Abstract
Signal transducers and activators of transcription (STATs) and interferon regulatory factors (IRFs) share common target genes. Here we show that the Irf7 gene is regulated by transcription factors STAT1 and IRF9 in response to the type II interferon (IFN) IFN-γ. IRF7 cooperated with STAT1 and IRF1 to stimulate the expression of a subset of IFN-γ-induced STAT1 target genes. IRF7-mediated control of the Gbp2 gene required the presence and basal activity of the S/T kinase TANK-binding kinase 1 (TBK1), whereas the binding of IRF7 to the Gbp2 promoter did not. Analysis of RNA polymerase II (Pol II) recruitment to the Gbp2 promoter revealed a role for IRF7 at later stages of the IFN-γ response. In support of the role of IRF7 in establishing an effective antibacterial response, IFN-γ-pretreated Irf7−/− macrophages showed an increased bacterial burden after infection with Listeria monocytogenes. Our data thus describe a biologically relevant basal activity of TBK1 and identify IRF7 as a novel player in the IFN-γ response.
INTRODUCTION
The interferon (IFN) family of antimicrobial cytokines consists of three distinct types. While the type I and type III IFN each contain several members, IFN-γ is the exclusive type II IFN. It is produced by activated T cells and NK cells and enhances cell-mediated immunity, most prominently by its contribution to macrophage activation. De novo gene induction is necessary for most—if not all—activities of IFN-γ in the immune system. Promoters of the vast majority of IFN-γ-induced genes contain an IFN-γ-activated site (GAS), a binding sequence for tyrosine-phosphorylated, dimerized STAT1. Association between Stat1 dimers and GAS sequences occurs as a consequence of Jak-Stat signaling by the IFN-γ receptor and, to a lesser extent, the type I IFN receptor (22).
Binding of the STAT1 dimer is sufficient to stimulate immediate transcription of primary response genes. However, a significant number of IFN-γ-induced genes show a delayed, secondary response. Many of the genes pertaining to this category contain a binding site for IFN regulatory factors (IRF), which is contained within the interferon-stimulated response element (ISRE) sequence (40, 45). The delayed response of these genes is due to the need to synthesize family member IRF1 within the primary transcriptional burst after stimulation with IFN-γ. A well-studied example is the Gbp2 gene, where STAT1 and IRF1 cause, respectively, the recruitment of HATs (histone acetyl transferases) and the RNA polymerase II (Pol II) complex (28, 36). GBP2 expression is absent in Stat1−/− fibroblasts, whereas Irf1−/− cells still show residual expression of GBP2 mRNA after treatment with IFN-γ (36). The studies shown below are based on the hypothesis that the incomplete shutdown of Gbp2 expression in IRF1 knockout cells may reflect the activity of another IRF family member and its interaction with IRF1. Support for this assumption comes from reports showing that IRF4 and IRF8 (IFN consensus sequence-binding proteins [ICSBP]) contribute to the control of genes induced by IFN-γ in lymphoid and myeloid cell types, suggesting that members of the IRF family other than IRF1 cooperate with STAT1 in gene regulation (16, 49).
IRF family members IRF3 and IRF7 are critically involved in the regulation of type I IFN genes (39). IRF3 is constitutively expressed in probably all cell types. In contrast, IRF7 needs to be synthesized in most cell types prior to its participation in gene regulation (29). The type I IFN IFN-α and IFN-β initiate Irf7 transcription by activating the ISGF3 complex, a tyrosine-phosphorylated STAT1-STAT2 heterodimer in combination with IRF9, which serves as a DNA-binding subunit. ISGF3 associates with an ISRE in the 5′ untranslated region (UTR) of the Irf7 gene (50).
Both IRF3 and IRF7 are crucial regulators of innate immunity to viral infections due to their essential contribution to the expression of all antiviral type I IFN genes (6, 10, 14, 25, 31). The activity of both IRF3 and IRF7 is regulated by phosphorylation of several clustered serine residues within their regulatory domains. TANK-binding kinase 1 (TBK1) and the IκB kinase-related IKKε were shown to exert this function (47). Whereas the main function of IRF3 and IRF7 is to regulate type I (and type III) IFN synthesis, both proteins are also able to stimulate the promoters of a subset of type I/type III IFN-inducible genes when introduced into cells as dominant active alleles (11, 40, 42). In contrast, an impact of IRF3 or IRF7 on gene regulation by IFN-γ has not been explored.
The studies summarized in this report show that the Irf7 gene responds to IFN-γ and that IRF7 contributes to the regulation of IFN-γ-induced genes by facilitating and maintaining the recruitment of RNA polymerase II. IRF7 function in this context depends on the presence and basal activity of TBK1 and/or IKKε. We thus describe a novel activity of the TBK1/IKKε-IRF7 pathway in the IFN-γ response and demonstrate its biological relevance by showing that IRF7 increased an antibacterial response in Listeria monocytogenes-infected bone marrow-derived macrophages (BMDM).
MATERIALS AND METHODS
Reagents and antibodies.
