Abstract
Analysis of the evolution of drug target genes under changing drug policy is needed to assist monitoring of Plasmodium falciparum drug resistance in the field. Here we genotype Pfcrt and Pfdmr1 of 700 isolates collected in French Guiana from 2000 (5 years after withdrawal of chloroquine) to 2008, i.e., the period when the artemether-lumefantrine combination was progressively introduced and mefloquine was abandoned. Gene sequencing showed fixation of the 7G8-type Pfcrt SMVNT resistance haplotype and near fixation of the NYCDY Pfdmr1 haplotype. Pfdmr1 gene copy number correlated with 50% inhibitory concentrations of mefloquine and halofantrine (r = 0.64 and 0.47, respectively, n = 547); its temporal changes paralleled changes in in vitro mefloquine susceptibility. However, the molecular parameters studied did not account for the regained in vitro susceptibility to chloroquine and showed a poor correlation with susceptibility to artemether, lumefantrine, or quinine. Identification of novel markers of resistance to these antimalarials is needed in this South American area.
INTRODUCTION
Plasmodium falciparum drug resistance is a major concern for malaria control. Mutations of the P. falciparum chloroquine resistance transporter (Pfcrt), a member of the drug metabolite transporter superfamily, have been identified as causal determinants of chloroquine resistance in vitro (17, 41) and associated with chloroquine treatment failures (13). Susceptibility to mefloquine is associated with single nucleotide polymorphisms (SNPs) and copy number variation of Pfmdr1 (P. falciparum multidrug resistance protein 1), a P-glycoprotein homolog (34, 35). Pfcrt and Pfmdr1 haplotypes interact to modulate in vitro sensitivity to amodiaquine and chloroquine; this interaction is particularly marked with alleles originating from South America (37). Furthermore, Pfcrt and Pfmdr1 haplotypes influence in vitro susceptibility to multiple antimalarials (23, 34, 36, 37, 39–41), probably by altering their intracellular transport (38).
Understanding of the evolution of drug target genes under changing drug policy is crucial for drug efficacy monitoring using molecular markers. In Malawi, withdrawal of chloroquine was followed by the expansion of chloroquine-sensitive parasites possessing a wild-type Pfcrt allele, apparently better fit than mutant parasites in the absence of chloroquine pressure (26, 32). A similar phenomenon was reported in China (44). Expansion of resistance to mefloquine following its deployment in Thailand was associated with a temporal increase of the frequency of parasites with an increased Pfmdr1 copy number and harboring specific SNPs (35). Here, we analyze the association of the Pfcrt and Pfmdr1 genotypes and Pfmdr1 gene copy number with the temporal changes of in vitro susceptibility in a series of isolates collected over the years 2000 to 2008 across French Guiana.
Chloroquine was abandoned as a first-line treatment for P. falciparum malaria in French Guiana in 1995 for the quinine-doxycycline combination, which was progressively replaced since 2002 by the artemether-lumefantrine combination. Mefloquine or halofantrine was used as a second-line treatment from 1990 to 2002, when it was replaced with atovaquone-proguanil (28). We reported that these changes impacted on in vitro drug sensitivity profiles. In vitro resistance to chloroquine declined in the years following its withdrawal to reach an approximately 50% frequency. Sensitivity to mefloquine decreased progressively after its introduction. Susceptibility to amodiaquine was correlated positively with susceptibility to chloroquine and more moderately with susceptibility to mefloquine or halofantrine (28).
We analyze here the temporal changes of Pfcrt and Pfmdr1 gene polymorphisms and of Pfmdr1 gene copy number during the years 2000 to 2008, i.e., the period starting 5 years after the withdrawal of chloroquine and encompassing the implementation of artemether-lumefantrine as a replacement for mefloquine. We report that Pfmdr1 copy number variation reflected the temporal evolution of in vitro mefloquine susceptibility. However, the molecular parameters studied did not account for the regained susceptibility to chloroquine and were poorly associated with responses to quinine, lumefantrine, and artemether.
MATERIALS AND METHODS
In vitro drug sensitivity assays.
The in vitro susceptibility to a panel of antimalarials of 513 P. falciparum isolates collected longitudinally during the years 2000 to 2005 by our reference center has been reported (28). Similar procedures were used for 187 isolates collected in the years 2006 to 2008. An aliquot of a patient isolate was cultured for 24 h in the absence of drug, frozen at −80°C, and subsequently processed for RNA and DNA extraction.
DNA extraction and genotyping.
DNA was extracted as described previously (27). Pfmsp1 block2 genotyping was carried out as described previously (23, 27). Pfcrt exon2 and exon4 amplification was performed as described previously (1, 19). Three overlapping PCR were used to amplify the full-length Pfmdr1 sequence. The conditions were 1.5 mM MgCl2, 2 μM each primer, and 200 μM deoxynucleoside triphosphate, 1.5 U AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA) in a final volume of 50 μl, with 1 cycle of 94°C for 8 min; 35 cycles of 94°C for 1 min, incubation at the annealing temperature for 1 min, and elongation at 72°C for 2 min; and final extension at 72°C for 10 min (for the primer sequence and annealing temperature, see Table S1 in the supplemental material). PCR products were analyzed on 2% agarose gels and stored at −20°C before being processed for direct sequencing.
mRNA extraction and cDNA synthesis.
