Abstract
Seventeen laboratories participated in a cooperative study to validate the regional susceptibility testing of Neisseria gonorrhoeae in The Netherlands. International reference strains were distributed. Each laboratory determined the MICs of ciprofloxacin, penicillin, and tetracycline, for each strain by Etest. To explore a more transparent assessment of quality and comparability, a statistical regression model was fitted to the data that accounted for the censoring of the MICs. The mean MICs found by all of the laboratories except three were closer than one 2-fold dilution step to the overall mean, and the mean MICs of each antimicrobial agent were close to the MICs for the international reference strains. This approach provided an efficient tool to analyze the performance of the Dutch decentralized gonococcal resistance monitoring system and confirmed good and comparable standards.
INTRODUCTION
Successful control of Neisseria gonorrhoeae over the past decades has been hampered by the rapid development of resistance against successive treatment regimes. Therefore, circulating strains need to be monitored carefully so the resistance levels are known and may be used for guidelines for appropriate treatment (5). This surveillance needs quality systems to ensure comparable and reliable data. In quality assurance exercises, MICs need to be compared between multiple antimicrobial agents against an appropriate number of reference strains, and between many laboratories. The statistical methods described here were used for the analysis of a quality control exercise in the laboratories which participate in monitoring the antimicrobial susceptibility of N. gonorrhoeae in The Netherlands (4).
Antimicrobial susceptibility of bacteria is often measured using dilution series methods and the last dilution inhibiting bacterial growth is registered as the MIC. However, in reality, the actual value of the MIC may be anywhere between the last dilution inhibiting growth and the first dilution not inhibiting growth. In statistics, such data are known as censored data, that is, data where the exact value of the outcome is not observed and only the boundaries below or above are and the value lies between them.
The question is how to deal with such data properly. Several methods have been proposed. For a specific strain and antimicrobial agent combination, one can determine the consensus MIC value (mode) among different laboratories (2, 9). However, how accurate is this modal MIC estimate, e.g., in terms of a 95% confidence interval? Another option is to replace the MICs with a value halfway between the lower and upper boundaries. A standard error can then be estimated (and thus a 95% confidence interval), but this is rather arbitrary.
In this paper, we describe a methodology that properly deals with censored MIC data. Analysis of censored data has a long history in statistics, especially in survival analysis, where death of biological organisms or failure of mechanical systems is studied (3). In that case, the analysis involves time-to-event (death, failure) data as a function of explanatory variables. These data are usually right censored because some subjects are still alive when they are lost to follow-up or when the study ends, meaning that the exact time of the event is unknown, but it is known to occur after the moment a subject is lost to follow-up or the end of the study. While modeling MICs, the actual MIC (the event) is unknown, and we only know that it is somewhere between two boundaries (interval censored). Moreover, similar to the time in survival analysis, the MIC may run off the scale and be higher than the highest concentration or lower than the lowest concentration of an Etest strip or a dilution series. So MIC values are left, interval, or right censored.
This methodology allows the fitting of a statistical model to censored MIC data. Subsequently, the results can be used to compare MICs between different strains and/or antimicrobial agents, including 95% confidence intervals. Furthermore, by allowing heterogeneity between different laboratories, we are able to consider lab performances. Finally, we compare our results to published MICs for reference strains.
MATERIALS AND METHODS
A hypothetical data example.
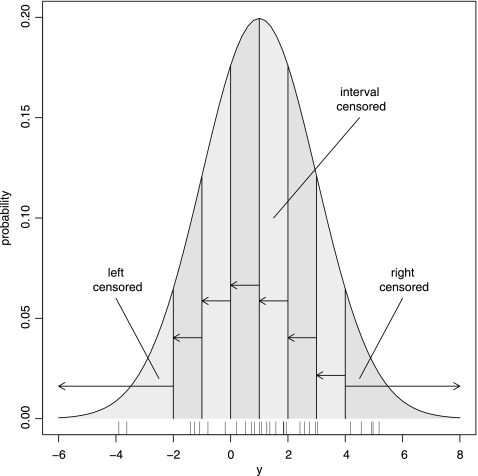
We first sketch the problem for which the method we propose was devised by giving a hypothetical example. We consider 30 observers (e.g., laboratories) and consider a random variable y (e.g., a log MIC). Each observer determines an actual value for y, which we call yact. The true mean of y equals 1, but due to random noise, the 30 observers all measure some value around 1. The true standard deviation equals 2. The random variable y has a normal distribution. For one particular case, the 30 yact values determined by the observers are shown in Table 1 and illustrated in Fig. 1 by the small vertical lines at the bottom.