The proteasome inhibitor MG132 was purchased from Calbiochem and used in a final concentration of 4 μM. IFN-γ was used in a final concentration of 10 ng/ml. IFNAR blocking antibody was used in a dilution of 1:1,000 (43). To generate the anti-IRF7 antibody, a rabbit was immunized with an IRF7-GST fusion protein containing amino acids 207 to 452 of the murine IRF7 protein. The antibody was used in a dilution of 1:1,000 for Western blotting. To generate the IRF9 antibody, a rabbit was immunized with a GST fusion protein containing the carboxy-terminal part of the murine IRF9 protein. STAT1 phospho-Y701 antibody was purchased from New England BioLabs (Beverly, MA) and used in a dilution of 1:2,000 for Western blotting. Antibodies against IRF1 were from Santa Cruz (Santa Cruz, CA) used in Western blots in a dilution of 1:1,000. Antibodies against ERK1/2 kinases (pan-ERK) was purchased from Becton Dickinson (Franklin Lakes, NJ) and used in a dilution of 1:2,000 for Western blots. Anti-Stat1-C-terminal antibody was described previously (20).
Cells and mice.
Mouse embryo fibroblasts (MEFs) derived from Irf1−/− mice (37) were kindly provided by J. Pavlovic (University of Zurich, Zurich, Switzerland); Irf3−/− and Irf7−/− MEFs were derived from recently described mice (14, 39). Tbk1-Ikbke−/− MEFs were kindly provided by S. Akira. MEFs were cultured in Dulbecco modified Eagle medium (DMEM) containing 10% fetal calf serum (FCS). Mice (wild-type [WT] C57BL/6, Irf1−/− [40], Irf7−/− [14], Irf9−/− [12]) were sacrificed for bone marrow between 7 to 10 weeks of age. Bone marrow-derived macrophages (BMDM) were obtained by culture of bone marrow in L-cell-derived colony-stimulating factor 1 as described previously (3).
RT-qPCR.
Primer for Gbp2 mRNA expression and quantitative real-time PCR (RT-qPCR) were described previously (36). Other mRNA primers used in this study were as follows: Tap1 forward, 5′-CTGGCAACCAGCTACGGGT-3′; Tap1 reverse, 5′-TGAGAATGAGGATGTGGTGGG-3′; Socs1 forward, 5′-ACTCCGTGACTACCTGAGTTCCTT-3′; Socs1 reverse, 5′-GCATCTCACCCTCCACAACCACT-3′; Irf7 forward, 5′-CTGGAGCCATGGGTATGCA-3′; Irf7 reverse, 5′-AAGCACAAGCCGAGACTGCT-3′.
Nuclear extraction.
Nuclear extracts of confluent MEFs in 10-cm tissue culture dishes were obtained as described in reference 7. Extraction was followed by a desalting step using PD Spin Trap G-25 desalting columns from GE-Healthcare (catalog no. 28-9180-04), according to the manufacturer's protocol.
2D gel electrophoresis.
Nuclear extracts were subjected to two-dimensional (2D) gel electrophoresis as described in reference 6.
Western blot analysis.
A protocol for this procedure was recently described (19). Cells used for Western blots with IRF7 antibodies were pretreated with MG132 for 30 min and during stimulation with IFN-γ. For Western blot analyses by the Odyssey infrared imaging system (Li-Cor Biosciences), secondary antibodies to mouse IgG (cat. no. 610-132-121) and rabbit IgG (cat. no. 611-132-122) were purchased from Rockland. Quantifications were performed using the Odyssey Infrared Imaging System Software.
ChIP and reChIP.
Chromatin immunoprecipitations (ChIPs) were performed following the protocol described in reference 30. Antibodies used were purchased from Santa Cruz (Santa Cruz, CA) and used at a 1:20 dilution (anti-RNA Pol II, anti-IRF1) or purchased from Bethyl (Montgomery, TX) and used in a dilution of 1:100 (anti-pS5 CTD Pol II). ChIP data were normalized to and expressed as a percentage of input. Primers used for PCR and qPCR of the Gbp2 promoter were described recently (36). Primers for the Irf7 promoter and enhancer were as follows: Irf7 promoter forward, 5′-GGTCGGGTGTAGTTTGAGGA-3′; Irf7 promoter reverse, 5′-GCCAAGGTGGCTGTAGATGT-3′; Irf7 enhancer forward, 5′-GCTTCTTGACCCAGCTGGAACA-3′; Irf7 enhancer reverse, 5′-ACAGTCAAGGGTTGTGTCCATCCT-3′. In the reChIP experiments, the immune complexes were eluted by adding 10 mM dithiothreitol (DTT) and incubation for 30 min at 37°C. The samples were diluted 40-fold in RIPA buffer and reimmunoprecipitated.
Plasmids and transfections.
Cells were transfected using the dual-luciferase reporter assay system (cat. no. E1910) from Promega (Promega Corporation, Madison, WI) according to the manufacturer's protocol, with IRF7 expressing plasmids as described in reference 6 and Gbp2 luciferase-reporter construct. Cells were analyzed 48 h after transfection.
Determination of CFU.
