marker; lanes 2 and 3, F1 and F2 from the peak eluted from SEC; lane 4, LF-Pg without SEC.
LF biofilm inhibitory activity.
When cultured in the presence of 5 μg/ml hemin, P. gingivalis produced significant biofilms with an optical density of 0.315 ± 0.054 after 24 h of incubation; however, in the absence of added hemin in the growth medium, there was a significant reduction in the biofilm mass (0.168 ± 0.048; P < 0.005). When LF was added to hemin-excess medium at concentrations above 0.05 mg/ml, it effectively abolished P. gingivalis biofilm formation, and there was a significant inhibitory effect at concentrations as low as 0.001 mg/ml (Fig. 8). In contrast, bovine serum albumin (BSA) and β-lactoglobulin (β-Lg) did not inhibit P. gingivalis biofilm formation at concentrations up to 10.0 mg/ml, and in fact it significantly enhanced biofilm formation (data not shown). Native LF, apo-LF, and holo-LF all had similar inhibitory effects on P. gingivalis biofilm formation at a concentration of 0.01 mg/ml, inhibiting biofilm formation by 87.2 ± 1.7, 88.1 ± 1.6, and 84.2 ± 5.5%, respectively. The gingipain inhibitor N-α-p-tosyl-l-lysine chloromethylketone (TLCK), at a concentration of 1 mM that inhibits Arg- and Lys-specific proteinase activities by >99%, reduced P. gingivalis biofilm formation after 24 h of incubation by 78.3 ± 5.9% from three biological replicates.
Fig 8.
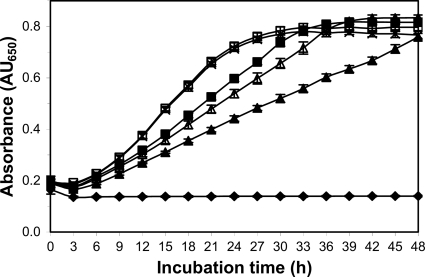
Effect of LF on P. gingivalis biofilm formation. P. gingivalis biofilm formation was determined in the presence of LF for 18 h (△), 24 h (▲), and 48 h (■). Each data point represents the mean and standard deviation of three replicates. Note the categorical scale on the x axis.
LF demonstrated a weak, dose-dependent inhibitory effect on P. gingivalis planktonic growth (Fig. 9). At concentrations below 0.5 mg/ml, there was little effect on P. gingivalis planktonic growth (Fig. 9).
Fig 9.
Effect of LF on planktonic growth of P. gingivalis in batch culture, LF concentrations (mg/ml): 0 (×), 0.5 (□), 2.5 (■), 5.0 (△), 10 (▲), uninoculated growth medium (♦). Each data point represents the mean and standard deviation of six biological replicates. LF concentration at 10 mg/ml, 5.0 mg/ml, and 2.5 mg/ml had a significant (P < 0.05) but moderate lowering of the growth rate but did not significantly effect final absorbance of the cultures.
Inhibitory activity of P. gingivalis-treated LF.
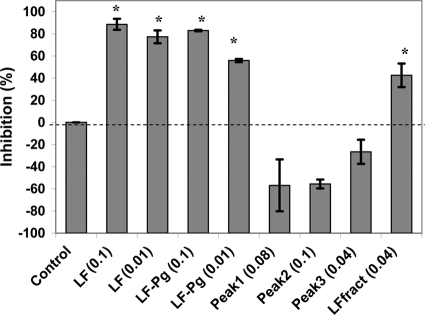
Native LF that had been incubated with P. gingivalis for 6 h (LF-Pg) at a concentration of 0.1 mg/ml inhibited P. gingivalis biofilm formation by 83%, while LF-Pg at 0.01 mg/ml inhibited biofilm formation by 56% (Fig. 10). Individual RP-HPLC fractions of LF-Pg had no biofilm inhibitory activity and in fact appeared to promote biofilm formation (Fig. 10). Native LF that was applied to RP-HPLC and eluted under the same conditions as that for LF-Pg (LFfract) still retained some activity, inhibiting P. gingivalis biofilm formation by 43% at a concentration of 0.04 mg/ml (Fig. 10). Hydrolysis of LF by trypsin (LF/trypsin = 5,000/1) (Fig. 3B) resulted in an almost complete loss of LF inhibitory activity (data not shown).