Table 1.
Hypothetical data created by 30 random samples from a normal distribution with mean 1 and standard deviation 2
| Observation | yact | yobs | Lower | Upper | Event | ySurv |
|---|---|---|---|---|---|---|
| 1 | −0.79 | 0 | −1 | 0 | 3 | [−1, 0] |
| 2 | 1.37 | 2 | 1 | 2 | 3 | [1, 2] |
| 3 | 4.18 | > 4 | 4 | Infa | 0 | 4+ |
| 4 | −1.26 | −1 | −2 | −1 | 3 | [−2, −1] |
| 5 | 0.84 | 1 | 0 | 1 | 3 | [0, 1] |
| 6 | 1.26 | 2 | 1 | 2 | 3 | [1, 2] |
| 7 | 2.42 | 3 | 2 | 3 | 3 | [2, 3] |
| 8 | 0.52 | 1 | 0 | 1 | 3 | [0, 1] |
| 9 | 4.97 | > 4 | 4 | Inf | 0 | 4+ |
| 10 | 0.72 | 1 | 0 | 1 | 3 | [0, 1] |
| 11 | 1.84 | 2 | 1 | 2 | 3 | [1, 2] |
| 12 | 2.96 | 3 | 2 | 3 | 3 | [2, 3] |
| 13 | 0.21 | 1 | 0 | 1 | 3 | [0, 1] |
| 14 | −1.08 | −1 | −2 | −1 | 3 | [−2, −1] |
| 15 | 4.56 | > 4 | 4 | Inf | 0 | 4+ |
| 16 | −3.62 | <−2 | −Inf | −2 | 2 | −2− |
| 17 | 2.76 | 3 | 2 | 3 | 3 | [2, 3] |
| 18 | 1.07 | 2 | 1 | 2 | 3 | [1, 2] |
| 19 | 3.03 | 4 | 3 | 4 | 3 | [3, 4] |
| 20 | 1.86 | 2 | 1 | 2 | 3 | [1, 2] |
| 21 | 5.18 | > 4 | 4 | Inf | 0 | 4+ |
| 22 | −1.40 | −1 | −2 | −1 | 3 | [−2, −1] |
| 23 | 4.18 | > 4 | 4 | Inf | 0 | 4+ |
| 24 | 4.91 | > 4 | 4 | Inf | 0 | 4+ |
| 25 | 1.01 | 2 | 1 | 2 | 3 | [1, 2] |
| 26 | −3.90 | <−2 | −Inf | −2 | 2 | −2− |
| 27 | 1.95 | 2 | 1 | 2 | 3 | [1, 2] |
| 28 | −0.19 | 0 | −1 | 0 | 3 | [−1, 0] |
| 29 | 2.58 | 3 | 2 | 3 | 3 | [2, 3] |
| 30 | 1.58 | 2 | 1 | 2 | 3 | [1, 2] |
Inf, infinity.
Fig 1.
Illustration of censored data. Variable y has a normal distribution with a mean of 1 and a standard deviation of 2. Thirty values are shown at the bottom. Values of <−2 are left censored, values of >4 are right censored, and values in between are interval censored.
Given these 30 values for yact, our task is to estimate the mean and the standard error of the mean. The estimate of the mean is simply calculated by taking the average of the yact values, while the standard error is calculated by dividing the estimated standard deviation by √30. In this example, the estimated mean is 1.46; its standard error 0.43.
Unfortunately, the actual y values cannot be observed. Observers can only determine the nearest integer value greater than yact (e.g., by dilution series). For example, yact = 2.42 will be observed as yobs = 3. To complicate things further, yact values lower than −2 and greater than 4 are off the strip; e.g., yact = 5.18 will be observed as yobs > 4. Such data are called interval-, left-, and right-censored data, respectively. These yobs are also shown in Table 1 and illustrated in Fig. 1 by the shaded areas.