CFU assays with L. monocytogenes (strain LO28)-infected macrophages were performed as described recently (17).
RESULTS
IFN-γ stimulates Stat1 association with the Irf7 promoter and induces IRF7 expression.
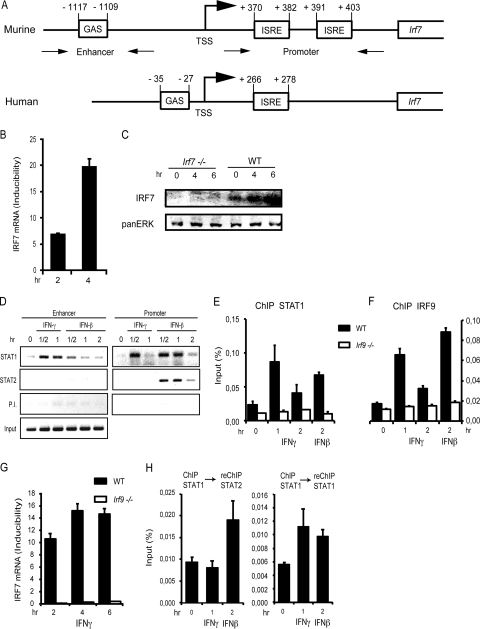
To study a possible contribution of IRF7 to the regulation of IFN-γ-induced genes, we tested whether the Irf7 gene itself responds to treatment of WT mouse embryonic fibroblasts (MEFs) with IFN-γ. Indeed, we could show an increase of IRF7 mRNA after IFN-γ treatment (Fig. 1B). Detection of IRF7 by direct Western blotting was possible in the presence of the proteasome inhibitor MG132 to increase stability of the protein. Under these conditions, a clear induction by IFN-γ was observed (Fig. 1C). Since IFN-γ-stimulated gene expression usually involves transcriptional activity of STAT1 dimers, we searched the 5′ region and intronic sequences of the murine Irf7 gene for the presence of a GAS. A perfect consensus sequence, TTCTCTGAA, was found 1,117 bp upstream of the transcription start site (TSS) (Fig. 1A). The human Irf7 gene promoter contains a GAS located between nucleotides −35 and −27 upstream of the TSS (TTCCCGGAA; Fig. 1A). This is consistent with a recent microarray experiment showing human IRF7 among IFN-γ-induced genes (38). To test whether the GAS of the murine Irf7 enhancer binds STAT1 homodimers in response to IFN-γ, we performed ChIP assay analysis in WT MEFs (Fig. 1D). In support of this assumption, STAT1 associated with the upstream GAS enhancer within 30 min after IFN-γ treatment. As expected, STAT2 was not found associated with the GAS sequence under these conditions. In addition to the upstream enhancer, STAT1 associated with the intronic ISRE (referred to as “Promoter” in Fig. 1) after 30 min of treatment with IFN-γ. The data therefore suggest that IRF7 mRNA expression in response to IFN-γ is regulated via binding of STAT1 to both the upstream and intronic response elements. STAT1 binding to the intronic sites appears to be transient compared to the recruitment to the upstream region.
Fig 1.
Kinetics of IRF7 mRNA expression and STAT1 recruitment to the Irf7 promoter, determined by qPCR and ChIP, respectively. (A) Schematic drawing of the murine and human Irf7 promoters. ISRE sites are located in the 5′ UTR of the murine Irf7 gene, and the newly defined GAS is located in the enhancer region 1.1 kb upstream of the TSS. (B and C) IRF7 mRNA (B) and protein expression (C). mRNA expression after treatment of WT MEFs with IFN-γ for the indicated time points was determined by qPCR and normalized to GAPDH levels. IRF7 protein was determined by Western blotting with IRF7 antibodies. To increase sensitivity, IRF7 was stabilized by treatment with the proteasome inhibitor MG132. (D) WT MEFs were stimulated with IFN-γ or IFN-β and processed for ChIP at the indicated time points. Antibodies for ChIP are shown on the left. P. I. indicates controls performed with preimmune sera. The precipitates were amplified by PCR with primers flanking the enhancer (GAS) or promoter (ISRE) region as indicated in panel A and were analyzed by gel electrophoresis. (E and F) Bone marrow-derived macrophages (BMDMs) of WT and Irf9−/− mice were treated with IFN-γ or IFN-β for the times indicated and processed for ChIP with the antibodies indicated on the top of each panel. (G) BMDMs derived from WT or Irf9−/− mice were treated with IFN-γ for the indicated time periods. IRF7 mRNA expression was analyzed by qPCR and normalized to GAPDH levels. (H) BMDMs of WT mice were treated with IFN-γ or IFN-β for the times indicated and processed for ChIP-reChIP. Antibodies for ChIP are shown on top of each panel. The precipitates were amplified with primers flanking the Irf7 promoter as shown in panel A. qPCR measurements were made in triplicate. All experiments were repeated at least three times.
To control for the specificity of STAT binding after IFN-γ treatment, cells stimulated with IFN-β were analyzed. Consistent with ISGF3 activation, both STAT1 and STAT2 bound the intronic ISRE sequences after treatment with IFN-β (Fig. 1D). In further accordance with expectations, a small amount of STAT1, but not STAT2, was found at the distal GAS after IFN-β treatment.
