Abstract
The mammalian Dcp2 mRNA-decapping protein functions primarily on a subset of mRNAs in a transcript-specific manner. Here we show that Dcp2 is an important modulator of genes involved in the type I interferon (IFN) response, which is the initial line of antiviral innate immune response elicited by a viral challenge. Mouse embryonic fibroblast cells with reduced Dcp2 levels (Dcp2β/β) contained significantly elevated levels of mRNAs encoding proteins involved in the type I IFN response. In particular, analysis of a key type I IFN transcription factor, IFN regulatory factor 7 (IRF-7), revealed an increase in both IRF-7 mRNA and protein in Dcp2β/β cells. Importantly, the increase in IRF-7 mRNA within the background of reduced Dcp2 levels was attributed to a stabilization of the IRF-7 mRNA, suggesting that Dcp2 normally modulates IRF-7 mRNA stability. Moreover, Dcp2 expression was also induced upon viral infection, consistent with a role in attenuating the antiviral response by promoting IRF-7 mRNA degradation. The induction of Dcp2 levels following a viral challenge and the specificity of Dcp2 in targeting the decay of IRF-7 mRNA suggest that Dcp2 may negatively contribute to the innate immune response in a negative feedback mechanism to restore normal homeostasis following viral infection.
INTRODUCTION
The interferon (IFN) response is a first line of defense against viral infection in mammals (1). Type I IFNs (IFN-α and IFN-β) are widely expressed cytokines that mediate the antiviral innate immune response (3). They are induced upon viral exposure and are extracellularly secreted to function on as-yet-uninfected cells. They lead to the activation of a global antiviral state in which virus replication is inhibited and components of the innate immune response are activated (11). Type I IFNs can be induced in most cell types and exhibit profound pleiotropic effects on many aspects of cellular functions, including gene transcription, protein translation, cell growth, and cell motility.
At the molecular level, the transcriptional activation of IFN-α/β genes is mediated by a complex array of pathways involving multiple transcription factors, including the IFN regulatory factors (IRFs) IRF-3 and IRF-7, which are crucial for mediating the induction of type I IFNs (31). IRF-3 is constitutively expressed in all cell types, while IRF-7 is usually induced by virus infection in many cell types. Plasmacytoid dendritic cells, which are the most potent IFN-α-producing cells, express high constitutive levels of IRF-7 (17). Most IFN-α subtypes require induced IRF-7, whereas IFN-β can be induced without IRF-7 by constitutively expressed IRF-3 (14).
Although transcriptional regulation of IFN gene expression in the activation of the antiviral innate immune response has been well characterized, the inherent instability of type I IFN mRNAs indicates that posttranscriptional regulation also contributes to the antiviral immune response (20). To date, much of the emphasis on the contribution of mRNA turnover within the immune response has focused on AU-rich element (ARE)-mediated mRNA decay of cytokine and chemokine mRNAs possessing AREs within their 3′ untranslated regions (UTRs) (32). Less is known regarding the nucleases involved and the contribution of non-ARE-mediated decay.
The regulation of mRNA turnover plays an important role in the control of gene expression and response to regulatory events. In eukaryotic cells, bulk mRNA decay typically initiates with the removal of the 3′ poly(A) tail, followed by degradation of the mRNA in a 5′-to-3′ direction or a 3′-to-5′ direction (12, 26). Degradation from the 3′ end is carried out by the cytoplasmic RNA exosome, which is a multisubunit complex possessing 3′-to-5′ exoribonuclease activity (27), and the resulting cap structure is hydrolyzed by the scavenger decapping enzyme DcpS (26). In the 5′-to-3′ decay pathway, the monomethyl guanosine (m7G) mRNA cap is cleaved first by a decapping enzyme and the monophosphate RNA is degraded progressively by the 5′-to-3′ exoribonuclease Xrn1 (8, 16). Dcp2 was the first decapping enzyme identified (9, 28, 36, 38), and more recently, Nudt16 was shown to be a second cytoplasmic decapping enzyme that can regulate the stability of specific mRNA substrates in mammalian cells (35). Furthermore, from a subset of mRNAs tested (23, 35), each decapping enzyme appears to target distinct transcripts, as well as overlapping transcripts.
To study the significance of decapping in global mRNA metabolism, we used mouse embryonic fibroblast (MEF) cells deficient in the Dcp2 or Nudt16 decapping enzyme for microarray analysis (35). Interestingly, we found that a group of antiviral genes were significantly upregulated in Dcp2 knockdown MEF cells (Dcp2β/β) but not in Nudt16 knockdown MEF cells. Further studies revealed that Dcp2 directly regulates the mRNA stability of IRF-7, a key transcription factor in the antiviral immune response. IRF-7 protein levels increased in Dcp2β/β MEF cells, which could subsequently lead to elevated expression of downstream antiviral effector genes. In addition, Dcp2 expression is also induced upon viral infection, providing the potential for a negative feedback regulatory network in the antiviral immune response.
MATERIALS AND METHODS
Plasmid constructs.