Following maturation in vitro, RNA was extracted at the late trophozoite stage using QIAamp RNA Blood (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. For each isolate, 200 μl of whole blood was treated. Five micrograms of total RNA was reverse transcribed using random hexamer priming and Moloney murine leukemia virus reverse transcriptase (Superscript First-Strand kit; Invitrogen, Cergy Pontoise, France). The full-length Pfcrt coding cDNA sequence was amplified by nested PCR as described previously (15).
Direct sequencing of PCR products.
PCR products were sequenced using ABI Prism BigDye Terminator chemistry as described previously (16). For the Pfmdr1 central region, specific internal primers were used to determine the whole sequence on both strands. Sequence analysis was done using the Phred Phrap package. Sequences with segments of ≥1,000 bp called with a quality over 20 per base were retained. Sequences of insufficient quality were either resequenced or rejected. Sequence assembly was done with Seqscape software v.2.0 (Applied Biosystems). SNPs were considered only if observed unambiguously.
Assessment of Pfmdr1 gene copy number and expression level.
Pfmdr1 (PFE1150w), β-tubulin (PF10_0084), and maebl (PF11_0486) copy numbers and mRNA expression were assessed by TaqMan real-time PCR (ABI 7300 real-time PCR systems; Applied Biosystems, Warrington, United Kingdom) (for primer sequences, see Table S2 in the supplemental material). The Pfmdr1 probes were FAM (6-carboxyfluorescein) labeled at the 5′ end, and the β-tubulin and maebl probes were VIC labeled (Table S2). All probes had a TAMRA (6-carboxytetramethylrhodamine) label at the 3′ end. All samples were assayed in triplicate in 96-well optical reaction plates in a 25-μl final volume containing 4 μl genomic DNA or cDNA, 1 μM each primer, and 0.25 μM fluorogenic probe in Universal PCR Master Mix (Applied Biosystems). The amplification conditions for all loci were 1 cycle at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min.
The relative mRNA levels were quantified and compared as described previously (30). The fold change in gene expression was normalized to the maebl or β-tubulin control gene.
Statistical analysis.
The association of the 50% inhibitory concentration (IC50) of each antimalarial, considered a quantitative variable with the Pfmdr1 genotype or gene copy number, was analyzed using the Pearson correlation test. The association of IC50 classes (see Table 1) with Pmdr1 copy number was examined using the Kruskal-Wallis test. The Spearman correlation coefficient was calculated. All tests were performed with SAS, version 9.1 (SAS Institute Inc., Cary, NC). Results were considered significant if the P value was below 0.05.
Table 1.
Overview of in vitro susceptibility of isolates from French Guianaa
| IC50 range | % (no.) of isolates |
||||||
|---|---|---|---|---|---|---|---|
| CQ | Quin | Mef | Hal | Art | Amo | Lum | |
| Total | 100 (700) | 100 (698) | 100 (684) | 100 (690) | 100 (600) | 100 (370) | 100 (280) |
| High | 53.4 (374) | 4.7 (33) | 20.2 (138) | 14.5 (100) | 4.8 (29) | 4.3 (16) | 27 (76) |
| Intermediate | 9.1 (64) | 12.8 (89) | 21.6 (148) | 25.8 (178) | 3.4 (20) | 5.3 (20) | 7.1 (20) |
| Low | 37.4 (262) | 82.5 (576) | 58.2 (398) | 39.7 (412) | 91.8 (551) | 90.4 (334) | 65.8 (184) |
Shown are percentages of isolates classified as resistant, intermediate, and susceptible based on the IC50 of each drug. Three classes were considered for each antimalarial. The IC50 cutoff for each class was as follows: CQ, chloroquine (high [resistant], IC50 of >100 nM; intermediate, 80 < IC50 < 100 nM; low [susceptible], IC50 of <80 nM); Quin, quinine (high [resistant], IC50 of >500 nM; intermediate, 300 < IC50 < 500 nM; low [susceptible], IC50 of <300 nM); Mef, mefloquine (high [resistant], IC50 of >30 nM; intermediate, 15 < IC50 < 30 nM; low [susceptible], IC50 of <15 nM); Hal, halofantrine (high, IC50 of >8 nM; intermediate, 4 < IC50 < 8 nM; low, IC50 of <4 nM); Art, artemether (high, IC50 of >12 nM; intermediate, 8 < IC50 < 12 nM; low, IC50 of <8 nM); Amo, monodesthyl-amodiaquine (high, IC50 of >60 nM; intermediate, 40 < IC50 < 60 nM; low, IC50 of <40 nM); Lum, lumefantrine (high, IC50 of >150 nM; intermediate, 100 < IC50 < 150 nM; low, IC50 of <100 nM).
RESULTS
Drug susceptibility and infection complexity.
A panel of 700 isolates was collected and tested for in vitro drug susceptibility by our reference center during the period 2000 to 2008 (Table 1). IC50s of mefloquine and halofantrine were correlated (r = 0.63; P < 0.0001); 88% of the isolates with a high IC50 of mefloquine (>30 nM) had an elevated IC50 of halofantrine. The IC50s of chloroquine and quinine, mefloquine, or doxycycline were unrelated.