Fig 10.
Comparison of the biofilm inhibitory activity of native LF, LF-Pg (6 h of incubation), and fractions obtained from RP-HPLC separation of LF-Pg. Each data point represents the mean of three biological replicates. Peaks 1 to 3 correspond to the three RP-HPLC fractions in Fig. 4. LFfract was native LF eluted under the same conditions as that for separation of LF-Pg sample. The number in each treatment represents the protein concentration at mg/ml. *, significantly (P < 0.001) inhibited biofilm formation.
DISCUSSION
The major virulence factors of P. gingivalis are its cysteine proteinases RgpA and Kgp and associated adhesins that form large complexes on the cell surface and cleave C-terminally to arginine or lysine residues (35). These proteinase-adhesin complexes hydrolyze a range of host regulatory proteins, peptides, and cell receptors, leading to dysregulation of the host immune response and subsequent tissue damage (19, 25, 35). The proteinases are essential for tissue invasion by P. gingivalis in an animal model (35, 38) that has been used extensively to determine the invasive characteristics of pathogenic bacteria (23, 50).
In this study, we have shown that LF inhibited the proteolytic activity of both RgpA and Kgp (Table 1). Shi et al. (42) have previously shown that human LF and bovine lactoferricin, the N-terminal domain of bovine LF, were able to bind to the P. gingivalis RgpA/Kgp proteinase-adhesin complexes and disrupt their quaternary structure, leading to the release of the hemoglobin-binding domain (Hbr). In our study, LF inhibition of the proteolytic activity of RgpB, which does not contain adhesin domains, indicated that the protein was interacting with the catalytic domain of the proteinases. Kinetic analyses demonstrated that LF inhibited the proteinases of P. gingivalis in a time-dependent manner, confirming that LF was interacting with the catalytic domain of the proteinases. The KI of LF for RgpA activity was 5.02 μM. This inhibition is consistent with the work of Ohashi et al. (36), who reported that LF inhibited the proteinases cathepsin and papain, although they did not examine the time-dependent nature or the mechanism of inhibition. The time dependency of the inhibition suggests that LF is a slow binding inhibitor with a low dissociation rate (49). The molecular dynamics simulation of the interaction between LF and RgpB (Fig. 2) provided a mechanism for the observed inhibition. The zinc ion-binding C-lobe of LF (21, 57) can bind the zinc ion in the active site of the RgpA/B and Kgp proteinases, ultimately forming a stable structure explaining the observed time-dependent inactivation of the proteinases.
Antimicrobial peptides and proteins in host secretions may have reduced efficacy against bacteria due to their susceptibilities to hydrolysis by cell-surface or secreted bacterial proteinases. P. gingivalis proteinases have been shown to degrade a range of host proteins, including human transferrin and hemoglobin (13, 17). The high level of P. gingivalis cell surface and extracellular proteolytic activity has been shown to hydrolyze antibacterial proteins and peptides, such as histatin, thereby reducing their efficacy (32). In this study, we showed that despite an abundance of arginine and lysine residues (Fig. 4), LF was relatively resistant to hydrolysis by P. gingivalis proteinases (Fig. 3). After 3 h of incubation of LF with P. gingivalis whole cells in a physiological buffer, only two major polypeptides (33 and 53 kDa) were detected, and these fragments resulted from cleavage at a single site. LF has been reported to be relatively resistant to degradation by both trypsin and chymotrypsin, and the N-linked glycosylation of LF has been shown to help protect the protein from trypsin hydrolysis (7, 53). In our study, LF was more extensively hydrolyzed by trypsin than the P. gingivalis proteinases (Fig. 3), which is most likely related to the ability of LF to inhibit the P. gingivalis proteinases.