The pLKO.1-puro, pCMV-VSV-G, and pSPAX2 plasmids used in the generation of lentiviral particles were purchased from Sigma-Aldrich (St. Louis, MO). To construct the retroviral plasmids pBMN-Dcp2 and pBMN-Dcp2 EE/QQ, which express wild-type (WT) or catalytically mutant Dcp2 proteins, respectively, plasmids pET-Dcp2 and pET-Dcp2 EE/QQ (38) were used as templates with PCR primers 5′-CAGTCTCGAGATGGAGACCAAACGGGTGG-3′ and 5′-CAGTGCGGCCGCTCAAAGGTCCAAGATTTT-3′. The PCR products were purified and digested by XhoI and NotI and inserted into pBMN-I-GFP (Addgene, Cambridge, MA).
MEF cells.
The generation of the primary MEF cells derived from WT and Dcp2β/β 12- to 14-day embryos obtained from the same litter of a heterozygous cross was reported by Song et al. (35). Briefly, decapitated and eviscerated embryos were minced and trypsinized to generate single cells that were subsequently transfected with simian virus 40 large T antigen and selected by culturing in growth medium containing 1% serum, and transformed clonal cell lines were isolated. Dcp2β/β MEF cells do not generate detectable levels of Dcp2 by Western analysis (35).
Microarray analysis.
WT and Dcp2β/β MEF cells were infected with lentiviruses expressing the control vector or short hairpin RNA (shRNA) against Nudt16. Total RNAs were harvested at 2 days postinfection with TRIzol reagent (Invitrogen, Carlsbad, CA) by following the manufacturer's instructions. RNA quality was assessed on an RNA Nano Chip using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). The mRNA was amplified with a TotalPrep RNA amplification kit with a T7-oligo(dT) primer according to the manufacturer's instructions (Ambion), and microarray analysis was carried out with the Illumina Sentrix MouseRef-8 24K Array at The Burnham Institute (La Jolla, CA).
Real-time PCR.
RNAs were reverse transcribed by Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA) with random primers according to the manufacturer's instructions. mRNA levels were quantified by real-time PCR using SYBR green PCR core reagent (Applied Biosystems, Foster City, CA), and the abundance of specific mRNAs was quantified using the standard-curve method according to the recommendations of the manufacturer. Values were normalized to the stable β-actin RNA. Each mRNA was amplified using the appropriate specific primers. The primers used for real-time PCR are listed in Table S2 in the supplemental material. Real-time PCR was carried out with an ABI Prism 7900HT sequence detection system, and the specificity of the amplified PCR products was assessed by a melting curve analysis after the last cycle by the manufacturer's suggested program.
Western blotting.
Cells were sonicated in phosphate-buffered saline, and protein extract was resolved by 12.5% SDS-PAGE. Affinity-purified Dcp2 polyclonal antibody (1:500 dilution) (38), IRF-7 polyclonal antibody (1:500 dilution; Sigma-Aldrich), and monoclonal anti-glyceraldehyde 3-phosphate dehydrogenase (anti-GAPDH) antibody (1:2,000 dilution; Abcam Inc., Cambridge, MA) were used for Western blot analysis and visualized using secondary antibodies coupled to horseradish peroxidase (Jackson ImmunoResearch, West Grove, PA) and chemiluminescence (ECL; GE Healthcare Bio-Sciences Corp., Piscataway, NJ).
In vitro RNA-decapping assay.
Generation of the STX7 RNA has previously been described (24). The chimeric IRF-STX RNA, containing the IRF-7 5′-terminal 57 nucleotides (nt) linked onto the 5′ end of the STX7 RNA, was generated by two PCR steps as follows. First, primers mIRF7-adaptpr-2 and STX7-M3 were used to amplify the STX7 RNA from the plasmid pGEM-Stx7 (24). The gel-purified PCR product was subsequently used as the template for PCR amplification with primers T7-mIRF7-F2 and STX7-M3, which contains a T7 promoter primer upstream of the chimeric IRF-7–STX7 sequences. All RNAs were generated from a PCR-derived template using T7 RNA polymerase and cap labeled at the gamma phosphate with 32P as previously described (39). In vitro RNA-decapping assays were carried out essentially as described previously (24), with 50 ng of 6×His-tagged human Dcp2 protein at 37°C for 15 min in IVDA-2 buffer (10 mM Tris-HCl at pH 7.5, 100 mM potassium acetate, 2 mM magnesium acetate, 0.5 mM MnCl2, 2 mM dithiothreitol). An aliquot of each sample was resolved by polyethyleneimine cellulose thin-layer chromatography (PEI-TLC), developed in 0.45 M (NH4)2SO4, and exposed to a PhosphorImager. Quantifications were carried out using a Molecular Dynamics PhosphorImager using ImageQuant 5 software. Percent decapping was determined as the level of m7GDP relative to the total RNA used in the reaction.
Antiviral activity assay.
In order to induce IFN production, Dcp2β/β or WT MEF cells cultured in a 24-well plate to a confluent monolayer (approximately 1 × 106/well) were infected with Newcastle disease virus (NDV; multiplicity of infection [MOI] = 0.5) or herpes simplex virus 1 (HSV-1; MOI = 1). After 24 h of stimulation, the supernatants were collected. To test for antiviral activity, WT MEF cells cultured in a 96-well plate to a confluent monolayer (approximately 1 × 104/well) were incubated with the prepared virus-stimulated supernatants for 2 h, followed by a challenge with HSV at an MOI of 5. After 24 h of infection, the cells were stained with crystal violet and cell viability was quantitated by the amount of crystal violet taken up by the cells as a function of the absorbance read at a wavelength of 595 nm (37).
