Abstract
We describe the discovery, purification, characterization, and expression of an antimicrobial peptide, epidermicin NI01, which is an unmodified bacteriocin produced by Staphylococcus epidermidis strain 224. It is a highly cationic, hydrophobic, plasmid-encoded peptide that exhibits potent antimicrobial activity toward a wide range of pathogenic Gram-positive bacteria including methicillin-resistant Staphylococcus aureus (MRSA), enterococci, and biofilm-forming S. epidermidis strains. Purification of the peptide was achieved using a combination of hydrophobic interaction, cation exchange, and high-performance liquid chromatography (HPLC). Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis yielded a molecular mass of 6,074 Da, and partial sequence data of the peptide were elucidated using a combination of tandem mass spectrometry (MS/MS) and de novo sequencing. The draft genome sequence of the producing strain was obtained using 454 pyrosequencing technology, thus enabling the identification of the structural gene using the de novo peptide sequence data previously obtained. Epidermicin NI01 contains 51 residues with four tryptophan and nine lysine residues, and the sequence showed approximately 50% identity to peptides lacticin Z, lacticin Q, and aureocin A53, all of which belong to a new family of unmodified type II-like bacteriocins. The peptide is active in the nanomolar range against S. epidermidis, MRSA isolates, and vancomycin-resistant enterococci. Other unique features displayed by epidermicin include a high degree of protease stability and the ability to retain antimicrobial activity over a pH range of 2 to 10, and exposure to the peptide does not result in development of resistance in susceptible isolates. In this study we also show the structural gene alone can be cloned into Escherichia coli strain BL21(DE3), and expression yields active peptide.
INTRODUCTION
The continued emergence of infections caused by multiresistant bacteria reinforces the urgent requirement for the identification and development of novel antibiotics. Bacteriocins are a group of agents that can show good activity against a range of human pathogens, including viruses, and have the potential to aid the fight against multiresistant bacteria (14, 33, 43).
Bacteriocins are ribosomally synthesized antimicrobial peptides produced to facilitate niche competition (26, 28). The inhibitory spectrum of these agents is predominantly directed toward bacteria that are closely related to the producer strain (22, 41). Due to their novel modes of action and activity against a range of multiresistant bacteria, these inhibitors could potentially be extremely useful in the treatment of multiple infections and have already been extensively investigated for their impact on prevention of food spoilage (41).
Bacteriocins exhibit a relatively narrow range of inhibition compared to the broad spectrum of classical antimicrobial agents; this, however, is thought to be a potential advantage regarding resistance development as a narrow-spectrum agent is thought to generate less pressure for resistance to develop (15).
Staphylococcus aureus and Staphylococcus epidermidis are prolific producers of bacteriocins, often referred to as staphylococcins (2). Many bacteriocins have been isolated from S. aureus, including staphylococcin C55 (34), type II bacteriocin aureocin A53 (36), and Bsa (bacteriocin of S. aureus) lantibiotic identified in community strains of S. aureus (7). Coagulase-negative staphylococci (CNS) also produce a number of bacteriocins, including the lantibiotics epilancin K7 (44), pep5 (24), epidermin (1), epilancin 15x (10), epicidin 280 (18) gallidermin (25), and warnericin (31, 45).
Gram-positive bacteria are still a major cause of infection with an increasing number of strains multiresistant to commonly used antimicrobials. It is therefore of great importance to identify and develop new agents that show activity toward these bacteria. Ideally, such agents should also show an alternative mode of action to that of the more traditional compounds. Here, we describe the peptide epidermicin NI01, which we have recently isolated from S. epidermidis strain 224. The peptide shows a broad spectrum of activity against a wide range of Gram-positive pathogens including methicillin-resistant S. aureus (MRSA) and vancomycin-resistant enterococci (VRE), both of which are a problem within the clinical environment. Analysis of the draft genome sequence of strain 224 indicated that the peptide is plasmid encoded and that the genetic locus involved in the production of the peptide does not fit with the conventional type II bacteriocin grouping system but does fit into the newly proposed group of class II-like peptides (21). We report the identification, characterization, genetic analysis, and recombinant expression of the peptide epidermicin, the first type II antimicrobial peptide to be isolated from S. epidermidis and the first of this family to be produced using a recombinant expression system.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
We previously determined that the skin isolate, S. epidermidis strain 224, produced an inhibitor, here described as epidermicin NI01, which was active against Gram-positive bacteria. A selection of indicator strains (Table 1) was used to determine the spectrum and level of activity of peptide NI01. Selected indicators (from an in-house collection), Micrococcus luteus, S. epidermidis strain ATCC 12228, and clinical S. epidermidis strain 156, a biofilm-forming isolate, were used to assess the activity of peptide recovered from strain 224 during purification.
Table 1.
Antibacterial spectrum of epidermicin NI01 against a range of multiresistant pathogens
| Straina | Epidermicin MIC |
|
|---|---|---|
| μg/ml | nM | |
| S. epidermidis 1 | 1 | 160 |
| S. epidermidis 2 | 4 | 658 |
| S. epidermidis 37 | 2 | 329 |
| S. epidermidis 42 | 2 | 329 |
| S. epidermidis 44 | 4 | 658 |
| S. epidermidis 60 | 2 | 329 |
| S. epidermidis 93 | 2 | 329 |
| S. epidermidis 156 | 0.0625 | 10.28 |
| S. epidermidis RP62A | 2–4 | 329–658 |
| S. saprophyticus | 0.5–1 | 82.3–160 |
| S. hominis | 1 | 160 |
| S. warneri | 1 | 160 |
| E. faecalis | 1 | 160 |
| VRE 1 | 1 | 160 |
| VRE 2 | 1 | 160 |
| VRE 3 | 1 | 160 |
| VRE 4 | 1–2 | 160–329 |
| VRE 5 | 0.5–1 | 82.3–160 |
| VRE 6 | 0.5 | 82.3 |
| MRSA s37 (EMRSA 15) | 1–2 | 160–329 |
| MRSA s41 (EMRSA 15) | 1–2 | 160–329 |
| MRSA s71 (EMRSA 15) | 2 | 329 |
| S. aureus 1195 | 2 | 329 |
S. hominis, Staphylococcus hominis; S. warneri, Staphylococcus warneri; E. faecalis, Enterococcus faecalis; EMRSA, epidemic methicillin-resistant S. aureus.