Bovine LF contains five N-linked glycosylation sites (Asn-233, -281, -368, -476, and -545) (55), and the majority of glycans are located in the N-terminal region of LF (33-kDa fragment); consequently, the variation in glycosylation could explain why this fragment eluted in two distinct peaks from RP-HPLC (Fig. 5). The glycosylation of both fragments made it impossible to use the measured masses of the peptide fragments to identify the cleavage site. ISD MS was therefore used to analyze the primary cleavage site of LF when exposed to P. gingivalis (Fig. 4 and 6). The R284-to-S285 cleavage site identified occurs on an exposed external hydrophilic loop of the LF molecule (2). Cleavage at this site is unlikely to cause dissociation of the two polypeptides such that the molecule would retain its tertiary structure. This was confirmed by SEC analysis of LF-Pg (6 h of incubation) that demonstrated that the two fragments (33 kDa and 53 kDa) eluted as a single peak with native LF (Fig. 7).
In this study, we also showed that LF inhibited P. gingivalis biofilm formation by >84% at concentrations above 0.01 mg/ml. At concentrations as low as 0.001 mg/ml, LF still significantly inhibited P. gingivalis biofilm formation by 50% (Fig. 10). The P. gingivalis biofilm inhibitory activity of LF was not a general protein effect, as BSA and β-Lg did not inhibit P. gingivalis biofilm formation and in fact enhanced it. This is consistent with the recent data of Wakabayashi et al. (54), who showed that LF inhibited P. gingivalis biofilm formation by ∼60% at a concentration of 0.008 mg/ml.
The biofilm inhibitory activity of LF against P. gingivalis was not attributed to its planktonic growth inhibitory activity, as LF displayed little growth inhibitory activity against planktonic cells (Fig. 9). LF reduced P. gingivalis planktonic growth at high concentrations while significantly increasing the mean generation time, indicating that LF was slowing growth rather than having a bactericidal action (Fig. 9). This is consistent with the findings of Wakabayashi et al. (54), who showed that native bovine LF had no significant effect on P. gingivalis viability when incubated in saline over a 4-h period. They also showed that apo-LF (iron free) had little effect on the planktonic growth of P. gingivalis, while high concentrations (8 mg/ml) of holo-LF (iron saturated) reduced growth by 60%, as determined using an ATP luminescence assay. These data are consistent with the findings of Singh et al. (45) and Singh (44), who showed that LF at 0.02 mg/ml effectively inhibited Pseudomonas aeruginosa biofilm formation but had no effect below 0.1 mg/ml on the growth rate of free-swimming bacterial cells (45). From the current study, the results suggest that the P. gingivalis biofilm inhibitory activity of LF may be attributable, at least in part, to its proteinase inhibitory activity, as the known proteinase inhibitor TLCK also inhibited P. gingivalis biofilm formation. Kontani et al. (24) have demonstrated that the Arg-specific proteinase of P. gingivalis can expose cryptic receptors to enhance binding, and this may explain why proteinase inhibition reduced biofilm formation.
Importantly, even after incubation with P. gingivalis proteinases, LF still retained its inhibitory activity. This further supports LF retaining its tertiary structure after cleavage at the R284-to-S285 site, as when it was purified using RP-HPLC the two major fragments of LF-Pg preparations individually had no inhibitory activity.
The proteolytic activity of P. gingivalis is central to its pathogenicity (33, 35, 38), and this study is the first to demonstrate that LF inhibits the proteinase activity of P. gingivalis in a time-dependent manner. It also shows that LF is relatively resistant to hydrolysis by these proteinases and that it displays sustained antibiofilm activity even when incubated with the bacterium. Therefore, LF may have an important role in gingival crevicular fluid and saliva in helping to prevent P. gingivalis-associated disease.
ACKNOWLEDGMENTS
We thank Gina Kusuma, Rishi Pathirana, Troy Attard, Huiling He, Dina Chen, and Katrina Walsh for their assistance during this study.