Given these 30 observed y values, we may set the same goal, i.e., to estimate the mean and its standard error. Several options are proposed, such as to take the mode of the 30 observed y values. Here the mode equals 2 (8 records). But how can the standard error of the mode be calculated? Another naive solution is to ignore the censoring and just calculate the mean of the observed y values and its standard error. Using these data, the estimated mean is 1.77 and its standard error 0.34, but is this appropriate?
Statistical analysis of censored data.
We propose a more sophisticated statistical method that properly estimates the mean and its standard error while dealing with the censored nature of the observations. We will first sketch the main idea in the context of the hypothetical example.
The idea is based on the maximum-likelihood principle (7): given a data set and a statistical model, for what model parameters do the observations become most likely? In our approach, the data set are the interval-censored data (lower and upper boundaries, Table 1), while the statistical model is an interval-censored normal distribution. This distribution has two parameters: the mean, which is the parameter of interest, and a standard deviation. The latter is a nuisance parameter, because the maximum-likelihood principle directly provides the standard error of the mean.
The model can easily be fitted using standard statistical software for survival analysis, for example, using the function survreg in R (8). R code is provided in the appendix. The lower and upper boundaries are provided in Table 1. Using this method, the estimated mean is 1.60 and its standard error 0.47.
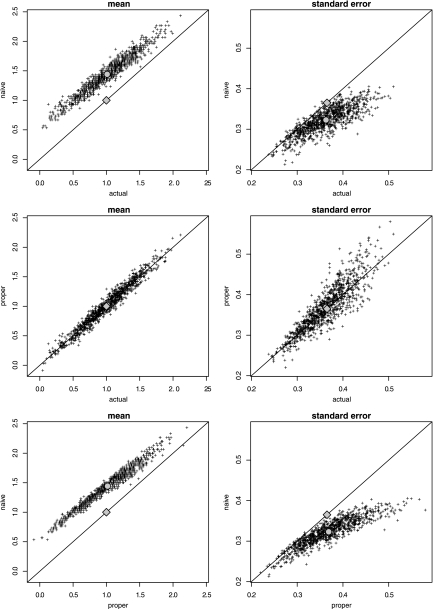
Figure 2 shows what happens if we repeat the above calculation 1,000 times (black dots). Each run provides 30 new samples of yact and yobs. For each run, the following values are being computed: the mean of yact (i.e., the mean that we would have obtained if we had observed the actual y values), the naively calculated mean of yobs (i.e., the mean that we obtain based on the observed y values, ignoring censoring) and the properly calculated mean of yobs (i.e., the mean that we obtain based on the observed y values, with censoring) (left panels). Standard errors are computed as well (right panels). Estimates differ between runs due to random variation of the samples. The true mean and standard error are indicated by diamonds. The means of the estimates are indicated by circles.
Fig 2.
One thousand runs of 30 samples where the mean (left panels) and standard error of the mean (right panels) were estimated for each run (black dots). The “actual” values are based on the actual values, yact; “naive” values are based on the observed values, yobs (ignoring censoring); and “proper” values are based on the observed values, yobs (accounting for censoring). The true mean (= 1) and standard error (= 2/√30) are indicated by diamonds. The means of the 1,000 estimates are indicated by circles.
Compared to the actual and proper estimate, the means of the naive estimate are clearly overestimated by about 0.5, while the standard errors are underestimated. The proper estimates are close to the actual estimates. The overestimation of the mean by 0.5 is due mainly to ignoring the interval censoring of the observed data, while the underestimation of the standard error is due mainly to ignoring the left and right censoring. Subtracting 0.5 from yobs would improve the estimate of the mean but not that of the standard error. In conclusion, the hypothetical example shows that we should analyze censored observations using proper statistical methods.
MIC data.