RESULTS
Expression of antiviral immune response genes is elevated in Dcp2β/β MEF cells.
In an effort to identify genes potentially regulated by the Dcp2 and/or Nudt16 decapping enzymes in cells, we recently reported a global analysis of mRNA decay following transcriptional silencing in MEF cell lines with reduced Dcp2 and/or Nudt16 protein levels (35). In a further evaluation of the original mRNA decay microarray data set, we now present an analysis of the global steady-state mRNA changes observed in the distinct decapping enzyme knockdown backgrounds (see Table S1 in the supplemental material). The data set consists of (i) WT MEF cells transduced with a control lentivirus (WT); (ii) Dcp2 knockdown MEF cells generated from a homozygous insertion of the β-galactosidase–neomyocin gene trap cassette, which results in >95% reduction of Dcp2 protein (Dcp2β/β) transduced with a control lentivirus; (iii) MEF cells transduced with a Nudt16-directed shRNA-expressing lentivirus (Nudt16KD); and (iv) Dcp2β/β MEF cells transduced with a Nudt16-directed shRNA-expressing lentivirus (Dcp2β/β/Nudt16KD). Compared to the WT MEF cells, 194 mRNAs were elevated more than 1.5-fold in Dcp2β/β cells (see Table S1A in the supplemental material), 323 were upregulated in Nudt16KD cells (see Table S1B), and 275 were upregulated in the double-knockdown cells (see Table S1C). Although an obvious array of mRNAs associated with a particular pathway was not detected in the Nudt16 knockdown list of genes, pathway analyses of the steady-state mRNA levels by Ingenuity Pathway Analysis software (Ingenuity Systems, CA) revealed a striking potential role for Dcp2 in antiviral innate immunity. An overrepresentation of mRNAs encoding proteins involved in the antiviral innate immune response was detected in Dcp2β/β cells or double knockdown cells but not in Nudt16KD cells (see Table S1A to S1C), indicating that they are specifically regulated by Dcp2. As shown in Fig. 1A, among the 35 genes involved in the antiviral immune response that were detected with confidence in the microarray, 14 were elevated >1.5-fold in Dcp2β/β cells. Three representative mRNAs, IRF-7, CXCL10, and OAS2, were chosen for further confirmation by real-time quantitative reverse transcription (qRT)-PCR. IRF-7 is a key transcription factor that induces the expression of type I IFNs and other cytokines and chemokines upon virus infection (21); CXCL10 is an IFN-inducible chemokine whose transcription is activated by IRF-7 (11), as well as directly by virus (30); OAS2 is an IFN-inducible gene which encodes 2′-5′-oligoadenylate synthetase, a critical effector in the innate immune response to viral infection (18). Consistent with the microarray, the expression of all three genes was upregulated from 2- to 5-fold in Dcp2β/β cells relative to that in WT cells, as shown by qRT-PCR analysis (Fig. 1B).
Fig 1.
A subset of genes involved in the antiviral immune response are upregulated in Dcp2β/β MEF cells. (A) WT and Dcp2β/β MEF cells were infected with lentivirus for 2 days. Total cellular RNAs were extracted and subjected to microarray analysis. Thirty-five genes involved in the cellular antiviral immune response with signals above the background were selected, and the n-fold change of each gene in Dcp2β/β MEF cells compared to the level in WT MEF cells is plotted. Data represent the average of two independent experiments, with error bars denoting the range of the two experiments. Dark bars indicate genes that were increased more than 1.5-fold in Dcp2β/β MEFs. (B) The levels of the indicated mRNAs were confirmed by real-time PCR analysis. The IRF-7, CXCL10, and OAS2 mRNAs were amplified with gene-specific primers and normalized to the β-actin mRNA level. The mRNA levels in the WT MEF cells were arbitrarily set to 100. The average of two independent experiments is shown with error bars indicating the range of the results in the two experiments.
The global mRNA analyses described above were carried out with cells infected with lentiviral particles. To examine whether the increased expression of the antiviral genes is a constitutive property of reduced Dcp2 in Dcp2β/β MEF cells or whether it requires viral infection, we determined the steady-state mRNA levels of four mRNAs encoding proteins involved in innate immunity. Importantly, steady-state IRF-7 mRNA levels were 2.2-fold higher in Dcp2β/β MEF cells than in WT cells prior to viral infection (Fig. 2A). Similarly, OAS2 mRNA levels were also elevated (2.5-fold) in Dcp2β/β MEF cells while no differences in CXCL10 mRNA levels were observed. IRF-3, which is also a critical transcription factor in the antiviral immune response but not upregulated in Dcp2β/β cells in the microarray, showed no significant difference between WT and Dcp2β/β MEF cells. These data indicate that the Dcp2 influence on IRF-7 and OAS2 mRNAs is not restricted to viral transduction.
Fig 2.