Pfmsp1 block2 genotyping of a subset of 61 isolates showed a very high rate (91.4%) of single infection, confirming previous studies (27).
Pfcrt genotype and cDNA sequence analysis.
The Pfcrt exon 2-exon 4 genotype was determined for 628 samples, including 212 chloroquine-sensitive samples. Unambiguous sequencing of 626 isolates showed the same haplotype at positions 72 to 76, 220 coding for SVMNT/S, associated with a monomorphic intron 4 microsatellite (TAAA)3(TA)14 in all isolates. Sequencing of reference alleles from laboratory lines amplified and run in the same experiments ruled out possible experimental contamination artifacts.
To determine whether additional mutations in the Pfcrt gene were present in the chloroquine-susceptible isolates, a complete cDNA sequence was determined for 53 samples (25 chloroquine sensitive and 28 chloroquine resistant). All but one cDNA had a C72S K76T A220S N326D I356L protein sequence haplotype, and 6 (11%) had the 72S residue encoded by AGT (i.e., the 7G8 allele, also called SMVNT1), while the other 47 sequences (89%) had a TCT codon (a 7G8 type also called SMVNT2). One susceptible sample, with a chloroquine IC50 of 22 nM, carried an additional C350R Pfcrt mutation. In brief, the Pfcrt genotype of the panel of isolates studied was unrelated to the observed IC50 of chloroquine or of any of the molecules from the panel of antimalarials tested.
Pfmdr1 sequencing.
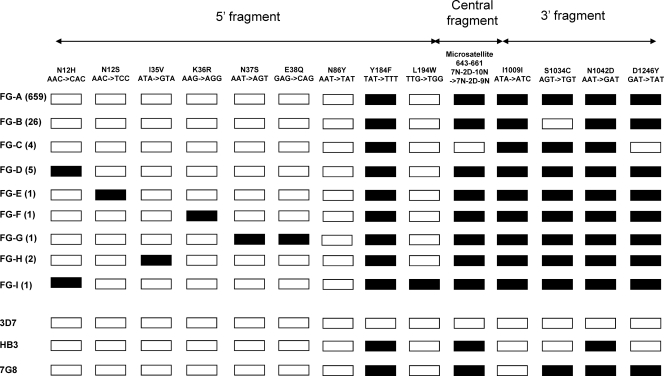
A full-length Pfmdr1 coding sequence was established for 700 isolates. Overall, nine alleles, called FG-A to FG-I, were observed (Fig. 1). Interestingly, all contained an as-yet-undescribed I1009I synonymous mutation. One highly dominant haplotype was observed (allele FG-A, 94% of the isolates) that differed from the 7G8-type haplotype at position 1009. A double mutant S1034C codon was observed in all samples, except 26 isolates harboring a wild-type codon 1034 (allele FG-B), which all originated from a single village (Papaïchton) and were collected in the years 2000 and 2001. The remaining seven alleles (FG-C to FG-I) were rare.
Fig 1.
Pfmdr1 sequence polymorphism of 700 isolates collected in French Guiana during the years 2000 to 2008. Polymorphic codons are indicated. Open symbols denote the wild-type (3D7-type) nucleotide sequence, and black symbols indicate the presence of the mutant sequence shown at the top. The reference alleles 3D7 (PFE1150w, http://www.plasmodb.org/plasmo/), HB3, and 7G8, (http://www.broad.mit.edu/annotation/genome/plasmodium_falciparum_spp/) are shown for comparison. The accession numbers of alleles FG-A to -I, respectively, are HQ215524 to HQ215532. The number of isolates observed for each allele is indicated in parentheses.
The Pfmdr1 allelic type was unrelated to the IC50 of any of the antimalarials tested. As all isolates but one harbored the same mutant Pfcrt haplotype, the combined Pfmdr1/Pfcrt genotype was also unrelated to any of the IC50s.
Pfmdr1 copy number and its relationship with in vitro resistance.
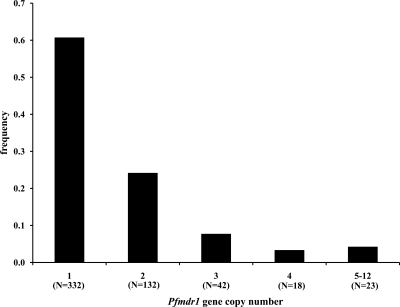
The Pfmdr1 copy number was determined in 547 samples using β-tubulin or Pfmaebl as a reference. Identical estimates were obtained for both. The Pfmdr1 copy number varied from 1 to 12 copies, and 39% of the samples had ≥2 copies (Fig. 2). Amplification was observed for the FG-A, FG-D, FG-G, FG-H, and FG-I alleles.
Fig 2.
Frequency distribution of Pfmdr1 gene copy numbers in 547 isolates collected in French Guiana during the years 2001 to 2008. The number of isolates per group is shown under each column. The 5-to-12 group includes 9, 8, 3, 1, 1, and 1 isolates with 5, 6, 7, 8, 9, and 12 copies, respectively.