Twenty regional laboratories participate in the surveillance of the antimicrobial susceptibility of N. gonorrhoeae in The Netherlands (4). In 2007 and 2010, the participating laboratories were requested to determine the susceptibilities of four and six reference strains, respectively, in order to determine quality and comparability of the decentralized testing results. Seventeen laboratories, including the reference laboratory, agreed to participate.
The reference strains used to assess the performance and comparability of the Dutch laboratories participating in the decentralized testing and monitoring of gonococcal surveillance were N. gonorrhoeae ATCC 49226; strains 5, 8, and 10 of the European Surveillance of Sexually Transmitted Infections; and WHO quality assessment strains 08-02 (WHO G), 09-02 (WHO F), 09-03 (WHO K), 09-04 (WHO L), 09-09 (WHO M), and 09-10 (WHO O), kindly provided by C. Ison (2, 9). The antimicrobial agents tested were ciprofloxacin, penicillin, and tetracycline. In addition, the strains were tested for cefotaxime but the cefotaxime data could not be included in this study because reference MICs were not available. The laboratories provided details about their laboratory methods. In 2007, all of the laboratories used the Etest (bioMérieux); in 2010, 8 of the 17 laboratories used MICE strips (Oxoid).
Statistical analysis of MIC data.
First the Etest values were log2 transformed. These log2-transformed MICs were treated as interval-censored data since only lower and upper boundary values can be measured. In this case, the upper boundary equals the measured log2 MIC and the lower boundary is the upper boundary value minus 1. The actual log2 MIC value lies between the upper and lower boundaries. Furthermore, we have to deal with left-censored MIC data (actual MIC lower than the lowest MIC on the strip) and right-censored MIC data (actual MIC higher than the highest MIC).
In order to compare the MICs between strains and antimicrobial agents, the above model was expanded to a more general regression model. First, the mean must become a function of the strain, antimicrobial agent, and agar medium. It is assumed, for any given combination of those variables, that the distribution of log2 MIC values is a censored normal distribution with a common variance, as is usually done in regression modeling.
The regression model was further expanded to a multilevel regression model. The strain, antimicrobial agent, and agar medium were treated as fixed effects, while the laboratory was treated as a random effect. The term “random,” usually “frailty” in survival analysis, is a random component designed to account for variability due to unobserved laboratory level factors that is otherwise unaccounted for by the fixed-effect terms in the model. These laboratory factors may contribute an extra layer of heterogeneity, leading to greater variability in MICs than may be expected under the model without the random component. An interaction term between the strain and the antimicrobial agent was added, since the antibiogram differs among the strains.
The multilevel regression model is specified symbolically as follows. The data set consists of n records. For each record i, the actual log2 MIC values are normally distributed, conditional on the mean μi and variance σ2. The distribution of the observed log2 MIC values is interval censored and normally distributed with lower and upper boundaries for each observation. In fact, these lower and upper boundary values are the observations: log2(MICi) ∼ Normal(μi, σ2)I(loweri, upperi).
Subsequently, the mean is a linear function of a model matrix X and regression parameters β. The model matrix represents the fixed-effect terms and is filled with ones and zeros, turning variables “on” and “off” for that specific record. In addition, a lab-specific random effect, blab, is added: μi = Xiβ + blab,j[i].
In turn, these random effects are assumed to be normally distributed with mean zero and variance σlab2: blab,j ∼ Normal(0, σlab2).
Fitting such a model is not straightforward using standard statistical software packages. We therefore reformulated the model into a Bayesian setting, which allowed us to implement it easily into the WinBUGS software package (6). Since a Bayesian approach requires specification of prior distributions on all unknown parameters, all (hyper)parameters were given noninformative prior distributions, meaning that the posterior parameter distributions were inferred from the observations only. The WinBUGS code is shown in the appendix.
We ran two independent MCMC (Markov chain Monte Carlo) chains, each containing 5,100 iterations. We used a “burn-in” period of 100 iterations and applied a thinning factor of 10 to prevent autocorrelation in the samples. For each parameter, this resulted in 1,000 samples from the posterior distribution.
A first analysis showed that the effects of the agar medium and test strip were not significant. Therefore, these factors were not taken into account in the further analyses. Next, for any given combination of antimicrobial agent and strain, the posterior distribution of the corresponding MIC can be calculated by adding up the corresponding regression parameters and taking the antilog. The posterior distributions of the laboratory-specific deviations were obtained directly from the model output.