IFN-mediated antiviral immune response was elevated in Dcp2β/β MEF cells following lentivirus infection. (A) Total RNAs of WT and Dcp2β/β MEF cells were extracted, and mRNA levels of specific genes were determined by qRT-PCR, normalized to β-actin mRNA. mRNA levels in WT cells were arbitrarily set to 1. Data represent the average of three independent experiments, with the standard deviation denoted by the error bar. **, P < 0.01 (Student's t test). (B) WT and Dcp2β/β MEF cells were treated with control lentivirus, and total RNAs were extracted at 0, 12, and 24 h postinfection. mRNA levels of specific genes were determined by qRT-PCR and normalized to β-actin mRNA. IRF-7, OAS2, CXCL10, and IRF-3 mRNA levels in WT cells at time zero were arbitrarily set to 1. IFN-α2 and IFN-β mRNA levels in WT cells at 12 h postinfection were arbitrarily set to 1 due to the very low levels of these two mRNAs before infection. Data represent the average of three independent experiments, with the standard deviation denoted by the error bar. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test).
To assess the impact of Dcp2 on the fate of a subset of mRNAs involved in antiviral innate immunity, mRNA levels were determined up to 24 h after lentiviral transduction. As shown in Fig. 2B, IRF-7 mRNA levels increased approximately 5-fold in Dcp2β/β MEF cells compared to those in WT cells at 12 and 24 h after viral infection. Similar results were observed for OAS2 mRNA, which had a 6-fold increase in Dcp2β/β cells. A modest (1.8-fold) increase in CXCL10 mRNA was evident in Dcp2β/β MEF cells 24 h after viral exposure. No significant difference between WT and Dcp2β/β MEF cell IRF-3 mRNA levels was detected following viral infection. Our previous finding that the stability of the ARE-containing IFN-α2 mRNA is regulated by Dcp2 (23), in conjunction with the fact that IRF-7 is a transcriptional activator of IFN genes (15), prompted us to test the expression of IFN-α and IFN-β in WT and Dcp2β/β cells upon viral infection. IFN-α2 and IFN-β mRNAs were below the limit of detection prior to infection but were both induced following a virus challenge. A 3-fold greater induction of IFN-α2 mRNA was observed in Dcp2β/β MEF cells than in WT cells, while a 2-fold higher induction of IFN-β was evident. These data indicate that mRNA levels for a subset of genes involved in the IFN-mediated antiviral response pathway are upregulated following viral infection in Dcp2β/β MEF cells.
Dcp2 regulates the mRNA stability and protein expression of IRF-7.
To determine whether the Dcp2-dependent increase in IRF-7 mRNA levels is a consequence of increased mRNA stability, we tested the half-life of the IRF-7 mRNA following transcriptional arrest. Consistent with our previous demonstration that the stability of IFN-α2 is regulated by Dcp2 (23), the stability of IRF-7 mRNA is controlled by Dcp2 (Fig. 3). WT and Dcp2β/β MEF cells were infected with lentiviral particles for 24 h and then treated with actinomycin D to block transcription, and mRNA decay was monitored by qRT-PCR over time. As shown in Fig. 3A, the IRF-7 mRNA was significantly stabilized under reduced Dcp2 levels. The half-life of the IRF-7 mRNA was 4 h in the WT MEF cells and appreciably greater than the 8-h time course of the experiment in the Dcp2β/β MEF. The significance and specificity of Dcp2 in IRF-7 mRNA stability was further confirmed in cells with reduced levels of the second decapping enzyme, Nudt16. Stability of IRF-7 mRNA was unaltered in a reduced Nudt16 background (Fig. 3B). Furthermore, the Dcp2-directed regulation of IRF-7 mRNA stability was dependent on the catalytic activity of Dcp2. Expression of comparable levels of either WT or catalytically inactive Dcp2 with glutamine substitutions at two conserved glutamic acid residues (147 and 148) within the NUDIX motif (38) was used to complement Dcp2 in the Dcp2β/β MEF cell background (Fig. 3C). As shown in Fig. 3D, Dcp2β/β MEF cells expressing the control vector lacking Dcp2 consistently demonstrated elevated levels of IRF-7 upon viral infection compared to WT MEF cells. Importantly, the expression of WT Dcp2 in the Dcp2β/β background greatly suppressed the expression of IRF-7 to levels comparable to that in WT MEF cells, while the expression of catalytically inactive Dcp2 did not have a significant effect on the expression of IRF-7 in Dcp2β/β MEF cells.
Fig 3.
Dcp2 modulates the stability of the IRF-7 mRNAs. (A) WT and Dcp2β/βMEF cells were infected with control lentivirus for 24 h and then treated with actinomycin D (ActD). Total RNAs were harvested at the indicated times posttreatment, and the decay of IRF-7 mRNA was determined by qRT-PCR and normalized to β-actin mRNA. The averages of three independent experiments are plotted, with standard deviations denoted by the error bars. IRF-7 mRNA was considerably stabilized in Dcp2β/β MEF cells. (B) WT and Nudt16KD MEF cells were infected with control lentivirus for 24 h and then treated with actinomycin D, and IRF-7 mRNA levels were determined as described for panel A. (C) Western blotting of Dcp2 expression. Dcp2 levels present in WT MEF cells transduced with a control (Con) retrovirus or Dcp2β/β MEFs stably transduced with either control retrovirus or retrovirus expressing WT human Dcp2 or catalytically inactive mutant Dcp2 (Dcp2 EE/QQ) are shown. Expression of Dcp2 protein was confirmed by Western blot analysis with GAPDH as the loading control. Comparable levels of WT and catalytically inactive Dcp2 are expressed in these cells. (D) MEF cells from panel C were infected with control lentivirus (LV). Total RNAs were extracted before infection and 12 h after infection, and the levels of the mRNAs indicated were determined by qRT-PCR and normalized to β-actin mRNA. The IRF-7 mRNA level in WT cells before infection was arbitrarily set to 1. Data represent the average of three independent experiments, with the standard deviation denoted by the error bar. A key listing the WT or Dcp2β/β cells tested and the particular exogenous protein expressed in the particular cells is shown at the bottom. Overexpression of WT but not the catalytically mutant Dcp2 reversed the increase in IRF-7 in Dcp2β/β MEF cells. *, P < 0.05 (Student's t test). (E) The IRF-7 mRNA 5′ end is selectively targeted by Dcp2. Decapping assays were carried out with 50 ng of bacterially expressed His-Dcp2 using cap-labeled STX7 or a chimeric RNA that harbors 57 nt of the IRF-7 5′ UTR at the 5′ end of STX7 (IRF-STX). The reaction products were resolved by PEI-TLC. The average decapping percentage for each RNA from three independent experiments is shown at the bottom.
