Abstract
We describe a novel Tn4401 variant (Tn4401d) in epidemic Klebsiella pneumoniae clone ST258, from which a partial blaKPC fragment has been excised along with ISKpn7 and a partial tnpA fragment. Nested-PCR experiments confirmed that this region can be removed from distinct Tn4401 isoforms in both K. pneumoniae and Escherichia coli. This study highlights that the region surrounding blaKPC is undergoing recombination and that Tn4401 itself is heterogeneous and highly plastic.
TEXT
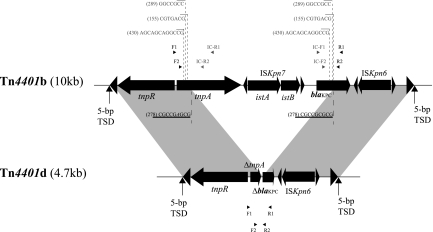
Klebsiella pneumoniae carbapenemase (KPC) is a molecular class A serine β-lactamase belonging to functional group 2f, which hydrolyzes β-lactams of several different classes, including carbapenems (1). The enzyme is encoded by the blaKPC gene, which maps to a transposon (Tn4401) that has been reported in a variety of transferable plasmids (10, 17). Tn4401 is approximately 10 kb in size, is delimited by two 39-bp imperfect inverted repeat sequences, and harbors insertion sequences ISKpn6 and ISKpn7 in addition to transposase and resolvase genes (Fig. 1) (5). Three isoforms (a, b, and c) of Tn4401 have been described, differing by a 100- to 200-bp sequence upstream of blaKPC (6, 17), while isoforms with 68-bp (13), 215-bp (GenBank accession no. DQ989640), and 255-bp (14) deletions were recently reported. Several descriptions of heterogeneous genetic environments have been reported for blaKPC, including the presence of other insertion sequences upstream of the blaKPC gene (7, 19, 20), suggesting that the region upstream of blaKPC is variable (5). Here we describe a novel truncated Tn4401 structure, tentatively named Tn4401d, with a 5.3-kb deletion encompassing more than half of the blaKPC gene, along with ISKpn7 and a partial tnpA gene fragment located upstream of Tn4401 (Fig. 1).
Fig 1.
Structure of truncated Tn4401 variant Tn4401d. Gray shading denotes regions of homology shared by Tn4401b and Tn4401d. Open reading frames (ORFs) are portrayed as large black arrows. Excision sites found in BK32529 are indicated by dotted lines beneath Tn4401b, with characteristic direct repeat (DR) sequences underlined. Imperfectly homologous sequences (with mismatches) within the two DRs are shown in italics. Numbers within parentheses denote the nested-PCR product sizes listed in Table 2. Additional potential excision sites determined by nested PCR are indicated by dotted lines above Tn4401b, with the corresponding direct repeat sequences shown in gray. Small black arrowheads represent the locations of primers used for nested PCR; small gray arrowheads indicate the locations of primers used for outward-directed nested PCR. Primers used for nested PCR were as follows: F1 (AATGCCCCATGTTTCTACGA), R1 (GGTCGTGTTTCCCTTTAGCC), F2 (TCACCAAGCATGAACGCTAC), and R2 (GCAGAGCCCAGTGTCAGTTT). Primers used for outward-directed nested PCR were as follows: IC-F1 (ATCGCCGTCTAGTTCTGCTG), IC-R1 (CAGCAGGTAGAGTTGGGTCTG), IC-F2 (CAGCTCATTCAAGGGCTTTC), and IC-R2 (CGTCGAGTTTAGGCAGCAGT). TSD, target site duplication.
K. pneumoniae isolate BK32529 was recovered from a urine culture from a patient with a urinary tract infection in a New York City hospital. Multilocus sequence typing (MLST) confirmed that the isolate belonged to the epidemic K. pneumoniae ST258 clone (9). Species identification and antibiotic susceptibility were initially determined using a Vitek-2 instrument (bioMérieux, Lyon, France). The isolate exhibited resistance to amikacin, aztreonam, cefepime, cefotaxime, ceftazidime, ceftriaxone, ciprofloxacin, ertapenem, gentamicin, levofloxacin, piperacillin, tigecycline, tobramycin, and trimethoprim-sulfamethoxazole but was susceptible to imipenem and meropenem. Additional susceptibility testing by broth microdilution (4) demonstrated that the isolate was resistant to ertapenem (MIC > 4 mg/liter), meropenem (MIC = 2 mg/liter), and doripenem (MIC = 2 mg/liter) but not imipenem (MIC ≤ 1 mg/liter) (Table 1).