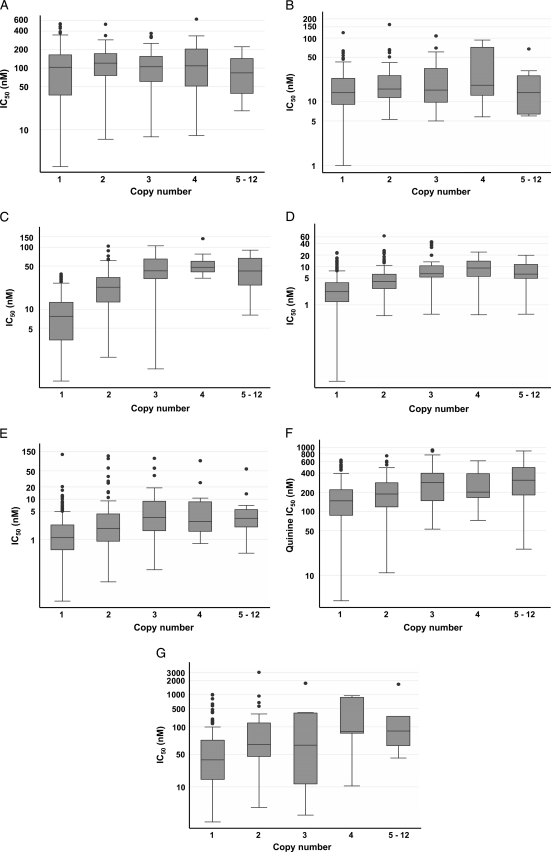
Figure 3 shows the distribution of the IC50s of the antimalarials by Pfmdr1 gene copy number. Chloroquine and amodiaquine susceptibility was unrelated to the Pfmdr1 copy number (P = 0.16 and 0.12, respectively, Kruskal-Wallis test), in contrast to the susceptibility to mefloquine, halofantrine, quinine, and lumefantrine (P = 0.0013, 0.026, 0.009, and 0.049, respectively). A low correlation with an artemether IC50 of >12 nM was also observed (P = 0.031). The presence of multiple (i.e., ≥2) Pfmdr1 copies positively correlated with in vitro resistance to mefloquine and halofantrine and was moderately associated with in vitro resistance to quinine, lumefantrine, and artemether (Table 2).
Fig 3.
Distribution of IC50s of chloroquine (A), amodiaquine (B), mefloquine (C), halofantrine (D), artemether (E), quinine (F), and lumefantrine (G) by Pfmdr1 gene copy number in 547 isolates collected in French Guiana during the years 2001 to 2008. The number of isolates per group is as in Fig. 2. Shown is a boxplot representation of the IC50 of each antimalarial. The boundaries of the boxes indicate the 25th (Q1) and 75th (Q3) percentiles, the line in each box indicates the median, and the whiskers indicate the IC50 range corresponding to Q1 − 1.5 × (Q3 − Q1) and Q3 + 1.5 × (Q3 − Q1) for the lower whisker and upper whisker, respectively. The outlying dots show values exceeding this range.
Table 2.
Spearman correlation coefficients of increased (≥2) Pfmdr1 copy number and in vitro resistance
| Molecule | Spearman correlation coefficient (r) | P value |
|---|---|---|
| Chloroquine | 0.06 | 0.17 |
| Quinine | 0.30 | <0.0001 |
| Mefloquine | 0.64 | <0.0001 |
| Halofantrine | 0.47 | <0.0001 |
| Artemether | 0.34 | <0.0001 |
| Atovaquone | −0.04 | 0.46 |
| Amodiaquine | 0.11 | 0.05 |
| Lumefantrine | 0.32 | <0.0001 |
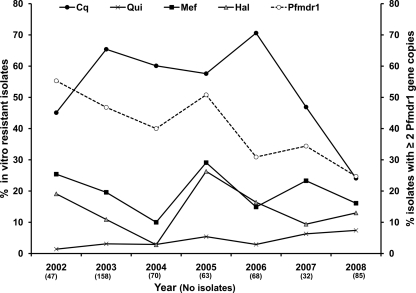
The association between Pfmdr1 amplification and elevated IC50s of mefloquine and halofantrine was also reflected by their parallel temporal fluctuation during the 2002 to 2008 period, corresponding to the progressive replacement of mefloquine for artemether-lumefantrine (Fig. 4). Interestingly, the proportion of isolates with multiple copies decreased in the years 2005 to 2008, when in vitro mefloquine resistance declined (P < 0.01).
Fig 4.
Temporal evolution of in vitro chloroquine (Cq), mefloquine (Mef), halofantrine (Hal), and quinine (Qui) resistance and frequency of Pfmdr1 gene amplification. The median chloroquine IC50s were 104, 118, 123, 123, 178, 70.5, and 80.1 nM in 2002, 2003, 2004, 2005, 2006, 2007, and 2008, respectively. The median quinine IC50s were 150.4, 179, 202, 194, 107.5, 163.5, and 170.6 nM in 2002, 2003, 2004, 2005, 2006, 2007, and 2008, respectively. The median mefloquine IC50s were 14.3, 13.9, 13.3, 14.4, 7.8, 4.3, and 2.5 nM in 2002, 2003, 2004, 2005, 2006, 2007, and 2008, respectively. The median halofantrine IC50s were 3.6, 3.4, 3.3, 3.8, 2.8, 2, and 1.9 nM in 2002, 2003, 2004, 2005, 2006, 2007, and 2008, respectively. The number of isolates studied each year is indicated in parentheses.