STAT1 association with intronic ISRE sequences in response to IFN-γ occurred in the absence of detectable STAT2. This rules out a participation of the transcriptional activator ISGF3. Complexes containing STAT1 and IRF9 are known to form in vitro. These offer the possibility of STAT2-independent association of STAT1 with the ISRE (4). In fact, STAT1-IRF9 was previously implied in the regulation of the Gbp2 gene by IFN-γ (48). The participation of IRF9 in Irf7 gene regulation by IFN-γ was examined in IRF9-deficient macrophages and appropriate control macrophages. Compared to fibroblasts, these cells show a less transient association of STAT1 with the intronic ISRE sequences (compare Fig. 1D, E, and F). Consistent with the assumption that IRF9 mediated association of STAT1 to the proximal ISREs, binding of IRF9 after IFN-γ treatment occurred with kinetics matching those of STAT1 (Fig. 1E and F). Furthermore, both STAT1 binding to these sites and IRF7 mRNA inducibility by IFN-γ were abolished in IRF9-deficient cells (Fig. 1E to G). To further rule out a participation of ISGF3 in Irf7 regulation by IFN-γ via the intronic ISRE, ChIP-reChIP analyses were performed. Figure 1H shows that STAT2 could be reprecipitated from an anti-STAT1 ChIP only after IFN-β and not after IFN-γ treatment (left panel). The presence of STAT1 in both ChIPs was confirmed by reChIP of the original anti-STAT1 precipitate with anti-STAT1 antibody (right panel). This clearly demonstrates that an ISGF3 complex associates with the intronic ISREs after IFN-β but not after IFN-γ treatment. Collectively the data show that IFN-γ regulates the Irf7 gene by associating a STAT1 dimer with the distal GAS sequence and by associating a STAT1/IRF9-containing, non-ISGF3 complex with the intronic ISREs.
Delayed transcriptional responses to IFN-γ require IRF7.
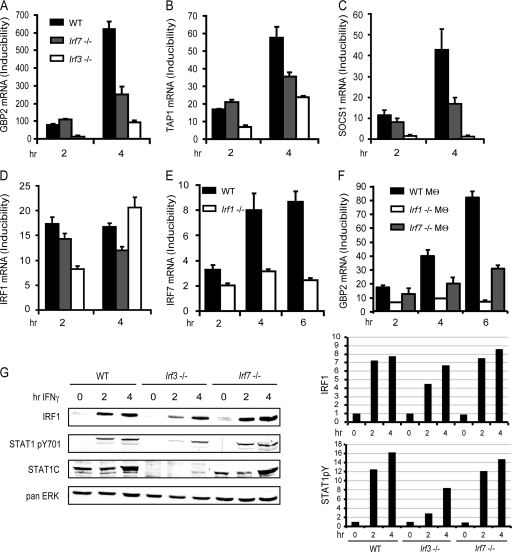
We asked whether IRF7 and/or IRF3 might contribute to IFN-γ-induced target gene expression and therefore cause the residual expression of GBP2 mRNA in Irf1−/− MEFs (36). We monitored the mRNA expression profile of several ISGs by qPCR. The chosen genes have well-characterized IFN-responsive promoters and respond to IFN-γ either immediately (Irf1) or with delay (Gbp2, Tap1, Socs1). This corresponds to their regulation by STAT1 only or a combination of STAT1 and IRF1.
WT MEFs or MEFs deficient for IRF7 or IRF3 were analyzed after treatment with IFN-γ (Fig. 2A to D). In line with the ability of IRF7 to bind ISRE sequences (50), mRNA expression of the ISRE-containing genes Gbp2 and Tap1 was reduced in the absence of IRF7, particularly at later time points after treatment. Expression of SOCS1 was also found to require IRF7 in accordance with a bona fide IRF binding site in an enhancer of the murine gene at position −1772 to −1764, which is conserved in humans (41). Expression of IRF1 mRNA showed virtually no IRF7 dependency, consistent with the regulation of IRF1 exclusively via STAT1 dimer association with a GAS. IRF7 mRNA induction by IFN-γ was reduced in Irf1−/− MEFs. Because IRFs bind to the core sequences of many ISRE, this suggests IRF1 control of Irf7 gene expression via the intronic ISRE sequences. It also provides an explanation for the particularly strong reduction of IFN-γ-induced gene expression in Irf1−/− MEFs because these cells show a strong loss of IFN-γ-induced IRF7 expression besides lacking IRF1 (Fig. 2E). Both WT and Irf7−/− MEFs used for these studies were derived from mice with identical genetic backgrounds. To exclude that the effect of IRF7 on the IFN-γ response was influenced by clonal variation of independent MEF isolates, primary bone marrow-derived macrophages were analyzed for IFN-γ-induced Gbp2 expression and showed a similar contribution of IRF7 to the transcriptional response (Fig. 2F).
Fig 2.