To further confirm a direct role for Dcp2 in IRF7 mRNA decapping, we determined whether the IRF7 5′ end was selectively targeted by Dcp2 for more efficient Dcp2 decapping. We recently reported that a subset of confirmed Dcp2 target substrate mRNAs possess a stem-loop structure positioned at the 5′ terminus termed the Dcp2 binding and decapping element (DBDE), which promotes Dcp2-mediated decapping (22, 24). The DBDE consists of a stem-loop structure positioned within 10 nt of the 5′ cap with a stem of at least 8 nt (22). Examination of the IRF-7 5′ UTR revealed the potential to form a stable 12-nt stem-loop structure within the first 10 nt of the mRNA. To test whether the IRF-7 5′ end promotes more-efficient Dcp2 decapping, a chimeric RNA consisting of the 5′-end 57 nt of the IRF-7 RNA was linked to a heterologous STX7 RNA. Consistent with the presence of a DBDE within the IRF-7 5′ UTR, Dcp2-directed decapping of the chimeric RNA was increased almost 2-fold relative to that of the same RNA lacking IRF-7 sequences (Fig. 3E). These results indicate that IRF-7 is a Dcp2 substrate and the induction of IRF-7 in Dcp2β/β MEF cells is primarily due to reduced levels of Dcp2.
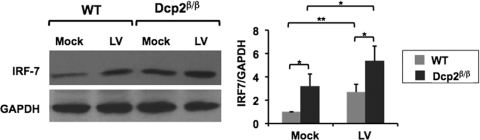
The correlation of increased IRF-7 mRNA with protein levels was tested next. Western blot analysis of IRF-7 protein revealed that loss of Dcp2 in Dcp2β/β MEF cells resulted in a 2.5-fold increase in IRF-7 protein compared to that in WT MEF cells prior to virus infection (Fig. 4). As expected, IRF-7 protein levels were further elevated in both cell backgrounds following viral infection and the differential higher levels of IRF-7 protein were maintained in Dcp2β/β MEF cells. Collectively, these data demonstrate that Dcp2 is a negative regulator of IRF-7 mRNA stability and protein accumulation and may function to attenuate IRF-7 expression.
Fig 4.
IRF-7 protein levels are elevated in Dcp2 knockdown cells. Total cell extract was harvested from WT or Dcp2β/β MEF cells either left uninfected (Mock) or infected with control lentivirus (LV) for 24 h. IRF-7 and GAPDH protein levels were determined from the indicated conditions by Western blotting. Quantitation of IRF-7 protein levels normalized to GAPDH is graphed on the right. The average of three independent experiments was plotted with the standard deviation denoted by the error bar. IRF-7 protein amounts were increased in Dcp2β/β MEF cells relative to WT MEF cells with or without virus infection. *, P < 0.05; **, P < 0.01 (Student's t test).
Lack of Dcp2 leads to enhanced IFN activity against a viral challenge.
To test whether Dcp2β/β MEF cells were more resistant to a viral challenge, WT and Dcp2β/β MEFs were treated with NDV or HSV-1 and the activity of IFN was determined by assessing the inhibition of an HSV-1-induced cytopathic effect (Fig. 5A). Twenty-four hours following exposure to NDV or HSV-1, culture medium was collected from WT or Dcp2β/β MEF cells and individually added to WT MEFs. After 2 h of incubation, MEF cells were infected with HSV-1 for 24 h. Cell survival was determined by crystal violet staining, in which absorbance at 595 nm (A595) directly correlates with viable cells. Uninfected cells displayed high cell viability, with an A595 of approximately 0.9 (data not shown). Culture medium from WT or Dcp2β/β MEF cells without virus treatment failed to protect cells from HSV-1 infection and displayed an A595 of approximately 0.1 at 24 h postinfection (Fig. 5B). Culture medium from WT MEFs pretreated with NDV or HSV-1 doubled the survival rate following a viral challenge, with an A595 of 0.2. Interestingly, culture medium from Dcp2β/β MEF cells quadrupled the survival rate, with an A595 of over 0.4, indicating that more cells were refractory to a viral challenge and virus-induced cytopathy. Consistently, expression of IRF-7 and IFN-α2 mRNAs was more responsive to NDV and HSV-1 treatment in Dcp2β/β MEF cells than in WT MEF cells, as determined by qRT-PCR (Fig. 5C). Collectively, these data demonstrate that the lack of Dcp2 results in enhanced type I IFN production and improved resistance to a viral challenge.