Pfmdr1 gene expression and its relationship with in vitro resistance.
The expression of Pfmdr1 was determined in 35 isolates harboring an FG-A haplotype (16 isolates with 1 copy and 19 isolates with 2 to 4 Pfmdr1 gene copies). The level of pfmdr1 mRNA of each isolate was compared to its level in 3D7, which harbors a single Pfmdr1 gene copy. The pfmdr1 mRNA levels of the various isolates did not linearly correlate with their Pfmdr1 gene copy numbers (Fig. 5A), with 11 isolates displaying discordant gene copy number to mRNA level ratios.
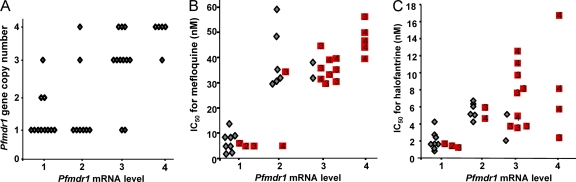
Fig 5.
Pfmdr1 gene expression levels (A) and their association with in vitro mefloquine (B) and halofantrine (C) resistance. Gray diamonds indicate isolates with one Pfmdr1 gene copy. Red squares indicate isolates with multiple gene copies (the number of gene copies is shown in each square).
Pfmdr1 gene expression showed a very strong correlation with the IC50 of mefloquine (Fig. 5B) (Spearman rank correlation coefficient r = 0.89; P < 0.0001). With the exception of one isolate with 4 gene copies, there was a perfect match between a ≥2-fold increased expression level and a resistance threshold of mefloquine IC50s above 30 nM in vitro. This correlation was stronger than the association with the Pfmdr1 copy number (r = 0.74 in this panel of isolates; P = 0.0001). A similar trend was observed for the in vitro response to halofantrine (r = 0.61; P = 0.0001), with a higher IC50 being associated with increased expression levels, but this was less clear-cut than for mefloquine (Fig. 5C). Pfmdr1 gene expression was unrelated to any of the other antimalarials tested (not shown).
DISCUSSION
The first remarkable finding of this work is the fixation of the SMNVT Pfcrt resistance haplotype and the quasifixation of a single, multiple mutant NYCDY Pfmdr1 haplotype in this parasite population. The most frequent polymorphism was the variation of Pfmdr1 gene copy number. Pfmdr1 copy number was associated with in vitro susceptibility to mefloquine and halofantrine, although the Pfmdr1 expression level was more informative than the copy number in the subset of isolates analyzed. Pfmdr1 copy number showed a modest association with susceptibility to quinine, lumefantrine, and artemether but no association with in vitro chloroquine or amodiaquine susceptibility profiles in this population. The restricted polymorphism of both loci indicates that additional loci modulate in vitro susceptibility to these antimalarials, which presented a relatively large dynamic range. Thus, Pfcrt and Pmdr1 gene/protein characteristics can no longer serve as molecular markers of in vitro susceptibility to chloroquine in this parasite population and moreover are of limited value for quinine, lumefantrine, and artemisinin derivatives. Novel markers need to be developed for improving monitoring of resistance to these antimalarials in this area.
In 1995, when chloroquine use for P. falciparum malaria was stopped in French Guiana, all isolates were resistant to chloroquine in vitro. In vitro resistance to chloroquine has declined since 2000 from 100% resistant isolates in 1999 to 24% resistant isolates in 2008 (28; this work). We show here that this temporal change cannot be attributed to the expansion of parasites carrying a wild-type Pfcrt allele, as previously observed in Africa (26, 32) and China (44). Indeed, as Pfcrt resistance alleles were fixed in this population at the time chloroquine was replaced, there were no wild-type alleles to compete against chloroquine resistance alleles and re-expand in the population. Furthermore, chloroquine has not been discontinued in the area, as it is still used to treat P. vivax malaria, which accounts for approximately half of the malaria cases in French Guiana (5). As a consequence, the bystander pressure on P. falciparum parasites may contribute to the maintenance of chloroquine-resistant Pfcrt in the population. Chloroquine sensitivity was unrelated to the Pfmdr1 sequence type, copy number, or gene expression level. In particular, the 86Y mutation associated with the in vitro resistance of field isolates to chloroquine in some studies (9, 14) was absent from the panel of isolates studied here. The 184F, 1034C, 1042D, and 1246D mutations associated with in vitro resistance to chloroquine of field isolates in some studies (18, 24) but not in allelic exchange experiments (36, 40) were almost at fixation and again unrelated to the IC50 of chloroquine. Therefore, neither the Pfcrt nor the Pfmdr1 locus accounts for the in vitro sensitivity of isolates collected in recent years across French Guiana. Importantly, such discordant genotypes were also observed in French Guiana isolates adapted to long-term in vitro culture (42). This indicates that additional loci govern the regained in vitro susceptibility to chloroquine in the parasite population from French Guiana. Although particularly striking in French Guiana due to the quite restricted polymorphism of the two loci reported to control resistance to chloroquine, the situation does not seem to be unique. Several recent studies reported a quasifixation of the Pfcrt resistance alleles in field populations from Southeast Asia, India, and East Africa and a lack of correlation with in vitro susceptibility to chloroquine and other antimalarials (3, 6, 7, 12).