For routine culture, bacteria were grown on Columbia blood agar (CBA) plates (Oxoid). Overnight cultures were prepared in tryptone soy broth (TSB) containing 4% yeast extract (Oxoid). Luria-Bertani broth (Sigma) was used for culturing E. coli BL21(DE3), and 2× yeast extract-tryptone (YT) broth (Melford Laboratories, United Kingdom) was used for culturing E. coli transformants. Unless stated otherwise, all cultures were incubated in an aerobic atmosphere at 37°C.
Assessment of antimicrobial activity using the well and spot inhibition assays.
Antimicrobial activity was assessed by the well inhibition method as previously described (38) with the following modifications: 50-μl drops of 2-fold dilutions of solutions for analysis were added into 4-mm wells that had been bored into the surface of CBA and sealed with 1.5% agarose. The surface of the agar plate was then exposed to chloroform vapor for 30 min. A suspension of an indicator strain equivalent to 1.5 × 108 CFU/ml was diluted 1/10 for staphylococci or used neat for M. luteus and was swabbed evenly over the surface. The plates were incubated for 18 to 24 h, after which the zones of inhibition were measured. The activity was defined as the reciprocal of the last dilution that produced a clear zone of inhibition and was expressed as arbitrary units (AU) per ml (38).
The spot-on-lawn method (30) was also used to determine the inhibitory activity of chromatography fractions. This involved inoculating the agar surface with a 15- to 20-μl drop of sample. The agar surface was then sterilized by exposure to chloroform vapors for 30 min before inoculation of the surface with indicator strains, as described above.
Purification of peptide epidermicin NI01.
S. epidermidis 224 was grown in overnight 20-liter fermentations in tryptic soy broth (TSB). These were then centrifuged at 10,000 × g for 15 min, and the supernatant was harvested. Epidermicin was purified by using Strata, a poly divinyl benzene hydrophobic resin (Phenomenex), during the initial capture phase, with activity eluted using 90% methanol at pH 2. The active fractions from this step were combined and applied to a cation exchange column (CIEX) containing Hi-Trap SP FF resin (GE Healthcare), separating the peptide based on its charge. A gradient of NaCl was used in combination with 20 mM MES to elute activity. Active fractions from CIEX were combined and further purified using a Proteo 90A (4-μm particle size) 250- by 10-mm (Phenomenex) reverse-phase high-performance liquid chromatography (RP-HPLC) column. The sample was then purified using an AKTA purifier HPLC machine (Amersham). Buffer A contained water plus 0.1% trifluoroacetic acid (TFA), and buffer B contained 100% MeCN and 0.1% TFA. A gradient of 10 to 60% buffer B was run over 120 min and recorded at 215 nm and 280 nm. Inhibitory activity was detected by using the spot-on-lawn assay against M. luteus.
MALDI-TOF.
Matrix-assisted laser desorption ionization (MALDI) was conducted to determine the purity of HPLC-separated samples. Briefly, a 1-μl sample was spotted onto the surface of a ground steel 96-well MALDI plate (Bruker) followed by 1 μl of matrix (10 mg/ml α-cyano-4-hydroxycinnamic acid [Sigma] dissolved in 1:1 acetonitrile-ethanol). The samples were then left to air dry at room temperature for 15 min prior to being analyzed using a Bruker Daltonics Ultraflex II tandem time of flight (TOF/TOF) instrument. Peptide calibration mix (Sigma) was used as an external calibration marker.
Mass spectrometric analysis of peptides and enzymatic digestion products.
HPLC-purified biologically active fractions were analyzed using nano-electrospray on a quadrupole time of flight (Q-TOF) mass spectrometer (Waters, Manchester, United Kingdom). A trypsin digest of the peptide was carried out by adding 15 μl of trypsin (Sigma) (0.01 mg/ml in 100 mM ammonium bicarbonate) to 2 μl of peptide with incubation at 37°C for 18 h. Chymotrypsin (Sigma) (25 μg/ml suspended in buffer containing 50 mM Tris-HCl–1 mM CaCl2) was used at an enzyme-to-substrate ratio of 1:50 with incubation at 27°C for 1 h prior to analysis by mass spectrometry.
Acetylation of the peptide was determined by adding 50 μl of 20 μl of acetic anhydride and 60 μl of methanol to 20 μl of the peptide solution (4 μl of peptide in 20 μl of 50 mM ammonium bicarbonate). The samples were then left at room temperature for 1 h before lyophilization and resuspension in 10 μl of 20% formic acid and mass spectrometry.
Determination of MIC.
The MICs of epidermicin NI01 against a range of bacteria were determined using 2-fold dilutions of antimicrobial agents in cation-adjusted Mueller-Hinton broth (MHB) (Oxoid) according to the broth microdilution guidelines set out by the Clinical and Laboratory Standards Institute ([CLSI] http://www.clsi.org).
Determination of enzyme, heat, and pH stability of peptide NI01.
Preparations of epidermicin of a known concentration (AU/ml) were incubated with various enzymes at 37°C for 4 h at the following concentrations: α-chymotrypsin, trypsin, protease, lipase, and α-amylase at 1 mg/ml; and lysozyme and proteinase K (Sigma) at 10 mg/ml. Following treatment, the activity in AU/ml was reassessed, using the well assay described above.
The stability of epidermicin to heat was examined by heating preparations to 80°C for 10, 15, 20, 30, and 60 min, and residual activity was assessed by measuring AU/ml. Similarly, the stability of epidermicin was assessed at pH 2, 4, 6, 8, and 10 by incubation in buffered solutions, followed by determination of residual AU/ml. Appropriate controls were used in all of these assays.
Hemolysis and cellular toxicity assays.
Estimation of the degree of hemolysis caused by epidermicin was conducted as previously described (11). Red blood cells (RBC) were treated with phosphate-buffered saline (PBS) and 1% Triton X-100 as indicators of 0 and 100% hemolysis, respectively. Following treatment, plates were centrifuged at 1,000 × g for 10 min, aliquots of the supernatant were diluted two times with PBS, and absorbance was measured at 560 nm. Assays were carried out in triplicate.