The fold differences between our MICs and the reference MICs were calculated as well. This was done as follows. For each antimicrobial agent and strain, we took the 1,000 samples from the posterior distribution of our calculated log2 MIC and subtracted 1,000 random uniformly distributed samples from the corresponding interval-censored reference log2 MIC. This resulted in 1,000 samples of log2 differences. Right-censored reference MICs were omitted, since differences from right-censored reference MICs are undefined. The differences were pooled for each antimicrobial agent separately, as well as for all antimicrobial agents together. Subsequently, the mean log2 differences and their 95% confidence intervals were determined. These log2 values were transformed back to the original scale (the difference becomes a fold difference, i.e., a ratio) by taking the antilog.
All data pre- and postprocessing were done in R (8).
RESULTS
Laboratory performance.
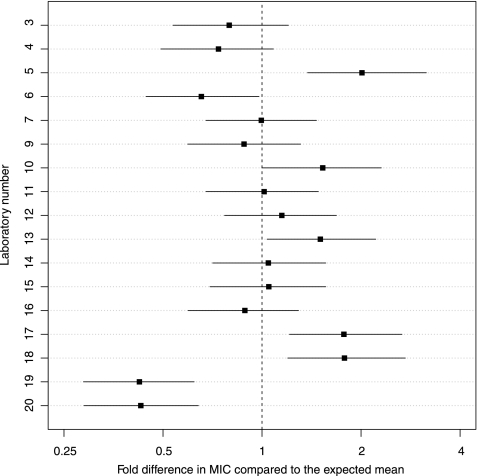
The model predicts laboratory-specific fold differences from the mean of all laboratories, which is the expected MIC value based on the strain and antimicrobial agent. Figure 3 shows the fold difference (and the 95% confidence interval) for each laboratory. This figure allows an objective rating of each laboratory. On the original scale, the differences are fold differences or ratios.
Fig 3.
Laboratory-specific fold differences (and 95% confidence intervals) from the expected mean MIC. A 1-fold difference is no difference [log2(1) = 0].
Most predicted MICs fell within a factor 2 (1 doubling dilution) of the overall mean. The deviation was significant only for laboratories 19 and 20, which reported MICs which were, on average, more than 2-fold lower than the overall mean. Laboratories 7, 9, 11, 12, 14, 15, and 16 were nearest to the overall mean.
Strain- and antimicrobial-agent-specific MICs compared to reference MICs.
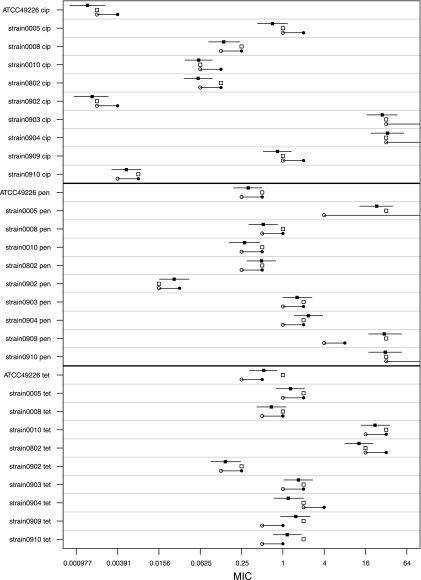
From the same analysis we obtained the predicted mean MIC and its 95% confidence interval for each combination of strain and antimicrobial agent (Fig. 4). The mode MICs are given for comparison. Although the original MIC data were interval censored, the model provides continuous mean MICs.
Fig 4.
Predicted mean MICs and 95% confidence intervals (solid squares), mode MICs (open squares), and reference MICs (solid circles), with their corresponding lower boundary accounting for interval censoring (open circles), for each combination of strain and antimicrobial agent.