Regulation of IFN-γ-induced genes by IRFs. (A to D) WT, Irf3−/−, and Irf7−/− MEFs were treated with IFN-γ for the indicated time periods, followed by determination of GBP2, TAP1, SOCS1, and IRF1 mRNA expression by qPCR. (E) WT and Irf1−/− MEFs were treated with IFN-γ for the indicated time periods, followed by determination of IRF7 mRNA expression by qPCR. (F) WT, Irf1−/− and Irf7−/− bone marrow-derived macrophages (Mθ) were treated with IFN-γ for the indicated time periods, followed by determination of IRF7 mRNA expression by qPCR. GBP2, TAP1, SOCS1, IRF1, and IRF7 mRNA expression was determined by qPCR and normalized to GAPDH levels. (G) IRF1 protein expression and STAT1 tyrosine phosphorylation were detected by Western blot analysis. Differences in STAT1 expression levels between WT MEFs and MEFs deficient for IRF3 or IRF7 were analyzed by reprobing the blot with an antibody against the STAT1 C terminus. The Western blot was quantified by densitometry of the antibody-mediated signal (panel G, right), normalizing IRF1 and pYSTAT1 to the pan-ERK signal. qPCR measurements were made in triplicate. All experiments were repeated at least three times.
mRNA expression of IFN-γ-induced genes in Irf3−/− MEFs was strongly diminished (Fig. 2A to D). Strikingly, the Irf1 gene, which does not contain a promoter binding site for IRFs, was affected by the absence of IRF3 at 2 h but not at 4 h after IFN-γ treatment. Consistent with this, STAT1 expression and tyrosine phosphorylation were severely reduced in Irf3−/− MEFs 2 h after treatment whereas 4 h after treatment STAT1 levels were increased and tyrosine phosphorylation was normal (Fig. 2G). This corresponded to WT levels of IRF1 at this time point (Fig. 2G). Gbp2, Socs1, and Tap1, which contain IRF binding sites, showed reduced expression also 4 h after IFN-γ treatment. This is most likely a direct consequence of delayed IRF1 synthesis in IRF3−/− cells (Fig. 2D and G). Together, the data obtained in IRF3-deficient cells suggest indirect effects of IRF3 in the regulation of ISGs and are consistent with the reported role of IRF3 in maintaining STAT1 expression through autocrine type I IFN production (44, 46).
In contrast with IRF3−/− MEFs, Irf7−/− MEFs displayed normal STAT1 expression and appearance of STAT1 phosphotyrosine upon IFN-γ treatment. In line with mRNA expression, IRF1 protein expression was not altered in Irf7−/− MEFs. A direct role for IRF7 in regulating gene expression in response to IFN-γ was further reported by our finding that, unlike IRF3, its regulatory function was confined to genes with IRF binding sites. mRNA expression profiles showed that IFN-γ-stimulated gene expression is reduced, but not completely abrogated, in Irf7−/− MEFs. This, in turn, points to an ancillary role for IRF7 in enhancing or prolonging target gene expression.
Basal TBK1 activity is needed for IRF7 function in the IFN-γ response.
In the context of viral infection, the transcriptional activity of IRF7 relies on the activity of the kinases TBK1 and/or IKKε. The two enzymes were shown to act redundantly for the activation of IRF3 and IRF7 and the production of type I IFN in antiviral responses of macrophages (34), although recent reports show differences at least in the mode of their activation (8, 14). Challenging this view of complete functional redundancy, antagonistic activity of TBK1 and IKKε, based on competition for a common adapter protein, was recently suggested by the group of J. Hiscott (33).
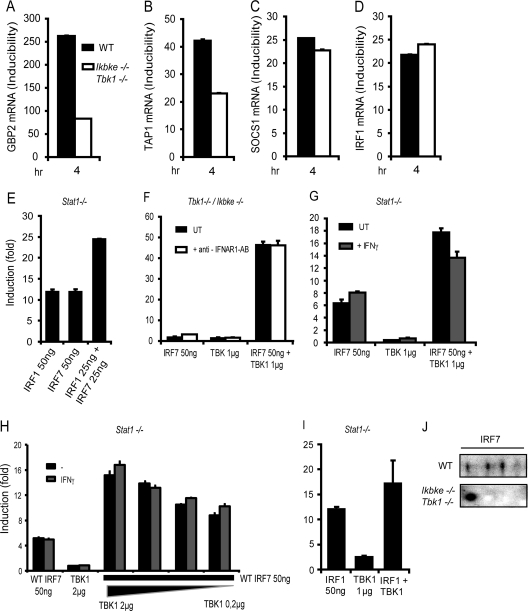
To determine whether TBK1 and IKKε kinase function is needed for the transcriptional regulation of IFN-γ-induced genes by IRF7, we performed qPCR analysis of IRF7 target genes, comparing WT or double-deficient Tbk1/Ikbke−/− MEFs. mRNA expression of the previously identified IRF7 target genes was differentially affected by the absence of the two kinases. We observed the strongest impact of Tbk1/Ikbke deficiency on IFN-γ induction of Gbp2 and Tap1, which showed a decrease in mRNA expression by more than 50% (Fig. 3A and B). In contrast, the absence of Tbk1/Ikbke did not reduce the ability of IFN-γ to induce expression of SOCS1 mRNA or to regulate the IRF7-independent Irf1 gene (Fig. 3C and D).