The toxicity of epidermicin against human fibroblast cell lines was determined at concentrations ranging from sub-MIC to 100× MIC using MTT [3-(4,5-dimethylthiazol-2-yl)2 2,5-diphenyl tetrazolium bromide] reduction and neutral red uptake assays.
Spontaneous mutation assays.
Studies were carried out as previously described (17). Briefly, single-step mutation studies were determined by plating out 0.1 ml of a 108 CFU/ml suspension of three biofilm-forming strains onto Mueller-Hinton (MH) agar plates containing epidermicin at concentrations of two, four, and eight times the MIC. The frequency of mutation was determined by colony counts at 24 and 48 h following inoculation.
The effect of growth in the presence of subinhibitory MIC concentrations was also assessed using semipure preparations of epidermicin. Strains were grown in Muller-Hinton broth (MHB) containing a subinhibitory concentration equivalent to half of the MIC. The cells were passaged daily into fresh MHB containing subinhibitory concentrations of epidermicin for 10 days, following which the MIC value for the strain was reassessed.
Examination of the kinetics of bacterial killing by epidermicin NI01.
Bacterial killing assays were based on previously described methods (36). Briefly, overnight cultures of the indicator strains M. luteus and clinical isolate 156, a biofilm-forming S. epidermidis strain, were diluted 100-fold in fresh MHB and incubated at 37°C for 4 h at 180 rpm. Epidermicin was then added at a concentration of 4× MIC. Samples were taken at regular intervals to determine the optical density at 600 nm (OD600). Samples were enumerated using the Miles and Misra method to obtain viable counts, and Gram staining was conducted on the cells to follow activity of the peptide against walls of target cells.
Draft genome sequence determination and preliminary annotation.
The genomic DNA of producer strain 224 was extracted using a QIAmp DNA Mini Extraction kit (Qiagen) as directed by the manufacturer but with prior incubation in lysis buffer (20 mM Tris [pH 8.0], 2 mM EDTA, and 1.2% Triton X-100) with 50 μl of lysostaphin (1 mg/ml), 50 μl of lysozyme (20 mg/ml), and 20 μl of mutanolysin (10,000 U/ml), followed by addition of proteinase K and RNase A. The integrity of the DNA was determined by visualization on an agarose gel, and the concentration was measured using a Qbit (Invitrogen) as per the manufacturer's instructions.
The draft genome sequence of strain 224 was determined at the University of Liverpool's Centre for Genome Research. A shotgun library of DNA fragments was prepared for analysis, and sequencing reactions were performed using a Roche 454 GS-FLX machine. Reactions were carried out using titanium chemistry on library fragments applied to one-quarter of an analysis plate. Raw data were assembled using GS De Novo Assembler, version 2.0.00.20 (Roche Diagnostics, Ltd., Lewes, England). Assembled contiguous sequences (contigs) were ordered with reference to the genome of S. epidermidis strain RP62A (accession number CP000029), and open reading frames were annotated following BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/) against staphylococcal genomes and data held in Swissprot, RefSeq, and Uniprot databases. Contig ordering and annotation were performed using SUGAR (a simple unfinished genome annotation resource) (M. Szubert and S. A. Beatson, unpublished; Perl scripts are available on request). A BLAST database of the draft genome sequence of strain 224 (produced by SUGAR) was queried using the tblastn algorithm, and a partial amino acid sequence was obtained for epidermicin NI01.
The DNA sequence encoding epidermicin was used to interrogate relevant DNA and protein databases and Bactibase (http://bactibase.pfba-lab-tun.org/main.php), a repository for bacteriocin sequences. Sequence alignment of peptides of interest was performed using the CLC Main Workbench program.
Cloning and expression of recombinant epidermicin NI01 in E. coli.
Total genomic DNA was used in PCR with primers NI01-F (5′-ATATATTACATATGGCAGCATTTATGAAGTTAATTCAG-3′) and NI01-R (5′-TACGTTCTCGAGTTATTATGCCCATAATTTTTTGATTTG-3′) target-ing the structural gene and including NdeI and XhoI restriction sites (underlined), respectively. The PCR product was cloned into an NdeI/XhoI-digested pET-29a vector using T4 DNA ligase and transformed into chemically competent BL21(DE3) E. coli cells using heat shock at 42°C for 45 s. Colony PCR and sequencing were conducted on transformants to determine validity of constructs. Colonies containing the insert were inoculated into 5 ml of 2× YT broth containing 15 μg/ml kanamycin and grown overnight at 37°C. These cultures were then used to inoculate 50 ml of 2× YT broth containing 15 μg/ml kanamycin and grown to an OD600 of 0.8 before induction with 50 μM/ml isopropyl-β-d-thiogalactopyranoside (IPTG) at 17°C overnight to reduce aggregation.
The following day, the cultures were centrifuged at 7,000 × g for 20 min. The supernatant was then discarded, and the pellet was resuspended in 3 ml of PBS. The cells were sonicated (Sonics Vibracell high-intensity ultrasonic processor unit) for 2 min with a microtip connector using the following parameters: pulse, 5 s; rest, 10 s; amplitude, 30. The lysate was centrifuged for 7,000 × g for 10 min, and the supernatant was acidified with 10% TFA prior to purification using a syringe-driven Sep-Pak cartridge with a MeCN step gradient applied from 10 to 90% MeCN, eluting in 1-ml fractions. Inhibitory activity of fractions against M. luteus was assessed using the spot-on-lawn method.
RP-HPLC was conducted as described above with fractions of 0.5 ml collected and assayed for activity using the spot-on-lawn assay. Antimicrobial activity was compared against fractions recovered from cultures of BL21 transformed with the pET-29a vector lacking the epidermicin structural gene, and MALDI-TOF was used to determine the presence of epidermicin, as described above.
Nucleotide sequence accession numbers.
The DNA and protein sequence data relating to the genes in the putative epidermicin NI01 locus in strain 224 have been submitted to GenBank under accessions numbers JQ025380 to JQ025388.
RESULTS
Determination of the draft genome sequence of S. epidermidis strain 224.
The draft genome sequence was assembled from a total of 275,997 reads with an average length of 407 bp (N50 = 148,627 [50% of the genome was contained in contigs of greater than 148,627 bp]), representing approximately 44-fold coverage. These were resolved into 58 contigs with an average length of 43,486 bp, yielding a draft genome of 2,539,959 bp, which putatively encodes 2,554 proteins.