The mean MICs of each strain and antimicrobial agent as predicted by the model were subsequently compared to the corresponding reference MICs (2, 9). The reference MICs are shown in Fig. 4 as intervals, since these MICs are interval censored as well. The fold differences of the mean MIC of the participating laboratories from the reference MIC for each antimicrobial agent and their 95% confidence intervals were 0.6 (0.3 to 1.4) for ciprofloxacin, 1.3 (0.5 to 7.0) for penicillin, 1.0 (0.3 to 2.9) for tetracycline, and 0.95 (0.33 to 4.73) for all antimicrobial agents. The wide confidence interval of the overall analysis was due mainly to the mean penicillin MIC of 32 mg/liter, which our laboratories found for strain 09-09, compared to the reference value of 8 mg/liter (9). When we left the MIC of penicillin for this strain out of the analysis, we found differences of 1.06 (0.46 to 2.44) for penicillin and 0.89 (0.34 to 2.53) for all antimicrobial agents.
DISCUSSION
Using statistical methods for the analysis of censored data, we were able to obtain mean MIC values with their confidence intervals for any given combination of explanatory variables, including laboratory (Fig. 3) and the combination of strain and antimicrobial agent (Fig. 4). The methods presented here are not new, but to our knowledge, this is the first time they have been applied in the field of antimicrobial susceptibility testing.
Although the basic model for censored data seems fairly simple, implementation of the multilevel regression model for censored data in standard statistical software is not straightforward. This especially applies to the “frailty” term in the model, which complicates the calculations. However, by putting the model into a Bayesian context, implementation in WinBUGS is relatively easy. Still, when doing MCMC inference, one has to inspect convergence and proper mixing of the chains. First, have both chains converged to the same posterior distribution? Second, is there no autocorrelation in the successive samples? Without these checks, results may be meaningless. Also the pre- and postprocessing of the data may be complicated. We are aware that our method may not be easy to implement without the help of a statistician. However, our R code for pre- and postprocessing of the data is available on request and the WinBUGS model is provided in the appendix.
An earlier analysis of a quality assurance exercise of antimicrobial susceptibility tests included counting the number of deviant results based on preestablished breakpoints (1). Our method allowed comparing the performances of the laboratories to each other (Fig. 3) and comparing the mean MICs for each antimicrobial agent and strain to the reference MICs of the WHO strains (Fig. 4). Both fold differences were within acceptable limits, i.e., one dilution step. The wide confidence interval for the mean fold difference of our mean penicillin MIC from the reference value was due mainly to strain 09-09 (WHO M). Our laboratories found a mean penicillin MIC of 32 mg/liter for this strain, whereas it is 8 mg/liter according to Unemo et al. (9).
All of the laboratories in this study except three scored a mean predicted MIC within one doubling dilution of the overall mean. We expected that the number of isolates tested per year in each laboratory, its experience, would have the greatest impact on its accuracy, but this effect was far from significant (data not shown). Overall, the participating Dutch laboratories performed well compared to each other and to international standards. Since 2011, strains have been routinely tested for ceftriaxone, spectinomycin, and azithromycin resistance and similar testing for resistance to these antimicrobial agents can be performed next year.
ACKNOWLEDGMENTS
We acknowledge Hendriek Boshuizen for her comments on the manuscript and the participating laboratories for their kind cooperation: Streeklaboratorium voor de Volksgezondheid GG&GD Amsterdam, Alysis Zorggroep Arnhem, Amphia Ziekenhuis Breda, Regionaal Laboratorium Medische Microbiologie Dordrecht, Laboratorium Microbiologie Twente-Achterhoek Enschede, Admiraal de Ruyterziekenhuis Goes, Laboratorium voor Infectieziekten Groningen, Streeklaboratorium Haarlem, Atrium Medisch Centrum Heerlen, Izore Leeuwarden, Academisch Ziekenhuis Maastricht, Canisius Wilhelmina Ziekenhuis Nijmegen, Laurentius Ziekenhuis Roermond, Erasmus Medisch Centrum Rotterdam, Jeroen Bosch Ziekenhuis 's Hertogenbosch, St. Elisabeth Ziekenhuis Tilburg, Universitair Medisch Centrum Utrecht, Stichting PAMM Veldhoven, and VieCuri Medisch Centrum Venlo.
APPENDIX
R code for calculating the mean of censored data.