Fig 3.
TBK1 is required for the regulation of IFN-γ-induced genes by IRF1 and IRF7. (A to D) Expression of GBP2, TAP1, SOCS1, and IRF1 mRNA in WT and Tbk1/Ikbke−/− MEFs after IFN-γ treatment for the times indicated was analyzed by qPCR and normalized to GAPDH mRNA levels. (E) Stat1−/− MEFs were transfected with either IRF1 or IRF7, and Gbp2-firefly luciferase reporter activity was measured. The values are expressed as fold induction relative to cells transfected only with reporter construct and normalization to a cotransfected, constitutively expressed, Renilla luciferase reporter. (F) Tbk1/Ikbke−/− MEFs were transfected with IRF7 or TBK1 alone or with a combination of both. MEFs were treated with IFNAR1 blocking antibody for the whole period of transfection or left untreated. (G) Stat1−/− MEFs were transfected with IRF7 or TBK1 alone or in combination. Transfected cells were stimulated overnight with IFN-γ or left untreated. (H) Stat1−/− MEFs were transfected with either WT IRF7 or TBK1 or a combination of both. IRF7 amounts were left constant, whereas TBK1 amounts were varied as shown. (I) Stat1−/− MEFs were transfected with IRF1 or TBK1 alone or in combination. In panels F to I, Gbp2 luciferase reporter activity was measured as described for panel A. (J) Requirement for TBK1/IKKε-mediated IRF7 phosphorylation determined by 2D gel electrophoresis. WT or Tbk1/Ikbke−/− MEFs were treated with IFN-γ for 4 h or left untreated. Nuclear extracts were prepared and subjected to 2D gel electrophoresis. IRF7 isoforms were analyzed by Western blotting. qPCR and luciferase measurements were made in triplicate. All experiments were repeated at least three times.
To further examine the role of TBK1 in the regulation of the transcriptional activity of IRF7 on the Gbp2 gene, we performed cotransfection experiments with IRF7 and TBK1 and a Gbp2 promoter-reporter gene. STAT1-deficient MEFs were used for these experiments to rule out an influence of STAT1. These experiments demonstrated the ability of IRF7 to stimulate the Gbp2 reporter construct in the presence of TBK1 and IKKε (Fig. 3E). In contrast, transfection of IRF7 into Tbk1/Ikbke−/− MEFs did not stimulate Gbp2 reporter gene expression, unless TBK1 was reintroduced by cotransfection (Fig. 3F). The addition of an antibody blocking the accessibility of the IFNAR for its ligands (43) had no effect on the expression of the reporter gene in this experiment. Hence we can rule out a contribution of type I IFN signaling to Gbp2 promoter activation by transfected TBK1/IRF7 (Fig. 3F).
Given the ability of IFN-γ to induce IRF7 mRNA expression and the TBK1 requirement for IRF7 activity, we examined whether IFN-γ treatment activates TBK1. To this end, we cotransfected IRF7 and TBK1 into Stat1−/− MEFs. Treatment of IRF7-transfected Stat1−/− MEFs with IFN-γ did not significantly enhance the reporter gene activity. In contrast, introduction of additional TBK1 activity via transfection further enhanced the ability of IRF7 to stimulate expression of the Gbp2 reporter gene in Stat1−/− MEFs, irrespective of prior treatment with IFN-γ (Fig. 3G). Reducing the amount of transfected TBK1 did not change this result (Fig. 3H), excluding the possibility that the lack of effect of IFN-γ on TBK1 activity is influenced by different amounts of the kinase. Unlike that of IRF7, the transcriptional activity of IRF1 is thought to occur without phosphorylation-mediated activation (40). In keeping with this notion, IRF1 and TBK1 cotransfection resulted in a modest increase of Gbp2 reporter gene expression. This increase is likely to reflect the activity of TBK1 on endogenous IRF7 substrates rather than on the cotransfected IRF1 (Fig. 3I).
Trying to reconstitute IRF7 kinase activity in Tbk1/Ikbke−/− MEFs by IKKε cotransfection failed. In fact, IKKε cotransfection inhibited the ability of TBK1 to provide IRF7 activity for the Gbp2 promoter (data not shown). At present we cannot explain this striking nonredundancy of the two kinases TBK1 and IKKε in the context of IRF7-mediated stimulation of IFN-γ-induced genes. It will be pursued in our future investigations.
Together the data in Fig. 3 suggest that IRF7 must be phosphorylated to efficiently drive the expression of GBP2 and that basal phosphorylating activity is present in WT or Stat1−/−, but not in Tbk1/Ikbke-deficient fibroblasts. To examine the IRF7 phosphorylation status, we performed 2D gel electrophoresis with nuclear extracts of WT and Tbk1/Ikbke−/− MEFs. The cells were treated with IFN-γ for 4 h to induce IRF7 expression, and nuclear proteins separated on a 2D gel were analyzed by Western blotting with polyclonal IRF7 antibody (6). Our results show the formation of at least two TBK1/IKKε-dependent IRF7 phosphoisoforms (Fig. 3J).