The inhibitor produced by S. epidermidis strain 224 can be purified to homogeneity and was identified as a novel peptide, designated epidermicin NI01.
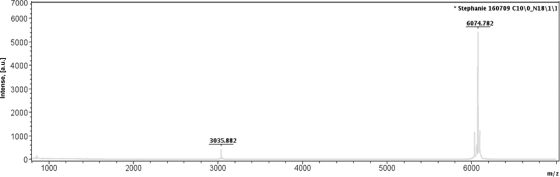
Epidermicin was purified by hydrophobic interaction, CIEX, and RP-HPLC, and the mass was determined to be 6,074 Da using MS (Fig. 1). No matches were obtained when data from trypsin digests were used to interrogate the MASCOT database, so the mass spectra were manually analyzed, leading to identification of the following sequence tags: (I/L)NAGQSFEW(I/L)YK (1,640.8 Da), (I/L)EGEGSSD(I/L)R (1,261.6 Da), and YEQ(I/L)AGAEAXXX or YEQ(I/L)AEQAXXX. These sequences were used to query the draft genome sequence of strain 224, and sequence tag NAGQSFEW was found to lie within an open reading frame (ORF) of 252 bp (Fig. 2). The ORF contained the coding sequence MAAFMKLIQFLATKGQKYVSLAWKHKGTILKWINAGQSFEWIYKQIKKLWA (sequence tag underlined) with a predicted mass of 6044.3 Da, approximately 30 Da less than the observed mass, indicating that the mature peptide carries an N-terminal formylated methionine.
Fig 1.
MALDI spectra for RP-HPLC-purified active fractions of peptide NI01. A mass of 6,074.782 Da was obtained for epidermicin. Other peaks represent multiple charged ions.
Fig 2.
Nucleotide sequence of a 252-bp ORF encoding the epidermicin structural gene, edcA. The deduced amino acid sequences are shown above the DNA sequence. Putative promoter elements are underlined at the −10 and −35 positions. Residues in italics show the PCR primers used for cloning and expression of recombinant NI01.
Epidermicin NI01 is stable at a wide range of pHs and with exposure to high temperatures and the activity of several enzymes.
Treatment of epidermicin with lysozyme, lipase, and α-amylase caused no effect on activity compared to the control treatment with molecular-grade water. Following treatment with proteinase K or protease, a 75% reduction in activity was observed (640 AU/ml to 160 AU/ml). Treatment with trypsin resulted in a 50% reduction of activity (to 320 AU/ml). There are nine potential cleavage sites for trypsin within epidermicin. These results provide evidence for the proteinaceous nature of epidermicin. As the peptide was not affected by α-amylase or lipase, it probably does not contain polysaccharide or lipid moieties (4, 39).
When examined in the absence of any of the above enzymes, epidermicin was highly stable between pH 2 and 10 and retained activity following exposure to 80°C for up to 60 min.
Epidermicin NI01 is active against a range of Gram-positive pathogens, and exposure to epidermicin does not encourage the development of resistance.
The MICs of epidermicin against a range of Gram-positive pathogens associated with clinical infections are listed in Table 1. In particular, epidermicin showed high activity against multidrug-resistant S. epidermidis strains causing biofilm-related infections, MRSA, and vancomycin-resistant enterococci (VRE). The MIC values recorded indicate that epidermicin is active against susceptible cells in the nanomolar range.
Single-step mutation tests were carried out at two, four, and eight times the MIC of epidermicin against S. epidermidis strain RP62A (a well-characterized, biofilm-forming isolate), strain 53, a gentamicin-resistant clinical isolate, and strain 156, a gentamicin-sensitive clinical isolate. No evidence of mutation to resistance was observed with these three strains (data not shown).
Twelve biofilm-forming clinical CNS isolates associated with prosthetic joint infections were passaged in subinhibitory concentrations of semipure epidermicin. Two strains showed a 2-fold increase in the MIC of epidermicin after 10 days of passage. All of the other strains tested exhibited no change in MIC values (data not shown).
Epidermicin NI01 is rapidly bactericidal and nonlytic against susceptible cells.
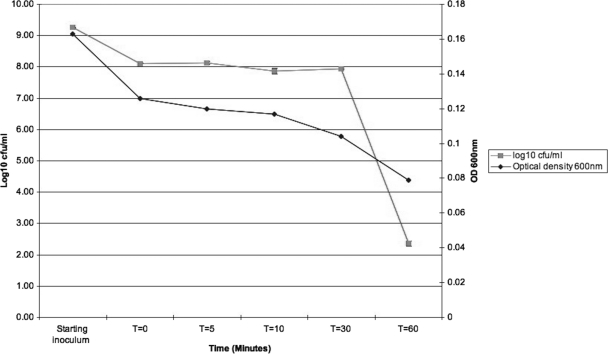
A bactericidal mode of action was observed against S. epidermidis strain 156; cell counts were reduced by almost 6 logs following a 30-min incubation. A decrease in OD600 of 0.085 was observed over 60 min, indicating limited cell lysis (Fig. 3).
Fig 3.
Effect of epidermicin NI01 on the viability and optical density of indicator strain S. epidermidis 156.
Assessment of the activity of epidermicin was also investigated by Gram staining susceptible cells at various time points following addition of the peptides. Prior to addition of peptide, at least 150 small cocci were observed per field of view, but addition of peptide led to a decrease to fewer than 20 cells per field of view, and ghost cells were present, indicating that the cells were no longer able to retain the stain. These observations indicate that the peptide causes membrane damage and disruption.
Epidermicin does not cause hemolysis or cell toxicity at 100× MIC.
Hemolysis assays were conducted against human erythrocytes using concentrations ranging from sub-MIC to 100× MIC of epidermicin. None of the concentrations assessed caused lysis of the erythrocytes (data not shown).
Toxicity studies were also conducted against human dermal fibroblast cell lines using concentrations ranging from sub-MIC to 100× MIC of epidermicin. None of the concentrations assessed caused any toxic effects against the cells when assessed by both MTT reduction and neutral red uptake.
Epidermicin shares homology with previously reported bacteriocins.