Table 1.
Characteristics of K. pneumoniae strain BK32529 and the Tn4401d-harboring E. coli DH10 transformant
| Isolate | InC type(s)a | β-Lactamasesb | MIC (mg/liter)c |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMP | MER | ERT | DOR | CAZ | CTX | CEF | ATM | AMI | GEN | TOB | TIC-CLAV | PIP-TAZ | CIP | LEV | SXT | TGC | COL | PLB | |||
| BK32529 | A/C, FIIs | TEM-1, SHV-12 | ≤1 | 2 | >4 | 2 | >16 | >32 | >16 | >16 | >32 | >8 | >8 | >128/2 | >64/4 | >2 | >8 | >4/76 | 1 | >4 | >4 |
| DH10B transformant | A/C | TEM-1, SHV-12 | ≤1 | ≤1 | ≤0.25 | ≤0.12 | >16 | 4 | ≤2 | >16 | >32 | 8 | >8 | 64/2 | <8/4 | ≤0.25 | ≤1 | >4/76 | 0.5 | 0.5 | 0.5 |
Plasmid incompatibility (InC) groups were determined using the multiplex-PCR method described by Carattoli et al. (2).
blaCTX-M, blaSHV, blaTEM, blaGES, blaNDM, blaVIM, blaIMP, blaOXA-48, blaBIC, blaSPM, blaGIM, blaAIM, blaSIM, and blaDIM, as well as AmpC β-lactamases blaACT-1, blaACC, blaBIL-1, blaCMY, blaDHA, blaFOX, blaLAT, blaMIR-1, and blaMOX, were identified by PCR using methods described elsewhere (8, 18).
MICs were determined using broth microdilution; resistance is indicated in boldface. IMP, imipenem; MER, meropenem; ERT, ertapenem; DOR, doripenem; CAZ, ceftazidime; CTX, cefotaxime; CEF, cefepime; ATM, aztreonam; AMI, amikacin; GEN, gentamicin; TOB, tobramycin; TIC-CLAV, ticarcillin-clavulanate; PIP-TAZ, piperacillin-tazobactam; CIP, ciprofloxacin; LEV, levofloxacin; SXT, trimethoprim-sulfamethoxazole; TGC, tigecycline; COL, colistin; PLB, polymyxin B.
A previously described molecular-beacon-based multiplex real-time PCR assay (3) used in routine surveillance of suspected carbapenem-resistant Enterobacteriaceae displayed an anomalous result suggestive of a novel blaKPC variant. Specifically, isolate BK32529 was positive only for the 716T target and negative for the other five single nucleotide polymorphisms (SNPs) associated with known blaKPC variants (147A, 308T, 308C, 716C, and 814C). However, we were unable to amplify the entire blaKPC region using previously published primers (16), suggesting the presence of a potential deletion or insertion within the blaKPC gene. The entire Tn4401 region was therefore amplified using long-range PCR with primers 4401-it-F1 (ACGTCGTGGCGATCGACGCA) and 4401-it-R2 (TTCCAGGTCCGCAATAGTTC), followed by DNA sequencing using primer walking. The results revealed the presence of a 5.3-kb deletion within Tn4401, from nucleotide (nt) 537 in tnpA to nt 553 in blaKPC, thereby encompassing all of ISKpn7 (istA and istB), 80% of tnpA, and 60% of blaKPC (Fig. 1). This truncated Tn4401 structure has been tentatively named Tn4401d (Tn4401 deletion). Further inspection of the deletion sites in Tn4401 indicated that the deleted region is bracketed by two 9-bp imperfectly matched direct repeats (DRs) in tnpA (CGCCGAGCG) and blaKPC (CGCCGCGCG), respectively (the mismatched nucleotides are underlined) (Fig. 1), suggesting that the deletion in Tn4401d may have arisen through DR-mediated slippage.