This project was supported by MG Nutritionals, Murray Goulburn Co-operative Ltd., Victoria, Australia.
Footnotes
Published ahead of print 3 January 2012
REFERENCES
- 1. Albandar JM, Brunelle JA, Kingman A. 1999. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988-1994. J. Periodontol. 70:13–29 [DOI] [PubMed] [Google Scholar]
- 2. Anderson BF, et al. 1987. Structure of human lactoferrin at 3.2-Å resolution. Proc. Natl. Acad. Sci. U. S. A. 84:1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Armfield J, Roberts-Thomson K, Spencer A. 2000. Australia's health 2000, vol 7. Catalogue no. AUS 19. Australian Institute of Health and Welfare, Canberra, Australia [Google Scholar]
- 4. Arnold RR, Russell JE, Champion WJ, Brewer M, Gauthier JJ. 1982. Bactericidal activity of human lactoferrin: differentiation from the stasis of iron deprivation. Infect. Immun. 35:792–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhogal PS, Slakeski N, Reynolds EC. 1997. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology 143:2485–2495 [DOI] [PubMed] [Google Scholar]
- 6. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 7. Brines RD, Brock JH. 1983. The effect of trypsin and chymotrypsin on the in vitro antimicrobial and iron-binding properties of lactoferrin in human milk and bovine colostrum. Unusual resistance of human apolactoferrin to proteolytic digestion. Biochim. Biophys. Acta 759:229–235 [DOI] [PubMed] [Google Scholar]
- 8. Brisson G, Britten M, Pouliot Y. 2007. Heat-induced aggregation of bovine lactoferrin at neutral pH: effect of iron saturation. Int. Dairy J. 17:617–624 [Google Scholar]
- 9. Chen YY, et al. 2002. CPG70 is a novel basic metallocarboxypeptidase with C-terminal polycystic kidney disease domains from Porphyromonas gingivalis. J. Biol. Chem. 277:23433–23440 [DOI] [PubMed] [Google Scholar]
- 10. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 11. Cvitkovitch DG, Li YH, Ellen RP. 2003. Quorum sensing and biofilm formation in streptococcal infections. J. Clin. Invest. 112:1626–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. D'Aiuto F, et al. 2006. Periodontal infections cause changes in traditional and novel cardiovascular risk factors: results from a randomized controlled clinical trial. Am. Heart J. 151:977–984 [DOI] [PubMed] [Google Scholar]
- 13. Dashper SG, et al. 2004. Hemoglobin hydrolysis and heme acquisition by Porphyromonas gingivalis. Oral Microbiol. Immunol. 19:50–56 [DOI] [PubMed] [Google Scholar]
- 14. Eichinger A, et al. 1999. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 18:5453–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ellison RT, III, Giehl TJ, LaForce FM. 1988. Damage of the outer membrane of enteric gram-negative bacteria by lactoferrin and transferrin. Infect. Immun. 56:2774–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farnaud S, Evans RW. 2003. Lactoferrin-a multifunctional protein with antimicrobial properties. Mol. Immunol. 40:395–405 [DOI] [PubMed] [Google Scholar]
- 17. Goulet V, Britigan B, Nakayama K, Grenier D. 2004. Cleavage of human transferrin by Porphyromonas gingivalis gingipains promotes growth and formation of hydroxyl radicals. Infect. Immun. 72:4351–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guha N, et al. 2007. Oral health and risk of squamous cell carcinoma of the head and neck and esophagus: results of two multicentric case-control studies. Am. J. Epidemiol. 166:1159–1173 [DOI] [PubMed] [Google Scholar]
- 19. Holt SC, Kesavalu L, Walker S, Genco CA. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168–238 [DOI] [PubMed] [Google Scholar]