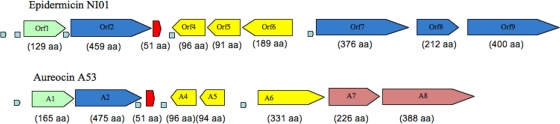
Analysis of genomic region containing the structural gene for epidermicin identified nine ORFs of potential significance (accessions numbers JQ025380 to JQ025388). A detailed description of ORFs and corresponding proteins is contained in Table 2 and Fig. 4. ORF 1 shares considerable similarity with the YdbS protein from Bacillus species, which is associated with resistance to antimicrobial agents (5), as is ORF 2, a homologue of the YdbT protein. These are followed by ORF 3, or edcA, the structural gene encoding epidermicin NI01. Downstream of edcA are ORFs 4 and 5, encoding hypothetical proteins also seen in the aureocin locus. ORF 6 is a homologue of a hypothetical protein in Staphylococcus saprophyticus (27), ORF 7 encodes an efflux transporter of the RND (resistance nodulation and cell division) family, and ORFs 8 and 9 are highly homologous to ABC transporters widely reported in staphylococci.
Table 2.
Genes neighboring edcA, the epidermicin structural gene
| ORF | Homolog description | Protein identity (%) | Function | Reference(s) |
|---|---|---|---|---|
| ORF 1 | YdbS-like protein | 34 | Self-protection (immunity) | 36 |
| ORF 2 | YdbT-like protein | 32 | Self-protection (immunity) | 36 |
| ORF 3 | Lacticin Z-like protein | 51 | Structural gene | 16, 22 |
| ORF 4 | Hypothetical protein | 47 | Unknown | 35 |
| ORF 5 | Hypothetical protein | 56 | Unknown | 35 |
| ORF 6 | Hypothetical protein | 36 | Unknown | 27 |
| ORF 7 | RND efflux transporter of S. warneria | 80 | Transport of peptide | 29 |
| ORF 8 | ABC transporter of S. epidermidis | 93 | Transport of peptide, immunity | 50 |
| ORF 9 | ABC transporter of S. epidermidis | 86 | Transport of peptide | 50 |
S. warneri, Staphylococcus warneri.
Fig 4.
Genetic organization of the putative epidermicin NI01 locus compared to the genes surrounding aureocin A53 (51 amino acids; not drawn to scale). Putative promoters are represented by small arrows, and terminators are represented by small boxes; numbers in parentheses indicate the number of amino acids encoded by each gene.
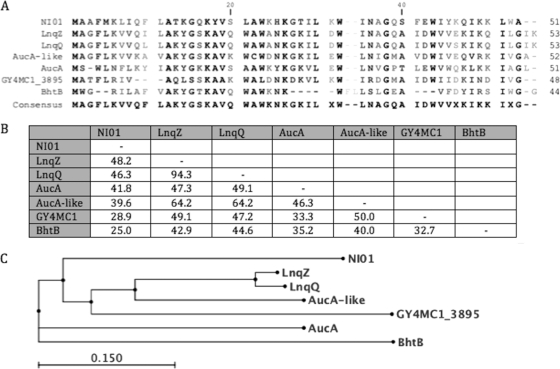
BLAST searches indicated that epidermicin displays homology to a number of previously described bacteriocins. Figure 5 displays the degree of amino acid sequence identity to lacticin Z (48% identity; accession number BAF75975) produced by Lactococcus lactis QU14 (21), lacticin Q (46% identity; BAF57910) produced by Lactococcus lactis QU5 (16), and plasmid-encoded aureocin A53 (41% identity; AF447813) produced by S. aureus (36). More distant homologues were an Auc-like hypothetical peptide (40%) encoded by Corynebacterium jeikeium plasmid pA501 (AY266269), a hypothetical protein (29%) encoded on Geobacillus plasmid pGY4MC101 (YP_003991117), and mutacin BhtB (25%), a bacteriocin produced by Streptococcus rattus strain BHT (DQ145753) (20). A ClustalW alignment was conducted to demonstrate sequence similarity between epidermicin and the above peptide sequences, and the relationship of all peptides is depicted in Fig. 5.
Fig 5.
ClustalW alignment of epidermicin NI01 with related peptides (A) with an indication of the percent amino acid identity between peptides of the type IIc/unmodified bacteriocin group (B) and a distance tree displaying the relationship of the peptides (C).
The genetic organization of epidermicin was compared to that of aureocin A53 to determine any similarities (Fig. 4). The genetic organizations of lacticin Q and Z have not yet been published. Analysis demonstrated that both aureocin 53 and epidermicin contained YdbT and YdbS homologues, possibly involved in immunity, in similar positions immediately upstream of the structural genes (Table 2). Downstream of the structural genes, synteny is largely maintained with the aureocin A53 locus with two short hypothetical proteins and genes that encode ABC transporters (36).
Recombinant epidermicin can be expressed in E. coli.
Cloning of edcA in E. coli strain BL21(DE3) and induction with 50 μM/ml IPTG led to expression of recombinant epidermicin, which was recovered from cells using sonication. Purified recombinant epidermicin was analyzed using MALDI-TOF MS demonstrating a peptide of 6,074 Da. This peptide also displayed activity against M. luteus and S. aureus strains in lawn assays, confirming recombinant expression of epidermicin NI01 (data not shown). This represents the first successful expression in E. coli of a bacteriocin from the type IIc, aureocin-like family.
DISCUSSION
According to the most recent classification system proposed by Heng and Tagg (19), bacteriocins can be separated into four classes. Class I includes the posttranslationally modified bacteriocins referred to as lantibiotics due to the presence of unusual posttranslationally modified amino acids such as lanthionine; class II contains small (<10 kDa) heat-stable membrane-active bacteriocins; class III inhibitors, which are often much larger (>30 kDa), are heat-labile bacteriocins; and class IV contains cyclic bacteriocins.
There are some striking differences between the small bacteriocins showing similarity to epidermicin and the conventional type II bacteriocins. These include stability at high pH, a lack of cysteine residues, absence of a conserved “pediocin box” containing the sequence YGNG(V/L), absence of a leader peptide, high tryptophan content, and high cationic charge. In line with findings for other leaderless bacteriocins (16), epidermicin carries an N-terminal formylated methionine. These peptides have been placed in type IIc (19), which encompasses a diverse family containing aureocin A53, lacticin Q, and lacticin Z. Given the homology to these peptides, we suggest that epidermicin is a new addition to this group.