As shown in Fig. 1, only 40% of the blaKPC gene is still present in Tn4401d. This finding calls into question the functionality of truncated blaKPC, as the clinical isolate is still resistant to three carbapenems (ertapenem, meropenem, and doripenem) (Table 1). In order to transfer the Tn4401-harboring plasmid into a different genetic background, plasmid DNA was extracted from BK32529 using a Qiagen plasmid maxi kit (Qiagen, Valencia, CA), followed by electroporation into Escherichia coli DH10B using a Gene Pulser II instrument (Bio-Rad Laboratories). Potential transformants were selected on LB agar plates containing 100 μg/ml ampicillin and then screened by multiplex real-time PCR for the presence of truncated blaKPC genes (positive only for target 716T, as described above) (3). Plasmid size and copy number were estimated by S1 nuclease digestion of plasmid DNA, followed by pulsed-field gel electrophoresis (PFGE) using a Bio-Rad CHEF-DR III variable-angle system. Transformants with single plasmids ∼70 kb in size were then selected and subjected to susceptibility testing according to CLSI guidelines (4).
In contrast to the parental strain, the transformant was susceptible to all four carbapenems tested (MICs were ≤1 mg/liter for imipenem and meropenem and ≤0.25 and ≤0.12 mg/liter for ertapenem and doripenem, respectively) but resistant to aztreonam, ceftazidime, tobramycin, and trimethoprim-sulfamethoxazole, suggesting that the truncated blaKPC enzyme is not functional. Further genotyping failed to identify additional carbapenem resistance determinants (blaVIM, blaIMP, blaNDM, blaOXA-48, etc.) or AmpC β-lactamase genes within the parental isolate (Table 1), while the modified Hodge test was negative for both the parental strain and the corresponding DH10B transformant (15). Porin genes ompK35 and ompK36 were then investigated using a PCR assay described elsewhere (12), followed by DNA sequencing. The results revealed an additional guanine (G) insertion at nt 121 within ompK35, resulting in a premature stop codon at amino acid position 89. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of outer membrane proteins (OMP) (12) from BK32529 confirmed the loss of OmpK35 (data not shown). Several nonsynonymous mutations were identified within the sequence of ompK36, but SDS-PAGE indicated that the full-length protein was still expressed (data not shown). The apparent loss of porin OmpK35, as well as the presence of other β-lactamases, such as TEM-1 and SHV-12 (Table 1), may contribute to the carbapenem resistance observed within strain BK32529, as suggested previously (11).
In order to investigate whether the deleted region (from tnpA to blaKPC) could be excised from Tn4401, nested PCR was performed using primer sets F1/R1 and F2/R2 as outer and inner primers, respectively (Fig. 1). In addition, an outward-directed nested PCR was designed to identify the extrachromosomal circular intermediate (CI) structure resulting from excision of Tn4401, using primer sets IC-F1/R1 and IC-F2/R2 as outer and inner primers, respectively.
For nested-PCR amplification, the initial PCR products from primers F1/R1 and IC-F1/R1 were diluted 1:1,000 with ultrapure water and then subjected to a second round of amplification with primers F2/R2 and IC-F2/R2. Plasmids from 10 K. pneumoniae and five E. coli isolates with different genotypic backgrounds and harboring different Tn4401 and blaKPC variants were subjected to nested and outward-directed PCR (Table 2). Several deletion structures, in addition to the deletion pattern found in strain BK32529 (corresponding to a 278-bp PCR product generated by primer set F2/R2), were identified by nested PCR (Table 2). Meanwhile, outward-directed nested PCR confirmed the existence of circular intermediates among these structures. Further sequence analysis revealed several potential spontaneous excision sites, including several DRs within tnpA and blaKPC, three of which are depicted in Fig. 1. However, no DRs were identified in some of the novel deletion structures (corresponding to 191-, 215-, 217-, and 255-bp PCR products generated by primer set F2/R2) (Table 2), indicating that there may be other deletion mechanisms besides DR-mediated slippage. No apparent associations were observed between the Tn4401 excision pattern and host species, MLST genotype, Tn4401 isoform, or blaKPC variant (Table 2).
Table 2.