In the panel of 700 isolates studied here, in vitro susceptibility to amodiaquine or chloroquine was not dependent on the interaction of the 7G8-type Pfcrt and 7G8-type Pfmdr1 alleles shown to modulate resistance to the two molecules in the 7G8 × GB4 genetic cross (37). Irrespective of their in vitro susceptibility profile, a large number of isolates harbored a 7G8-type Pfcrt haplotype associated with a Pfmdr1 allele identical to the 7G8 sequence, except for one synonymous mutation at position 1009. Although the possibility exists that the I1009I mutation has phenotypic repercussions, as observed for the human MDR protein (25), its fixation in French Guiana rules out an impact on the in vitro susceptibility profile. Our conclusion is that, in this parasite population, the interaction between the 7G8-type Pfcrt (SMVNT1 or SMVNT2), and 7G8-type Pfmdr1 haplotypes does not account for the in vitro susceptibility patterns to chloroquine. Amodiaquine IC50s varied over a 2-log concentration range, although 90% of the isolates were classified as presenting low or no resistance. This relative phenotypic homogeneity regarding amodiaquine is consistent with the restricted Pfcrt and Pfmdr1 polymorphism, although Sá et al. (37) reported that the specific 7G8 Pfcrt/ 7G8 Pfmdr1 haplotype combination and the 7G8 Pfmdr1 haplotype (NFCDY) were associated with resistance to amodiaquine. Reduced amodiaquine sensitivity in French Guiana was rather associated with an increased Pfmdr1 copy number, although the correlation was modest. It is worth noting that the 86Y Pfmdr1 mutation, associated with amodiaquine resistance in other settings (11, 21, 33), was totally absent from the French Guiana isolates studied here and that the NFY Pfmdr1 haplotype has been associated with an adequate treatment response in African settings where the Pfcrt allele was present at a very high frequency (11, 21). Unfortunately, most of the field studies conducted so far to investigate the relationship between amodiaquine susceptibility and Pfmdr1 polymorphisms did not investigate the full gene sequence and ignored polymorphic positions such as 1034 and 1042, precluding definitive comparisons with the data presented here. Nevertheless, our data indicate that apart from the Pfmdr1 gene copy number variation, no single mutation, haplotype, or combination showed a strong association with the observed variation of the amodiaquine IC50.
In vitro susceptibility to mefloquine and, to a lesser extent, halofantrine was associated with an increased Pfmdr1 copy number, which itself was modestly correlated with susceptibility to quinine, lumefantrine, or artemether. This is consistent with reports from Southeast Asia (7, 29) and with data from laboratory lines selected for resistance to mefloquine in vitro (10, 34). The temporal trends of Pfmdr1 amplification paralleled in vitro susceptibility to mefloquine, and in particular, the mean copy number decreased in the years following the switch from mefloquine monotherapy to the artemether-lumefantrine combination. These data are consistent with Pfmdr1 gene amplification driven by mefloquine monotherapy, deamplification upon cessation of mefloquine use, and reversal to susceptibility to mefloquine as observed by others (8).
The pilot study performed on a set of isolates harboring the FG-A allele outlined a very strong correlation between the IC50 of mefloquine and the Pfmdr1 transcript level. Elevated Pfmdr1 mRNA levels were observed in all isolates displaying mefloquine IC50s of >30 nM, commonly used as the cutoff for mefloquine resistance. Furthermore, the association with mefloquine resistance was stronger for an increased expression level than for an increased gene copy number. This suggests that increased Pfmdr1 transcript abundance indeed contributes to in vitro resistance to mefloquine in these isolates, in contrast to a trend toward higher Pfmdr1 expression levels in mefloquine-sensitive isolates in Thailand (6). We did not observe a linear relationship between transcript levels and gene copy numbers in the field isolates studied here, consistent with data from Thailand (6). This differs from observations of laboratory lines (10, 34). Further studies are needed to identify the factors governing Pfmdr1 mRNA expression and/or stability in field isolates and how they impact on Pfmdr1 protein levels and/or cellular localization and on profiles of resistance to mefloquine, halofantrine, and possibly lumefantrine.