All of the agents in this bacteriocin family are rich in tryptophan, with NI01 and lacticin Q and Z containing four and aureocin A53 containing five tryptophan residues. Tryptophan is an amphiphilic amino acid, which is thought to be important in the interaction between antimicrobial peptides and bacterial membranes (12). Using fluorescence spectroscopy, the C-terminal tryptophan in pediocin PA-1 and mesentericin Y105 has been shown to be essential for antimicrobial activity (13). The location of the tryptophan has also been shown to affect the extent to which penetration occurs (6, 9, 32). In the synthetic peptide cecropin A(1-8)-magainin 2(1-12) (consisting of residues 1 to 8 of cecropin A and residues 1 to 12 o f magainin 2), tryptophan has been proposed as a potential anchor site on the membrane (37). Epidermicin, aureocin A53, and lacticin Z are highly cationic with a net charge of +8, and lacticin Q has a net charge of +6. Previous studies have indicated that some highly cationic peptides showing antimicrobial activity exhibit extremely low levels of hemolytic activity (23, 42).
Lacticins Q and Z have been shown to exhibit a good range of activity against Gram-positive bacteria, with lacticin Q showing superior activity to nisin in the inhibition of lactococci and bacilli (16). Lacticin Q, however, is reported to have poor activity against staphylococci and streptococci though activity against only a limited number of isolates has been assessed (21). Aureocin A53 is active in the nano- to micromolar range against a range of Gram-positive bacteria, including lactic acid bacteria, listeria, and staphylococci (36).
Epidermicin was selected by our group for its ability to inhibit CNS and S. aureus and has been shown to inhibit a range of clinically relevant pathogens (Table 1). S. epidermidis strain 224 was isolated from a skin swab, and it is therefore not surprising that it exhibits activity against other staphylococci and other organisms that may pose a threat to survival within the human commensal skin flora (41).
Similar to results observed for lacticin Q and aureocin A53, which show no hemolysis on sheep and horse blood agar plates (16, 36), epidermicin did not show any significant homology to hemolysins and exhibited no hemolysis of human erythrocytes at 100× MIC.
High levels of conserved residues between epidermicin and lacticin Q and Z were observed between Met1 and Gln37 with 57% of residues being identical. However, the C-terminal sequences were quite variable, sharing only conserved residues Trp41, Ile46, and Lys47, therefore suggesting that the presence of these residues at specific positions could be important for maintaining the structure and activity of this group of peptides.
Lysine is the most predominant amino acid in epidermicin, making up 17.6% of the peptide. Lysine-enriched peptides display strong antimicrobial activity and reduced hemotoxicity and cytotoxicity (40), which may explain the low toxicity of epidermicin NI01. No cytotoxicity studies have been published for aureocin A53 or lacticin Q and Z, but it is known that these peptides are not hemolytic and that they all contain a high proportion of lysine residues, with aureocin A53 containing 10 (19%) and lacticin Q and Z containing 8 (15%).
Epidermicin shows high stability under neutral, acidic, and alkaline conditions, a feature also observed for aureocin 53 (35), lacticin Z (21), and lacticin Q (16). This is an advantage over other common bacteriocins, and it has been shown that nisin is most susceptible to heat treatment under alkaline conditions (8).
A bactericidal mode of action was observed against S. epidermidis 156 with nearly a 6-log reduction in growth following a 30-min incubation. No specific targets are involved in the activity of aureocin A53 (36) and lacticin Q (47), which bind to both negatively charged and neutral liposomes (35). In a recent study, Yoneyama et al. (49) suggested that the mechanism of action of lacticin Q is more comparable to that of magainin 2 (isolated from the skin of the African clawed frog Xenopus laevis) (51) than that of other bacteriocins, including nisin and lacticin 3147, whose antimicrobial activities rely on the presence of docking molecules such as the peptidoglycan precursor lipid II. Lacticin Q was shown to act in the absence of a docking molecule, exerting its antimicrobial activity via the formation of huge toroidal pores (HTP), which are between 4.6 and 6.6 nm in diameter, representing the largest HTP ever described (48). Toroidal pore formation is thought to allow lacticin Q to achieve high antimicrobial activity at nanomolar concentrations (48). Pore formation is initiated via electrostatic interaction with the phospholipid bilayer. The peptide then forms and amphiphilic α-helical structure, and a pore is formed, accompanied by a rapid lipid trans-bilayer movement, referred to as lipid “flip-flop” (48), and leakage of cellular macromolecules. Lacticin Q has been shown to bind to the cell membranes of Gram-negative bacteria, but it does not go on to form pores in the membranes of these bacteria because of the physicochemical properties of the outer membranes (49). Preliminary NMR 1H/1H spectra for epidermicin show excellent signal dispersion and line widths consistent with formation of a monomer (J. P. Derrick, personal communication). Combined with in silico prediction, these analyses provide evidence that epidermicin forms a globular α-helical structure, suggesting that epidermicin will behave in a similar manner to lacticin Q, but further work is required to elucidate the mode of action. The nonspecific activity of these peptides may explain the lack of spontaneous mutation to resistance that is observed. This is obviously a favorable trait with respect to the clinical application of bacteriocins.
The edcA locus is carried on a plasmid, as seen for a number of other homologues in this family. Streptococci are known to carry multiple bacteriocins of the lantibiotic class on plasmids (46), but many staphylococcal bacteriocins are chromosomally encoded (44). The evolutionary significance of carriage of these unmodified bacteriocins on plasmids is not clear.
The genes surrounding edcA share notable synteny with the aureocin A53 locus, with both gene clusters located on plasmids. These regions contain homologues of YdbS and YdbT, which have previously been found to confer resistance to antimicrobial compounds produced by Bacillus amyloliquefaciens FZB42 (5). The region containing edcA also contains an RND efflux transporter, which is known to be involved in the transport of lipophilic or amphiphilic molecules (29). In keeping with the transport systems associated with nonlantibiotics (50), the RND transporter may be important for the transport of epidermicin in association with two ABC transporters that are possibly cotranscribed.