Fig 5.
Enhanced resistance to a viral challenge in Dcp2β/β MEF cells. (A) Diagram of experimental protocol. WT and Dcp2β/β MEFs were treated with NDV or HSV-1 for 24 h, and medium, which would contain secreted cytokines, was collected and transferred to new naive WT MEF cells. Following a 2-h incubation, the cells were infected with HSV-1 for 24 h and cell viability was determined. (B) Cytopathic effect at 24 h after infection. Cell viability was determined by crystal violet staining 24 h after HSV-1 infection. A595 directly correlates with viable cells. Averages of three independent experiments were plotted with error bars denoting the standard deviations. **, P < 0.01. (C) WT and Dcp2β/β MEFs were treated with NDV or HSV-1 for 24 h, and the levels of IRF-7 and IFN-α2 mRNAs were determined by qRT-PCR and normalized to β-actin mRNA. Averages of three independent experiments were plotted, with error bars denoting the standard deviations. *, P < 0.05 (Student's t test).
Dcp2 expression is induced following viral infection.
We next determined whether the expression of Dcp2 itself, as a potential modulator of type I IFN and antiviral immune response gene mRNAs, is altered during viral infection. WT MEF cells were infected with lentiviral particles, and Dcp2 expression was followed over time by qRT-PCR and Western blot analysis. Dcp2 mRNA levels increased upon viral infection, with 2.5- and 4-fold induction at 12 and 24 h postinfection, respectively (Fig. 6A). A comparable 3.5-fold induction was also observed when cells were treated with the viral double-stranded RNA mimic poly(I · C). Dcp2 mRNA levels were unchanged in control cells treated with growth medium lacking virus (mock-infected cells), suggesting that Dcp2 expression is induced by viral infection. Consistent with the mRNA data, Dcp2 protein levels increased approximately 3-fold upon poly(I · C) treatment or lentiviral infection (Fig. 6B). To further confirm that the induction of Dcp2 was not restricted to poly(I · C) viral mimic and lentiviral challenges, the level of Dcp2 mRNA in WT MEF cells was determined 24 h after NDV or HSV-1 infection. As shown in Fig. 6C, both viruses increased Dcp2 levels >3-fold relative to those in mock-infected cells. Collectively, these data imply that upon activation of the antiviral immune response, Dcp2 expression is also induced and possibly functions to avert a hyperactive immune response.
Fig 6.
Dcp2 expression increases upon viral infection and double-stranded RNA treatment. (A) WT MEF cells were treated with lentivirus (LV), poly(I · C) (50 μg/ml), or growth medium containing no virus (Mock) for 24 h. Total RNAs were harvested before and after treatment, and Dcp2 mRNA levels were determined by qRT-PCR and normalized to β-actin mRNA. Dcp2 mRNA levels in untreated MEF cells were arbitrarily set to 1. The average of three independent experiments was plotted with the standard deviation denoted by the error bar. Dcp2 mRNA significantly increased with viral infection or poly(I · C) treatment. (*, P < 0.05; **, P < 0.01). (B) Dcp2 protein was induced upon viral infection and poly(I · C) treatment. WT MEF cells were treated for 24 h with lentivirus (LV), poly(I · C) (50 μg/ml), or growth medium containing no virus (Mock). Total cell extracts were harvested before and after treatment, and Dcp2 protein levels were determined by Western blot analysis with GAPDH as the internal control. Quantitation of Dcp2 protein levels from three independent experiments normalized relative to GAPDH is shown on the right, with the level of Dcp2 in untreated cells set arbitrarily to 1. *, P < 0.05 (Student's t test). (C) WT MEFs were treated with NDV (MOI = 0.5) or HSV-1 (MOI = 1) for 24 h. Dcp2 mRNA levels were analyzed by real-time RT-PCR and normalized to β-actin mRNA levels. *, P < 0.05; **, P < 0.01 (Student's t test).
DISCUSSION
In this study, we report a novel role for the Dcp2 decapping enzyme in regulating the type I IFN response in antiviral innate immunity. Microarray assays revealed that a group of genes involved in the antiviral immune response were upregulated in Dcp2β/β MEF cells upon virus infection. Further mechanistic analysis identified at least IRF-7 mRNA as a direct target of Dcp2 decapping where reduced levels of Dcp2 resulted in greater stability of IRF-7 mRNA. Increased IRF-7 mRNA corresponded to higher levels of IRF-7 protein in Dcp2β/β MEF cells than in WT MEF cells. The relative increase in IRF-7 mRNA was observed in the absence of viral stimulation and was further accentuated upon viral infection. The central role of IRF-7 in the transcriptional activation of type I IFNs (15) indicates that modulators of IRF-7 expression would have profound consequences on the antiviral immune response, as has been demonstrated by the translational regulation of IRF-7 mRNA (6). Regulation of IRF-7 at the level of mRNA stability defines a previously unknown mechanism of regulating the immune response upon viral infection.