Critical role of serines (S425-S426 and S437-S438) of the IRF7 regulatory domain as phosphoacceptors for the stimulation of IRF7 activity by TBK1.
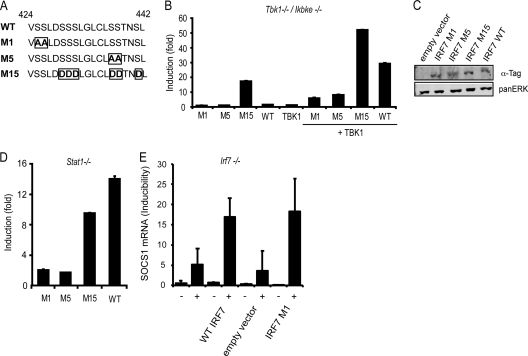
IRF7 can be activated by phosphorylation of several serine residues, located in the carboxy-terminal regulatory domain (6, 26). For in-depth analysis of the serine residues required for IRF7 transcriptional activity at the Gbp2 promoter we performed transfection experiments in Stat1−/− MEFs and Tbk1/Ikbke−/− MEFs with a set of IRF7 phosphomutants (Fig. 4A) (6).
Fig 4.
Transcriptional activity of IRF7 mutants. Transactivation of the Gbp2 promoter by different mutants of IRF7. Stat1−/− MEFs (D) or Tbk1/Ikbke−/− MEFs (B) were transfected with WT IRF7 or the IRF7 mutants indicated (A). Fifty nanograms of IRF7 constructs were transfected alone or cotransfected with 1 μg TBK1 (B) as indicated. Gbp2-firefly luciferase reporter gene expression is indicated as fold induction relative to cells transfected only with reporter construct and normalization to cotransfected, constitutively expressed Renilla luciferase reporter. (C) Protein expression of IRF7 M1, M5, M15, and WT in Stat1−/− MEFs was detected by Western blot analysis and probing the blot with anti-tag antibodies recognizing Flag-tagged IRF7 M1, M5, and M15 or HA-tagged WT IRF7. Pan-ERK levels were analyzed as a normalization control. (E) Irf7−/− MEFs were transfected with 2 μg of a WT IRF7 construct, an empty vector control, or the IRF7 M1 mutant and treated for 6 h with IFN-γ (+) or left without treatment (−), followed by determination of SOCS1 mRNA expression. qPCR and luciferase measurements were made in triplicate. All experiments were repeated at least six times.
Transfection of WT IRF7 as well as the serine-to-alanine mutants M1 (S425A and S426A) and M5 (S437A and S438A) in Tbk1/Ikbke−/− MEFs failed to induce Gbp2 reporter gene expression, whereas transfection of the serine-to-aspartate mutant M15 (all serines except S425 and S426 are mutated to phosphomimetic aspartate) upregulated the Gbp2 reporter gene activity very efficiently (Fig. 4B). Cotransfection of TBK1, to reconstitute kinase activity, strongly increased reporter gene activity in the cases of both WT IRF7 and the M15 mutant. TBK1 cotransfection with M1 and M5 mutants led to a moderate increase of Gbp2 reporter gene expression. Differences between WT IRF7 and the M1, M5, and M15 mutants did not result from different expression levels as shown by Western blot analysis (Fig. 4C).
All mutants produced similar results when transfected into Stat1−/− MEFs, with the two notable exceptions that reporter gene expression stimulated by the M15 mutant did not exceed the levels obtained with WT IRF7 and that the inducibility of the reporter gene by transfected IRF7 was much lower than in Tbk1/Ikbke−/− MEFs (Fig. 4D).
Taken together, these results indicate that a combination of the serines mutated in M1 and M5 is needed for full transcriptional activity of IRF7. The importance of the more N-terminal serines 425 and 426 is emphasized by the results with the M15 mutant. S425 and S426 are the only serines not phosphomimetic in M15 and must therefore be responsible for the increased activity of M15 after cotransfection with TBK1 into Tbk1/Ikbke−/− MEFs.
In contrast to that of the Gbp2 gene, IRF7-dependent expression of the Socs1 gene after IFN-γ treatment was unaffected by the absence of TBK1 and IKKε (Fig. 3C). To confirm TBK1/IKKε-independent IRF7 activity on the Socs1 gene we transfected WT IRF7 or the M1 mutant into IRF7-deficient fibroblasts, followed by IFN-γ treatment and examination of SOCS1 mRNA expression. In these experiments the M1 mutant was as capable of restoring the full responsiveness of the Socs1 gene to IFN-γ as WT IRF7 (Fig. 4E). This is in agreement with the notion that the need for basal TBK/IKKε activity for IRF7-mediated regulation of the IFN-γ response is gene specific.
Cooperation between IRF1 and IRF7 in the control of GBP2 expression.