It was originally suggested that the genes surrounding AucA in S. aureus were unlikely to be required for production (and export) of a functioning inhibitor (36). However, given the degree of similarity between the loci for aureocin A53 and epidermicin NI01, we suggest that it is likely that the proteins encoded in the edcA locus are all required for production and export of epidermicin and for producer self-protection in S. epidermidis strain 224. A full functional analysis of the other genes in the ecdA locus is warranted and will clarify the role of each protein in export of and immunity to epidermicin. Such studies are outside the scope of the current report but could be achieved through heterologous expression in a staphylococcal or lactococcal host.
We have demonstrated that expression of the structural gene alone is sufficient for production of epidermicin NI01 by E. coli, but this involves recovery of the peptide from the cytoplasm of E. coli, which is suboptimal. When we consider larger-scale production of epidermicin, it will be important to examine heterologous expression in a genetically pliable S. epidermidis strain or a different staphylococcal background, where peptide can be recovered from culture supernatants. Additional systems for expression in E. coli should also be considered, including the SUMO fusion system, which has recently facilitated high-yield production of antimicrobial peptides (3).
In summary, the characteristics of peptide epidermicin NI01 described in the current study demonstrate that this novel peptide may be a promising candidate for use within the clinical environment. It displays potent inhibitory activity against a range of multiresistant Gram-positive bacterial pathogens, and preliminary toxicity data indicate it has no adverse effects on human blood cells or on human fibroblast cell lines, suggesting a wide therapeutic window. The peptide has high-heat stability, which may be beneficial in recombinant expression and purification and may allow a diversity of formulations, and the low rate of mutation to resistance that was observed is encouraging with respect to clinical applications. Further studies are warranted to expand the toxicological data set relating to epidermicin NI01 and to elucidate the mode of action employed by this novel peptide.
ACKNOWLEDGMENTS
We thank Emmy Hoyes for assistance with MS analysis and the staff at the University of Manchester Biomolecular Analysis Facility for further peptide analysis. We gratefully acknowledge the contributions of Marek Szubert and Scott Beatson (University of Queensland, Brisbane, Australia) for running the SUGAR genome annotation pipeline.
Footnotes
Published ahead of print 12 December 2011
REFERENCES
- 1. Allgaier H, Jung G, Werner RG, Schneider U, Zahner H. 1986. Epidermin: sequencing of a heterodet tetracyclic 21-peptide amide antibiotic. Eur. J. Biochem. 160:9–22 [DOI] [PubMed] [Google Scholar]
- 2. Bastos MC, Ceotto H, Coelho ML, Nascimento JS. 2009. Staphylococcal antimicrobial peptides: relevant properties and potential biotechnological application. Curr. Pharm. Biotechnol. 10:38–61 [DOI] [PubMed] [Google Scholar]
- 3. Bommarius B, et al. 2010. Cost-effective expression and purification of antimicrobial and host defense peptides in Escherichia coli. Peptides 31:1957–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brink M, Todorov SD, Martin JH, Senekal M, Dicks LM. 2006. The effect of prebiotics on production of antimicrobial compounds, resistance to growth at low pH and in the presence of bile, and adhesion of probiotic cells to intestinal mucus. J. Appl. Microbiol. 100:813–820 [DOI] [PubMed] [Google Scholar]
- 5. Butcher BG, Helmann JD. 2006. Identification of Bacillus subtilis W-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by bacilli. Mol. Microbiol. 60:765–782 [DOI] [PubMed] [Google Scholar]
- 6. Chikindas ML, et al. 1993. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl. Environ. Microbiol. 59:3577–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daly KM, et al. 2010. Production of the Bsa lantibiotic by community-acquired Staphylococcus aureus strains. J. Bacteriol. 192:1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. 1996. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69:193–202 [DOI] [PubMed] [Google Scholar]
- 9. Drider D, Fimland G, Hechard Y, McMullen LM, Prevost H. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ekkelenkamp MB, et al. 2005. Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis. FEBS Lett. 579:1917–1922 [DOI] [PubMed] [Google Scholar]
- 11. Fernandez-Lopez S, et al. 2001. Antibacterial agents based on the cyclic d,l-[alpha]-peptide architecture. Nature 412:452–455 [DOI] [PubMed] [Google Scholar]
- 12. Fimland G, Eijsink VGH, Nissen-Meyer J. 2002. Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry 41:9508–9515 [DOI] [PubMed] [Google Scholar]
- 13. Fleury Y, et al. 1996. Covalent structure, synthesis, and structure-function studies of mesentericin Y 10537, a defensive peptide from Gram-positive bacteria Leuconostoc mesenteroides. J. Biol. Chem. 271:14421–14429 [DOI] [PubMed] [Google Scholar]
- 14. Fontana MB, de Bastos Mdo C, Brandelli A. 2006. Bacteriocins pep5 and Epidermin inhibit Staphylococcus epidermidis adhesion to catheters. Curr. Microbiol. 52:350–353 [DOI] [PubMed] [Google Scholar]
- 15. Fox JL. 2006. The business of developing antibacterials. Nat. Biotechnol. 24:1521–1528 [DOI] [PubMed] [Google Scholar]
- 16. Fujita K, et al. 2007. Structural analysis and characterization of lacticin Q, a novel bacteriocin belonging to a new family of unmodified bacteriocins of gram-positive bacteria. Appl. Environ. Microbiol. 73:2871–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haas CE, Nix DE, Schentag JJ. 1990. In vitro selection of resistant Helicobacter pylori. Antimicrob. Agents Chemother. 34:1637–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heidrich C, et al. 1998. Isolation, characterisation and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl. Environ. Microbiol. 64:3140–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heng NCK, Tagg JR. 2006. What's in a name? Class distinction for bacteriocins. Nat. Rev. Microbiol. doi:10.1038/nrmicro1273-c1 [Google Scholar]