blaKPC excision patterns in additional Tn4401-bearing strains
| Strain | Species | InC type(s)a | MLSTb | KPC | Tn4401 variantc | Size(s) of Tn4401 deletion pattern(s) (bp)d |
|---|---|---|---|---|---|---|
| 1 | K. pneumoniae | N, FIIs | 63 | KPC-4 | b | 160, 278, 430 |
| 2 | K. pneumoniae | Unk | 76 | KPC-3 | a | 155, 217 |
| 3 | K. pneumoniae | FIIs | 234 | KPC-2 | a | 289 |
| 4 | K. pneumoniae | FIIs | 258 | KPC-2 | a | 278, 430 |
| 5 | K. pneumoniae | FIIs | 258 | KPC-3 | b | 186, 278, 430 |
| 6 | K. pneumoniae | N, A/C | 258 | KPC-2 | a | 215, 278, 430 |
| 7 | K. pneumoniae | Unk | 258 | KPC-2 | a | 191, 278, 430 |
| 8 | K. pneumoniae | FIIs | 258 | KPC-3 | b | 278, 430 |
| 9 | K. pneumoniae | FIIs | 258 | KPC-3 | −68 | 255 |
| 10 | K. pneumoniae | A/C | 486 | KPC-2 | b | 278, 430 |
| 11 | E. coli | L1, N, Y, FIA, FIB, FIIs | 2 | KPC-2 | b | 278, 430 |
| 12 | E. coli | L1, N, Y, FIA, FIB, FIIs | 2 | KPC-3 | b | 278, 430 |
| 13 | E. coli | L1, FIA, FIB, FIIs | 43 | KPC-2 | a | 278, 430 |
| 14 | E. coli | FIB, A/C, Frep | 33 | KPC-2 | a | 278, 430 |
| 15 | E. coli | A/C | 35 | KPC-3 | b | 278 |
Plasmid replicon typing was performed using a method previously described by Carattoli et al. (2). Unk, unknown.
MLST for K. pneumoniae was performed using the method described by Diancourt et al. (9); MLST for E. coli was performed using the Institut Pasteur MLST scheme (http://www.pasteur.fr/recherche/genopole/PF8/mlst/EColi.html).
Tn4401 variants were identified by PCR (6), followed by DNA sequencing. “−68” indicates the previously described variant with a 68-bp deletion upstream of blaKPC (13).
Tn4401 deletion patterns were identified by nested PCR and are expressed as the PCR product length resulting from amplification with primer set F2/R2.
Taken together, these results demonstrate that blaKPC can be partially excised from Tn4401, thereby causing a loss of carbapenemase activity. The observation of different excision patterns highlights the notion that the region surrounding blaKPC is undergoing recombination. Further study of the mechanisms underlying excision may therefore be helpful in controlling the spread of blaKPC in K. pneumoniae and other Enterobacteriaceae.
Nucleotide sequence accession number.
The nucleotide sequence of Tn4401d from K. pneumoniae strain BK32529 was deposited in GenBank as accession no. JN974188.
ACKNOWLEDGMENTS
This study was supported by a grant (to B.N.K.) from the National Institutes of Health (1R01AI090155-01A1). The Veterans Affairs Merit Review Program, Geriatric Research Education and Clinical Care (GRECC), and the National Institutes of Health (grant RO1 AI063517-01) supported R.A.B.
Footnotes
Published ahead of print 27 December 2011
REFERENCES
- 1. Bush K, Fisher JF. 2011. Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 65:455–478 [DOI] [PubMed] [Google Scholar]
- 2. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 3. Chen L, et al. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (blaKPC) variants. J. Clin. Microbiol. 49:579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CLSI 2010. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement (June 2010 update). M100-S20-U CLSI, Wayne, PA [Google Scholar]
- 5. Cuzon G, Naas T, Nordmann P. 2011. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob. Agents Chemother. 55:5370–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cuzon G, et al. 2010. Worldwide diversity of Klebsiella pneumoniae that produce β-lactamase blaKPC-2 gene. Emerg. Infect. Dis. 16:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cuzon G, et al. 2011. Wide dissemination of Pseudomonas aeruginosa producing β-lactamase blaKPC-2 gene in Colombia. Antimicrob. Agents Chemother. 55:5350–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 9. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gootz TD, et al. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 53:1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jacoby GA, Mills DM, Chow N. 2004. Role of β-lactamases and porins in resistance to ertapenem and other beta-lactams in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:3203–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin phoe. Antimicrob. Agents Chemother. 50:3396–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kitchel B, et al. 2010. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54:4201–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kitchel B, et al. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee K, et al. 2001. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 7:88–91 [DOI] [PubMed] [Google Scholar]
- 16. Lomaestro BM, Tobin EH, Shang W, Gootz T. 2006. The spread of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae to upstate New York. Clin. Infect. Dis. 43:e26–e28 [DOI] [PubMed] [Google Scholar]
- 17. Naas T, et al. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123 [DOI] [PubMed] [Google Scholar]
- 19. Shen P, et al. 2009. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53:4333–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolter DJ, et al. 2009. Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob. Agents Chemother. 53:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]