The population characteristics of French Guiana isolates differ from those of isolates from neighboring areas, suggesting different gene flow dynamics. Although fixed as in other South American areas (reference 20 and references therein), the Pfcrt 76T mutation was harbored by two synonymous alleles, in contrast to neighboring Guyana or Brazil, where up to five distinct alleles were reported (4, 31, 43). The 7G8 type (also called SMVNT1) is rare in French Guiana (11.3%) and in Brazil (6.9%), where the synonymous SMVNT2 allele predominates (4, 31), in contrast to one of two studies from Guyana (4, 31). The other resistance haplotypes described in South and Central America (see reference 20) were totally absent from French Guiana. Regarding Pfmdr1, the frequency of the 7G8 Pfmdr1 (FG-A) and FG-B alleles resembles the distribution described in Brazil (19, 31) and Guyana (31), where the rare FG-E, -F, -G, -H, and -I alleles have not been observed (4, 19, 31). In contrast, minor alleles harboring the wild type N1042 codon (4, 19) or the mutant 86Y codon (19, 31) observed in the neighboring countries were not observed in our sample of French Guiana isolates. Interestingly, the FG-B allele observed here with a 3.7% frequency had a 10-fold higher frequency in Venezuela (20) and allele FG-C has been repeatedly observed in Peru (2). Pfmdr1 gene amplification was observed at a higher rate in French Guiana (a mean of 40%) than, e.g., the 12% rate observed in Venezuela during a period where mefloquine monotherapy was used (20), in stark contrast to the absence of Pfmdr1 gene amplification in the Peruvian Amazon region, where artesunate-mefloquine has been the first line treatment since 2001 (2). As amodiaquine and the artemether-lumefrantrine combination have been reported to exert opposite selective pressures on the Pfmdr1 gene (22), we are currently pursuing monitoring of the Pfmdr1 gene copy number to investigate whether the Pfmdr1 gene copy number continues to decline as the number of years of artemether-lumefantrine use increases.
In conclusion, withdrawal of chloroquine in French Guiana was not followed by expansion of wild-type parasites, as reported elsewhere (26, 32, 44), and none of the molecular features studied here accounted for the regained susceptibility to chloroquine seen during the last decade. The proposed interaction between the specific Pfcrt and Pfmdr1 7G8-type alleles for determining susceptibility to chloroquine and amodiaquine (37) did not apply to the parasite population of French Guiana. Cessation of mefloquine monotherapy was rapidly followed by reduction of the prevalence of isolates with multiple Pfmdr1 copies without impacting on the haplotype frequency. The strong association of mefloquine susceptibility with the Pfmdr1 expression level is interesting and certainly deserves further studies to evaluate its usefulness as a molecular marker of susceptibility to mefloquine and possibly other antimalarials, including halofantrine, quinine, lumefantrine, or artemisin derivatives. It is worth noting that about 90% of isolates seem to have good sensitivity to artemether and amodiaquine. The sensitivity profile of lumefantrine, with only 65.8% of isolates susceptible, is a concern and needs to be actively monitored, as the artemether-lumefantrine combination is being used as a first-line treatment for malaria in the area. Novel molecular markers of susceptibility to chloroquine and lumefrantrine are urgently needed in this area. These could be identified using whole-genome sequencing or expression profiling. The limited polymorphism of the French Guiana parasites will facilitate the identification of loci modulating susceptibility to these molecules and understanding of the molecular basis of parasite evolution following changes in treatment policy.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the French Ministry of Health (InVS Agency, Paris, France); by the Prix Louis D., French Academy of Sciences; and by the European Commission (contract QLK2-CT20021-1503-ResMalChip).
We have no competing interests to declare. The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 9 January 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Ariey F, et al. 2006. Invasion of Africa by a single pfcrt allele of South East Asian type. Malar. J. 5:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bacon DJ, et al. 2009. Dynamics of malaria drug resistance patterns in the Amazon basin region following changes in Peruvian national treatment policy for uncomplicated malaria. Antimicrob. Agents Chemother. 53:2042–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baliraine FN, et al. 2011. Limited ability of Plasmodium falciparum pfcrt, pfmdr1, and pfnhe1 polymorphisms to predict quinine in vitro sensitivity or clinical effectiveness in Uganda. Antimicrob. Agents Chemother. 55:615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Best Plummer W, Pinto Pereira LM, Carrington CV. 2004. Pfcrt and pfmdr1 alleles associated with chloroquine resistance in Plasmodium falciparum from Guyana, South America. Mem. Inst. Oswaldo Cruz 99:389–392 [DOI] [PubMed] [Google Scholar]
- 5. Carme B, et al. 2009. Update on the epidemiology of malaria in French Guiana. Med. Trop. 69:19–25 [PubMed] [Google Scholar]
- 6. Chaijaroenkul W, et al. 2011. Sequence and gene expression of chloroquine resistance transporter (pfcrt) in the association of in vitro drugs resistance of Plasmodium falciparum. Malar. J. 10:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaijaroenkul W, Wisedpanichkij R, Na-Bangchang K. 2010. Monitoring of in vitro susceptibilities and molecular markers of resistance of Plasmodium falciparum isolates from Thai-Myanmar border to chloroquine, quinine, mefloquine and artesunate. Acta Trop. 113:190–194 [DOI] [PubMed] [Google Scholar]
- 8. Chen N, et al. 2010. Deamplification of pfmdr1-containing amplicon on chromosome 5 in Plasmodium falciparum is associated with reduced resistance to artelinic acid in vitro. Antimicrob. Agents Chemother. 54:3395–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cojean S, et al. 2006. Lack of association between putative transporter gene polymorphisms in Plasmodium falciparum and chloroquine resistance in imported malaria isolates from Africa. Malar. J. 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cowman AF, Galatis D, Thompson JK. 1994. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross resistance to halofantrine and quinine. Proc. Natl. Acad. Sci. U. S. A. 91:1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Danquah I, et al. 2010. Selection of pfmdr1 and pfcrt alleles in amodiaquine treatment failure in north-western Burkina Faso. Acta Trop. 114:63–66 [DOI] [PubMed] [Google Scholar]