Of the 35 genes involved in antiviral immunity detected with confidence by microarray screening of WT and Dcp2β/β MEF cells, 40% were elevated >1.5-fold in Dcp2β/β MEF cells. The demonstration that Dcp2 regulates IRF-7 and IFN-α mRNA stabilities suggests a model where reduced levels of Dcp2 result in increased levels of IRF-7 and IFN-α mRNAs, which consequently result in elevated type I IFN production upon virus infection. As a result, these activities evoke an increase in IFN-inducible antiviral effector genes. Consistent with this premise, IRF-7 protein levels were upregulated in uninfected Dcp2β/β MEF cells (Fig. 4), and the expression of the Oas2, IFN-α, and IFN-β mRNAs was enhanced (Fig. 2B) in Dcp2β/β MEF cells upon exposure to virus. Elevation of IRF-7 mRNA was dependent on the decapping activity of Dcp2, since exogenous expression of WT but not catalytically inactive mutant Dcp2 abolished its enhanced expression in Dcp2β/β MEF cells (Fig. 3D). The enhanced antiviral response in Dcp2β/β MEF cells was not restricted to lentivirus infection but was also responsive to at least two pathogenic viruses, NDV and HSV-1 (Fig. 5C). More importantly, culture medium from virus-infected Dcp2β/β MEF cells contained more IFN activity than that from WT MEF cells, which protected better against an HSV-1-induced cytopathic consequence (Fig. 5B). Therefore, Dcp2 could modulate a broad antiviral immune response by, at least in part, targeting a critical upstream transcription factor.
IRF-7 is an important regulator of antiviral immunity, and an IRF-7 deficiency in mice results in increased susceptibility to viral infection and lethality in addition to a blunted systemic type I IFN response (7). Conversely, mice with a disrupted gene encoding the translational repressor 4E-BP and an increase in IRF-7 levels are highly resistant to infection by a variety of viruses (6). Interestingly, the enhanced type I IFN response in 4E-BP knockout mice is caused by an increase in IRF-7 mRNA translation and subsequent higher levels of IRF-7 protein, further demonstrating the critical function of IRF-7 in mobilizing the immune response (6). In this study, we demonstrated negative regulation of IRF-7 by the Dcp2 decapping enzyme at the level of mRNA stability. The half-life of IRF-7 mRNA increased significantly in Dcp2β/β MEF cells (Fig. 3) compared to that in WT MEF cells, indicating that IRF-7 mRNA degradation proceeds through Dcp2-mediated decapping. Importantly, IRF-7 protein levels were >3-fold elevated in the Dcp2β/β background. The striking feature of this observation is that the observed level was similar to what was detected in WT cells following a viral challenge (Fig. 4) and was further induced in Dcp2β/β cells. The comparable levels of IRF-7 elevation in virus-challenged WT cells and unchallenged Dcp2β/β cells strongly implicate Dcp2 as a critical regulator of IRF-7 expression.
The importance of regulated mRNA stability within the control of cytokine and chemokine mRNAs during an immune response has long been established (32). These mRNAs usually contain AREs in their 3′ UTRs which confer rapid degradation by an ARE-mediated decay mechanism. Various ARE-binding proteins are involved in regulating the stability of these mRNAs. The tristetraprolin protein binds to AREs in various mRNAs (such as tumor necrosis factor alpha and granulocyte-macrophage colony-stimulating factor) and can promote the deadenylation, as well as decapping and subsequent degradation, of these transcripts (4, 5, 10, 29). More recently, the physiological consequence of an ARE-binding protein in the regulation of type I IFN genes in innate immunity was demonstrated. Cells from mice containing a disrupted KSRP gene encoding the KSRP ARE-binding protein were more resistant to a viral challenge, further demonstrating the significance of posttranscriptional regulation in innate immunity (25). Our studies indicate that modulation of IRF-7 mRNA stability is also an important contributor to the type I IFN response. The IRF-7 mRNA has a short 3′ UTR that does not contain an obvious ARE and contains a considerably longer mRNA half-life (4 h) than ARE-containing mRNAs, which is typically less than 1 h (13, 19, 23, 33). Therefore, IRF-7 represents an example of non-ARE-containing mRNA that undergoes regulation of mRNA stability in the immune system. However, it is likely that the influence of Dcp2 on innate immunity genes extends beyond a singular modulation of IRF-7, since the stability of ARE-containing IFN-α2 is also directly influenced by Dcp2 (23), implicating Dcp2 as a broader antiviral response regulator.
Our data indicate that following viral infection, Dcp2 levels are elevated and IRF-7 is a direct target of Dcp2 decapping. We propose that the functional consequence of increased Dcp2 would be a reduction of IRF-7 mRNA to temper the antiviral immune response. One significant outcome of such a mechanism could be to prevent excess production of IFNs to curtail the manifestation of autoimmune pathologies such as those that have been observed with therapeutic type I IFN administration (2). Whether negative regulation of the antiviral immune response by Dcp2 could be a protective mechanism against deleterious overexpression of IFNs remains to be determined. Conversely, acute inhibition of Dcp2 decapping could also be envisioned as an adjuvant therapy for an antiviral response. The isolation of a bioavailable compound that selectively binds to and inhibits the activity of the scavenger-decapping enzyme DcpS (34) provides a precedent for such an approach. Future efforts to obtain Dcp2 conditional knockout mice will enable a more systemic analysis of Dcp2 and its regulation in the IFN-mediated immune response.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Moore, P. Xie, L. Covey, and members of the Kiledjian lab for technical assistance and helpful discussions.
This work was supported by NIH grants AI026806 to P.F.-B. and GM67005 to M.K.