- 20. Hujoel PP, Drangsholt M, Spiekerman C, Weiss NS. 2003. An exploration of the periodontitis-cancer association. Ann. Epidemiol. 13:312–316 [DOI] [PubMed] [Google Scholar]
- 21. Jabeen T, Sharma S, Singh N, Bhushan A, Singh TP. 2005. Structure of the zinc-saturated C-terminal lobe of bovine lactoferrin at 2.0-Å resolution. Acta Crystallogr. D Biol. Crystallogr. 61:1107–1115 [DOI] [PubMed] [Google Scholar]
- 22. Kanyshkova TG, Buneva VN, Nevinsky GA. 2001. Lactoferrin and its biological functions. Biochemistry 66:1–7 [DOI] [PubMed] [Google Scholar]
- 23. Kesavalu L, Holt SC, Ebersole JL. 1998. Virulence of a polymicrobic complex, Treponema denticola and Porphyromonas gingivalis, in a murine model. Oral Microbiol. Immunol. 13:373–377 [DOI] [PubMed] [Google Scholar]
- 24. Kontani M, Kimura S, Nakagawa I, Hamada S. 1997. Adherence of Porphyromonas gingivalis to matrix proteins via a fimbrial cryptic receptor exposed by its own arginine-specific protease. Mol. Microbiol. 24:1179–1187 [DOI] [PubMed] [Google Scholar]
- 25. Lamont RJ, Jenkinson HF. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lönnerdal B. 2003. Lactoferrin, p 449–466 In Fox PF, McSweeney PL. (ed), Advanced dairy chemistry, vol 1 Proteins Kluwer Academic/Plenum Publishers, London, United Kingdom [Google Scholar]
- 27. Malkoski M, et al. 2001. Kappacin, a novel antibacterial peptide from bovine milk. Antimicrob. Agents Chemother. 45:2309–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maurer T, Fung HL. 2000. Comparison of methods for analyzing kinetic data from mechanism-based enzyme inactivation: application to nitric oxide synthase. AAPS PharmSci. 2:E8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Michaud DS, Joshipura K, Giovannucci E, Fuchs CS. 2007. A prospective study of periodontal disease and pancreatic cancer in US male health professionals. J. Natl. Cancer Inst. 99:171–175 [DOI] [PubMed] [Google Scholar]
- 30. Milner P, Batten JE, Curtis MA. 1996. Development of a simple chemically defined medium for Porphyromonas gingivalis: requirement for alpha-ketoglutarate. FEMS Microbiol. Lett. 140:125–130 [DOI] [PubMed] [Google Scholar]
- 31. Molloy MP, Herbert BR, Williams KL, Gooley AA. 1999. Extraction of Escherichia coli proteins with organic solvents prior to two-dimensional electrophoresis. Electrophoresis 20:701–704 [DOI] [PubMed] [Google Scholar]
- 32. O'Brien-Simpson NM, Dashper SG, Reynolds EC. 1998. Histatin 5 is a substrate and not an inhibitor of the Arg- and Lys-specific proteinases of Porphyromonas gingivalis. Biochem. Biophys. Res. Commun. 250:474–478 [DOI] [PubMed] [Google Scholar]
- 33. O'Brien-Simpson NM, et al. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527–7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Brien-Simpson NM, et al. 2005. An immune response directed to proteinase and adhesin functional epitopes protects against Porphyromonas gingivalis-induced periodontal bone loss. J. Immunol. 175:3980–3989 [DOI] [PubMed] [Google Scholar]
- 35. O'Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. 2003. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr. Protein Pept. Sci. 4:409–426 [DOI] [PubMed] [Google Scholar]
- 36. Ohashi A, et al. 2003. New functions of lactoferrin and beta-casein in mammalian milk as cysteine protease inhibitors. Biochem. Biophys. Res. Commun. 306:98–103 [DOI] [PubMed] [Google Scholar]
- 37. Pan Y, Rowney P, Guo P, Hobman P. 2007. Biological properties of lactoferrin: an overview. Aust. J. Dairy Technol. 62:31–42 [Google Scholar]