In R, we must first create a so-called Surv object for the data. This object contains the lower and upper boundaries, based on the observed y values. In order to do that, we must know what type of censoring we are dealing with. This should be translated to an event code as follows: 3 = interval-censored data, 2 = left-censored data, 0 = right-censored data. After identifying the event code for each record, a Surv object is created as follows:
library(survival)
- y.Surv <- Surv(
- time = ifelse(event == 3, y.obs-1, y.obs),
- time2 = y.obs,
- event = event,
- type = “interval”)
The result is shown in Table 1. The mean and standard error of the mean are subsequently estimated by the survreg function as follows:
y.mean <- survreg(y.Surv ∼ 1, dist = “gaussian”)
summary(y.mean)
WinBUGS code for fitting the multilevel model with interval-censored response variable.
The data set consists of n records. Conditional on the mean μ and precision τ, the observations are normally distributed (note that WinBUGS works with precisions [= variance−1] instead of variances). The I operator indicates that the distribution is interval censored with a lower and an upper boundary. The mean is a function of the model matrix X and regression coefficients β. The blab term is a lab specific deviation. These deviations are normally distributed with mean zero and precision τlab. All (hyper)parameters are given noninformative priors.
- model {
- for (i in 1:n) {
- y[i] ∼ dnorm(mu[i], tau)I(lower[i], upper[i])
- mu[i] <- inprod2(X[i, ], beta[]) + b.lab[lab[i]]
- }
- for (j in 1:n.lab) {
- b.lab[j] ∼ dnorm(0.0, tau.lab)
- }
- for (k in 1:n.beta) {
- beta[k] ∼ dnorm(0.0, 1.0E-4)
- }
- tau <- pow(sigma, −2)
- sigma ∼ dunif(0.01, 100)
- tau.lab <- pow(sigma.lab, −2)
- sigma.lab ∼ dunif(0.01, 100)
}
Footnotes
Published ahead of print 9 January 2012
REFERENCES
- 1. Bronzwaer S, et al. 2002. Comparability of antimicrobial susceptibility test results from 22 European countries and Israel: an external quality assurance exercise of the European Antimicrobial Resistance Surveillance System (EARSS) in collaboration with the United Kingdom National External Quality Assurance Scheme (UK NEQAS). J. Antimicrob. Chemother. 50:953–964 [DOI] [PubMed] [Google Scholar]
- 2. Ison CA, Martin IM, Lowndes CM, Fenton KA. 2006. Comparability of laboratory diagnosis and antimicrobial susceptibility testing of Neisseria gonorrhoeae from reference laboratories in western Europe. J. Antimicrob. Chemother. 58:580–586 [DOI] [PubMed] [Google Scholar]
- 3. Kleinbaum DG, Klein M. 2006. Survival analysis, a self-learning text, 2nd ed Springer, New York, NY [Google Scholar]
- 4. Koedijk FD, van Veen MG, de Neeling AJ, Linde GB, van der Sande MA. 2010. Increasing trend in gonococcal resistance to ciprofloxacin in The Netherlands, 2006-8. Sex. Transm. Infect. 86:41–45 [DOI] [PubMed] [Google Scholar]
- 5. Lewis DA. 2010. The gonococcus fights back: is this time a knock out? Sex. Transm. Infect. 86:415–421 [DOI] [PubMed] [Google Scholar]
- 6. Lunn DJ, Thomas A, Best N, Spiegelhalter D. 2000. WinBUGS—a Bayesian modelling framework: concepts, structure, and extensibility. Stat. Comput. 10:325–337 [Google Scholar]
- 7. Pawitan Y. 2001. In all likelihood: statistical modelling and inference using likelihood. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 8. R Development Core Team 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.r-project.org/ [Google Scholar]
- 9. Unemo M, Fasth O, Fredlund H, Limnios A, Tapsall J. 2009. Phenotypic and genetic characterization of the 2008 WHO Neisseria gonorrhoeae reference strain panel intended for global quality assurance and quality control of gonococcal antimicrobial resistance surveillance for public health purposes. J. Antimicrob. Chemother. 63:1142–1151 [DOI] [PubMed] [Google Scholar]