Footnotes
Published ahead of print 17 January 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. [DOI] [PubMed] [Google Scholar]
- 2. Biggioggero M, Gabbriellini L, Meroni PL. 2010. Type I interferon therapy and its role in autoimmunity. Autoimmunity 43:248–254 [DOI] [PubMed] [Google Scholar]
- 3. Borden EC, et al. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6:975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carballo E, Lai WS, Blackshear PJ. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891–1899 [PubMed] [Google Scholar]
- 5. Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001–1005 [DOI] [PubMed] [Google Scholar]
- 6. Colina R, et al. 2008. Translational control of the innate immune response through IRF-7. Nature 452:323–328 [DOI] [PubMed] [Google Scholar]
- 7. Daffis S, et al. 2008. Interferon regulatory factor IRF-7 induces the antiviral alpha interferon response and protects against lethal West Nile virus infection. J. Virol. 82:8465–8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Decker CJ, Parker R. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7:1632–1643 [DOI] [PubMed] [Google Scholar]
- 9. Dunckley T, Parker R. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18:5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. 2005. Multiple processing body factors and the ARE-binding protein TTP activate mRNA decapping. Mol. Cell 20:905–915 [DOI] [PubMed] [Google Scholar]
- 11. Fensterl V, Sen GC. 2009. Interferons and viral infections. Biofactors 35:14–20 [DOI] [PubMed] [Google Scholar]
- 12. Garneau NL, Wilusz J, Wilusz CJ. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8:113–126 [DOI] [PubMed] [Google Scholar]
- 13. Herrick DJ, Ross J. 1994. The half-life of c-myc mRNA in growing and serum-stimulated cells: influence of the coding and 3′ untranslated regions and role of ribosome translocation. Mol. Cell. Biol. 14:2119–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Honda K, Takaoka A, Taniguchi T. 2006. Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349–360 [DOI] [PubMed] [Google Scholar]
- 15. Honda K, et al. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777 [DOI] [PubMed] [Google Scholar]
- 16. Hsu CL, Stevens A. 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13:4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Izaguirre A, et al. 2003. Comparative analysis of IRF and IFN-alpha expression in human plasmacytoid and monocyte-derived dendritic cells. J. Leukoc. Biol. 74:1125–1138 [DOI] [PubMed] [Google Scholar]
- 18. Justesen J, Hartmann R, Kjeldgaard NO. 2000. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell. Mol. Life Sci. 57:1593–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kabnick KS, Housman DE. 1988. Determinants that contribute to cytoplasmic stability of human c-fos and beta-globin mRNAs are located at several sites in each mRNA. Mol. Cell. Biol. 8:3244–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khabar KS, Young HA. 2007. Post-transcriptional control of the interferon system. Biochimie 89:761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levy DE, Marie I, Smith E, Prakash A. 2002. Enhancement and diversification of IFN induction by IRF-7-mediated positive feedback. J. Interferon Cytokine Res. 22:87–93 [DOI] [PubMed] [Google Scholar]
- 22. Li Y, Ho ES, Gunderson SI, Kiledjian M. 2009. Mutational analysis of a Dcp2-binding element reveals general enhancement of decapping by 5′-end stem-loop structures. Nucleic Acids Res. 37:2227–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Y, Song M, Kiledjian M. 2011. Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA 17:419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Y, Song MG, Kiledjian M. 2008. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol. Cell. Biol. 28:939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin WJ, et al. 2011. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol. Cell. Biol. 31:3196–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu H, Kiledjian M. 2006. Decapping the message: a beginning or an end. Biochem. Soc. Trans. 34:35–38 [DOI] [PubMed] [Google Scholar]
- 27. Liu Q, Greimann JC, Lima CD. 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127:1223–1237 [DOI] [PubMed] [Google Scholar]
- 28. Lykke-Andersen J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22:8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lykke-Andersen J, Wagner E. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19:351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Megjugorac NJ, Young HA, Amrute SB, Olshalsky SL, Fitzgerald-Bocarsly P. 2004. Virally stimulated plasmacytoid dendritic cells produce chemokines and induce migration of T and NK cells. J. Leukoc. Biol. 75:504–514 [DOI] [PubMed] [Google Scholar]
- 31. Nguyen H, Hiscott J, Pitha PM. 1997. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 8:293–312 [DOI] [PubMed] [Google Scholar]
- 32. Schott J, Stoecklin G. 2010. Networks controlling mRNA decay in the immune system. Wiley Interdiscip. Rev. RNA 1:432–456 [DOI] [PubMed] [Google Scholar]
- 33. Shyu AB, Greenberg ME, Belasco JG. 1989. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 3:60–72 [DOI] [PubMed] [Google Scholar]
- 34. Singh J, et al. 2008. DcpS as a therapeutic target for spinal muscular atrophy. ACS Chem. Biol. 3:711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Song MG, Li Y, Kiledjian M. 2010. Multiple mRNA decapping enzymes in mammalian cells. Mol. Cell 40:423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Dijk E, et al. 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21:6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vinkemeier U, Meyer T. 2005. Antiviral activity of oligomerization-deficient Stat1. Genome Inform. 16:44–48 [PubMed] [Google Scholar]
- 38. Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. U. S. A. 99:12663–12668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Z, Kiledjian M. 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107:751–762 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.