- 38. Pathirana RD, O'Brien-Simpson NM, Veith PD, Riley PF, Reynolds EC. 2006. Characterization of proteinase-adhesin complexes of Porphyromonas gingivalis. Microbiology 152:2381–2394 [DOI] [PubMed] [Google Scholar]
- 39. Pihlanto A, Korhonen H. 2003. Bioactive peptides and proteins. Adv. Food Nutr. Res. 47:175–276 [DOI] [PubMed] [Google Scholar]
- 40. Rainard P. 1986. Bacteriostatic activity of bovine milk lactoferrin against mastitic bacteria. Vet. Microbiol. 11:387–392 [DOI] [PubMed] [Google Scholar]
- 41. Reich E. 2001. Trends in caries and periodontal health epidemiology in Europe. Int. Dent. J. 51:392–398 [DOI] [PubMed] [Google Scholar]
- 42. Shi Y, Kong W, Nakayama K. 2000. Human lactoferrin binds and removes the hemoglobin receptor protein of the periodontopathogen Porphyromonas gingivalis. J. Biol. Chem. 275:30002–30008 [DOI] [PubMed] [Google Scholar]
- 43. Shimazaki K. 2000. Lactoferrin: a marvelous protein in milk? Animal Sci. J. 71:329–347 [Google Scholar]
- 44. Singh PK. 2004. Iron sequestration by human lactoferrin stimulates P. aeruginosa surface motility and blocks biofilm formation. Biometals 17:267–270 [DOI] [PubMed] [Google Scholar]
- 45. Singh PK, Parsek MR, Greenberg EP, Welsh MJ. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552–555 [DOI] [PubMed] [Google Scholar]
- 46. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134–144 [DOI] [PubMed] [Google Scholar]
- 47. Spahr A, et al. 2006. Periodontal infections and coronary heart disease: role of periodontal bacteria and importance of total pathogen burden in the Coronary Event and Periodontal Disease (CORODONT) study. Arch. Intern. Med. 166:554–559 [DOI] [PubMed] [Google Scholar]
- 48. Stolzenberg-Solomon RZ, et al. 2003. Tooth loss, pancreatic cancer, and Helicobacter pylori. Am. J. Clin. Nutr. 78:176–181 [DOI] [PubMed] [Google Scholar]
- 49. Szedlacsek SE, Duggleby RG. 1995. Kinetics of slow and tight-binding inhibitors. Methods Enzymol. 249:144–180 [DOI] [PubMed] [Google Scholar]
- 50. Toh EC, et al. 2011. Porphyromonas gingivalis cysteine proteinase inhibition by kappa-casein peptides. Antimicrob. Agents Chemother. 55:1155–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tripos International 2008. SYBYL 8.1. Tripos International, St. Louis, MO [Google Scholar]
- 52. van Hooijdonk AC, Kussendrager KD, Steijns JM. 2000. In vivo antimicrobial and antiviral activity of components in bovine milk and colostrum involved in non-specific defence. Br. J. Nutr. 84(Suppl 1):S127–S134 [DOI] [PubMed] [Google Scholar]
- 53. van Veen HA, Geerts ME, van Berkel PH, Nuijens JH. 2004. The role of N-linked glycosylation in the protection of human and bovine lactoferrin against tryptic proteolysis. Eur. J. Biochem. 271:678–684 [DOI] [PubMed] [Google Scholar]
- 54. Wakabayashi H, et al. 2009. Inhibitory effects of lactoferrin on growth and biofilm formation of Porphyromonas gingivalis and Prevotella intermedia. Antimicrob. Agents Chemother. 53:3308–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wei Z, Nishimura T, Yoshida S. 2001. Characterization of glycans in a lactoferrin isoform, lactoferrin-a. J. Dairy Sci. 84:2584–2590 [DOI] [PubMed] [Google Scholar]
- 56. Wen ZT, Burne RA. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yamini S, et al. 2011. Crystal structure of C-lobe of bovine lactoferrin complexed with Nabumetone at 1.7–Å resolution. PDB ID 3TAJ. National Center for Biotechnology Information, Bethesda, MD: http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=3TAJ [Google Scholar]