- 12. Das MK, et al. 2010. High chloroquine treatment failure rates and predominance of mutant genotypes associated with chloroquine and antifolate resistance among falciparum malaria patients from the island of Car Nicobar, India. J. Antimicrob. Chemother. 65:1258–1261 [DOI] [PubMed] [Google Scholar]
- 13. Djimdé A, et al. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 344:257–263 [DOI] [PubMed] [Google Scholar]
- 14. Duraisingh MT, et al. 1997. Evidence for selection for the tyrosine-86 allele of the pfmdr 1 gene of Plasmodium falciparum by chloroquine and amodiaquine. Parasitology 114:205–211 [DOI] [PubMed] [Google Scholar]
- 15. Durrand V, et al. 2004. Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and their relationship to chloroquine resistance in vitro. Mol. Biochem. Parasitol. 136:273–285 [DOI] [PubMed] [Google Scholar]
- 16. Ekala MT, et al. 2007. Sequence analysis of Plasmodium falciparum cytochrome b in multiple geographic sites. Malar. J. 6:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fidock DA, et al. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6:861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Foote SJ, et al. 1990. Several alleles of the multidrug resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature 345:255–258 [DOI] [PubMed] [Google Scholar]
- 19. Gama BE, et al. 2010. Brazilian Plasmodium falciparum isolates: investigation of candidate polymorphisms for artemisinin resistance before introduction of artemisinin-based combination therapy. Malar. J. 9:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Griffing S, et al. 2010. pfmdr1 amplification and fixation of pfcrt chloroquine resistance alleles in Plasmodium falciparum in Venezuela. Antimicrob. Agents Chemother. 54:1572–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holmgren G, et al. 2006. Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76T and pfmdr1 86Y. Infect. Genet. Evol. 6:309–314 [DOI] [PubMed] [Google Scholar]
- 22. Humphreys GS, et al. 2007. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob. Agents Chemother. 51:991–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson DJ, et al. 2004. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol. Cell 15:867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khim N, et al. 2005. Countrywide survey shows very high prevalence of Plasmodium falciparum multilocus resistance genotypes in Cambodia. Antimicrob. Agents Chemother. 49:3147–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kimchi-Sarfaty C, et al. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315:525–528 [DOI] [PubMed] [Google Scholar]
- 26. Laufer MK, et al. 2010. Return of chloroquine-susceptible falciparum malaria in Malawi was a reexpansion of diverse susceptible parasites. J. Infect. Dis. 202:801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Legrand E, et al. 2005. Molecular analysis of two local falciparum malaria outbreaks on the French Guiana coast confirms the msp1 B-K1/varD genotype association with severe malaria. Malar. J. 4:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Legrand E, Volney B, Meynard JB, Mercereau-Puijalon O, Esterre P. 2008. In vitro monitoring of Plasmodium falciparum drug resistance in French Guiana: a synopsis of continuous assessment from 1994 to 2005. Antimicrob. Agents Chemother. 52:288–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lim P, et al. 2009. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar. J. 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 31. Mehlotra RK, et al. 2008. Discordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob. Agents Chemother. 52:2212–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mita T, et al. 2004. Expansion of wild type allele rather than back mutation in pfcrt explains the recent recovery of chloroquine sensitivity of Plasmodium falciparum in Malawi. Mol. Biochem. Parasitol. 135:159–163 [DOI] [PubMed] [Google Scholar]
- 33. Nsobya SL, et al. 2010. In vitro sensitivities of Plasmodium falciparum to different antimalarial drugs in Uganda. Antimicrob. Agents Chemother. 54:1200–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peel SA, Bright P, Yount B, Handy J, Baric RS. 1994. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 51:648–658 [DOI] [PubMed] [Google Scholar]
- 35. Price RN, et al. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906–909 [DOI] [PubMed] [Google Scholar]
- 37. Sá JM, et al. 2009. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc. Natl. Acad. Sci. U. S. A. 106:18883–18889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanchez CP, Dave A, Stein WD, Lanzer M. 2010. Transporters as mediators of drug resistance in Plasmodium falciparum. Int. J. Parasitol. 40:1109–1118 [DOI] [PubMed] [Google Scholar]
- 39. Sidhu AB, et al. 2006. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J. Infect. Dis. 194:528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sidhu AB, Valderramos SG, Fidock DA. 2005. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol. Microbiol. 57:913–926 [DOI] [PubMed] [Google Scholar]
- 41. Sidhu AB, Verdier-Pinard D, Fidock DA. 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298:210–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Valderramos SG, et al. 2010. Identification of a mutant PfCRT-mediated chloroquine tolerance phenotype in Plasmodium falciparum. PLoS Pathog. 6:e1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vieira PP, et al. 2004. pfcrt polymorphism and the spread of chloroquine resistance in Plasmodium falciparum populations across the Amazon basin. J. Infect. Dis. 190:417–424 [DOI] [PubMed] [Google Scholar]
- 44. Wang X, et al. 2005. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People's Republic of China. Am. J. Trop. Med. Hyg. 72:410–414 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.