- 20. Hyink O, Balakrishnan M, Tagg JR. 2005. Streptococcus rattus strain BHT produces both a class I two-component lantibiotic and a class II bacteriocin. FEMS Microbiol. Lett. 252:235–241 [DOI] [PubMed] [Google Scholar]
- 21. Iwatani S, Zendo T, Yoneyama F, Nakayama J, Sonomoto K. 2007. Characterisation and structure analysis of a novel bacteriocin, lacticin Z, produced by Lactococcus lactis QU14. Biosci. Biotechnol. Biochem. 71:1984–1992 [DOI] [PubMed] [Google Scholar]
- 22. Jack RW, Tagg JR, Ray B. 1995. Bacteriocins of Gram-positive bacteria. Microbiol. Rev. 59:171–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang Z, et al. 2008. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic α-helical cationic antimicrobial peptides. Biopolymers 90:369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaletta C, et al. 1989. Pep5, a new lantibiotic: structural gene isolation and prepeptide sequence. Arch. Microbiol. 152:16–19 [DOI] [PubMed] [Google Scholar]
- 25. Kellner R, et al. 1988. Gallidermin: a new lanthionine-containing polypeptide antibiotic. Eur. J. Biochem. 177:53–59 [DOI] [PubMed] [Google Scholar]
- 26. Klaenhammer TR. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39–85 [DOI] [PubMed] [Google Scholar]
- 27. Kuroda M, et al. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. U. S. A. 102:13272–13277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Majeed H, Gillor O, Kerr B, Riley MA. 2011. Competitive interactions in Escherichia coli populations: the role of bacteriocins. ISME J. 5:71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marquez B. 2005. Bacterial efflux systems and efflux pumps inhibitors. Biochimie 87:1137–1147 [DOI] [PubMed] [Google Scholar]
- 30. Mayr-Harting A, Hedges AJ, Berkeley RCW, Norris JR, Ribbons DW. 1972. Methods for studying bacteriocins, p 315–422 In Norris JR, Ribbons DW. (ed), Methods in microbiology. Academic Press, New York, NY [Google Scholar]
- 31. Minamikawa M, Kawai Y, Inoue N, Yamazaki K. 2005. Purification and characterization of Warnericin RB4, anti-Alicyclobacillus; bacteriocin, produced by Staphylococcus warneri; RB4. Curr. Microbiol. 51:22–26 [DOI] [PubMed] [Google Scholar]
- 32. Morisset D, Berjeaud J-M, Marion D, Lacombe C, Frere J. 2004. Mutational analysis of mesentericin Y105, an anti-listeria bacteriocin, for determination of impact on bactericidal activity, in vitro secondary structure, and membrane interaction. Appl. Environ. Microbiol. 70:4672–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nascimento JS, et al. 2006. Bacteriocins as alternative agents for control of multiresistant staphylococcal strains. Lett. Appl. Microbiol. 42:215–221 [DOI] [PubMed] [Google Scholar]
- 34. Navaratna M, Sahl HG, Tagg JR. 1998. Two-component Anti Staphylococcus aureus lantibiotic activity produced by Staphylococcus aureus C55. Appl. Environ. Microbiol. 64:4803–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Netz DJ, Bastos Mdo C, Sahl HG. 2002. Mode of action of the antimicrobial peptide aureocin A53 from Staphylococcus aureus. Appl. Environ. Microbiol. 68:5274–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Netz DJ, et al. 2002. Biochemical characterisation and genetic analysis of aureocin A53, a new, atypical bacteriocin from Staphylococcus aureus. J. Mol. Biol. 319:745–756 [DOI] [PubMed] [Google Scholar]
- 37. Oh D, et al. 2000. Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A(1-8)-magainin 2(1-12) and its analogues, on their antibiotic activities and structures. Biochemistry 39:11855–11864 [DOI] [PubMed] [Google Scholar]
- 38. Parente E, Hill C. 1992. A comparison of factors affecting the production of two bacteriocins from lactic acid bacteria. J. Appl. Microbiol. 73:290–298 [Google Scholar]
- 39. Risøen PA, Rønning P, Hegna IK, Kolstø AB. 2004. Characterization of a broad range antimicrobial substance from Bacillus cereus. J. Appl. Microbiol. 96:648–655 [DOI] [PubMed] [Google Scholar]
- 40. Sato H, Feix JB. 2008. Lysine-enriched cecropin-mellitin antimicrobial peptides with enhanced selectivity. Antimicrob. Agents Chemother. 52:4463–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tagg J, Dajani A, Wannamaker L. 1976. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40:722–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tencza SB, et al. 1999. Lentivirus-derived antimicrobial peptides: increased potency by sequence engineering and dimerization. J. Antimicrob. Chemother. 44:33–41 [DOI] [PubMed] [Google Scholar]
- 43. Todorov SD, et al. 2010. Chracterisation of an antiviral pediocin-like bacteriocin produced by Enterococcus faecium. Food Microbiol. 27:869–879 [DOI] [PubMed] [Google Scholar]
- 44. van de Kamp M, et al. 1995. Elucidation of the primary structure of the lantibiotic epilancin K7 from Staphylococcus epidermidis K7 cloning and characterisation of the epilancin-K7-encoding gene and NMR analysis of mature epilancin K7. Eur. J. Biochem. 230:587–600 [DOI] [PubMed] [Google Scholar]
- 45. Verdon J, Berjeaud J-M, Lacombe C, Héchard Y. 2008. Characterization of anti-Legionella activity of warnericin RK and delta-lysin I from Staphylococcus warneri. Peptides 29:978–984 [DOI] [PubMed] [Google Scholar]
- 46. Wescombe P, et al. 2006. Megaplasmids encode differing combinations of lantibiotics in Streptococcus salivarius. Antonie Van Leeuwenhoek 90:269–280 [DOI] [PubMed] [Google Scholar]
- 47. Yoneyama F, et al. 2009. Lacticin Q, a lactococcal bacteriocin, causes high-level membrane permeability in the absence of specific receptors. Appl. Environ. Microbiol. 75:538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yoneyama F, et al. 2009. Peptide-lipid huge toroidal pore, a new antimicrobial mechanism mediated by a lactococcal bacteriocin, lacticin Q. Antimicrob. Agents Chemother. 53:3211–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoneyama F, et al. 2011. Lacticin Q-mediated selective toxicity depending on physicochemical features of membrane components. Antimicrob. Agents Chemother. 55:2446–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Young J, Holland IB. 1999. ABC transporters: bacterial exporters-revisited five years on. Biochem. Biophys. Acta 1461:177–200 [DOI] [PubMed] [Google Scholar]
- 51. Zasloff M. 1987. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. U. S. A. 84:5449–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]