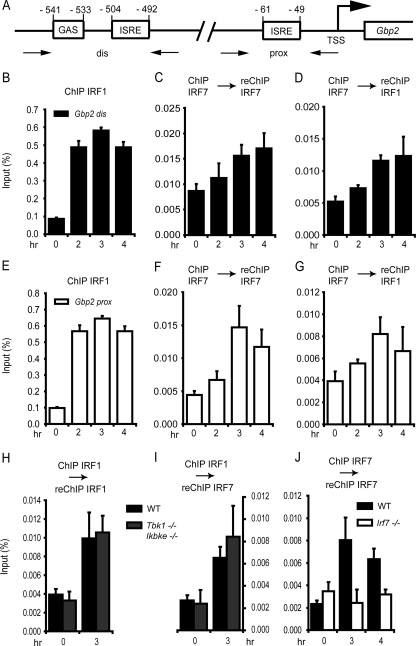
IRF7 is able to form either homodimers or heterodimers with other IRF family members. For example, physical interaction with IRF3 occurs in the context of IFN-β and IFN-α4 gene expression after pathogen exposure (1, 2, 24). IRF1 physically and functionally interacts with IRF8 (21) and participates in the IRF3- and IRF7-mediated regulation of human IFN-α genes (1). Since all of the IFN-γ-inducible genes identified in our experiments as IRF7 target genes are known targets for IRF1, we wondered whether IRF7 has the capability to functionally interact with IRF1 in the control of IFN-γ-induced genes. The possible interplay of IRF7 with IRF1 in the regulation of GBP2 mRNA expression was first tested in reporter gene assays. Transfection of equal amounts of Irf1 or Irf7 genes in the context of an otherwise identical expression plasmid demonstrated nearly identical abilities of the two IRFs, taken singly, to stimulate the Gbp2 promoter-luciferase reporter gene. Cotransfection of the same copy number of combined Irf1 and Irf7 genes produced a 2-fold-higher activity of the Gbp2 promoter (Fig. 3E). In this experiment, Stat1−/− MEFs were transfected to avoid indirect stimulation of the Gbp2 reporter through IRF7-stimulated type I IFN expression. Together with our analysis of IFN-γ-induced genes in IRF1- and IRF7-deficient fibroblasts, these results suggest that IRF1 and IRF7 interact functionally to control the expression of genes containing IRF binding sites in the context of an IFN-γ response. We therefore investigated whether IRF1 and IRF7 colocalize on the promoter of the Gbp2 gene following treatment of cells with IFN-γ. For this purpose, we performed ChIP-reChIP assay analysis and monitored the binding of IRF1 and IRF7 to the Gbp2 gene promoter by qPCR. The Gbp2 promoter can be divided into a distal and a proximal region, containing either a GAS and ISRE site or only one ISRE site, respectively (Fig. 5A). Both regions were shown to bind IRF1 efficiently after IFN-γ treatment (36). Consistently, we observed an increased binding of IRF1 to both promoter regions with virtually indistinguishable kinetics (Fig. 5B and E). IRF7 binding alone was similarly analyzed by ChIP-reChIP in order to reduce the background obtained in single-round ChIPs with our IRF7 antibody. These experiments revealed IRF7 association with both IFN response regions of the Gbp2 promoter. The binding kinetics of IRF7 at the Gbp2 promoter were consistent with the IRF7 mRNA expression profile after IFN-γ treatment (Fig. 1B). IRF7 association with the distal and the proximal regions was slightly different (Fig. 5C and F), being more sustained with the distal Gbp2 promoter region. Sequential ChIP with an antibody against IRF7 and reChIP with an IRF1-specific antibody revealed a pattern for IRF1 and IRF7 cooccupancy at both promoter regions in agreement with the one observed for IRF7 association (Fig. 5D and G).
Fig 5.
IRF1 and IRF7 recruitment to the Gbp2 promoter after treatment with IFN-γ. (A) Schematic drawing of the GAS located in the distal region and ISRE sites located in both the distal and proximal promoter regions of the Gbp2 gene. (B to J) WT MEFs, Tbk1/Ikbke−/− MEFs (H and I), or Irf7−/− MEFs were treated with IFN-γ for the time periods indicated. The cells were processed for ChIP (B and E) or ChIP-reChIP (C, D, and F to J) with the antibodies shown on top of the panels. The precipitates were amplified with primers flanking the distal (B to D) or proximal (E to J) Gbp2 promoter and were analyzed by qPCR. Data are expressed as percentages of precipitate relative to input DNA. qPCR measurements were made in triplicate. All experiments were repeated at least three times.
To test whether phosphorylation of IRF7 is important for its association with the Gbp2 promoter, we assessed IRF7 and IRF1 recruitment to the proximal Gbp2 promoter in WT and Tbk1/Ikbke−/− MEFs by sequential ChIP assays. ChIP for IRF1 and reChIP for IRF1 revealed that IRF1 binding to the Gbp2 promoter is similar in the two genotypes. IRF7 recruitment was unimpeded in Tbk1/Ikbke−/− MEFs, as revealed by ChIP of IRF1 and reChIP of IRF7 (Fig. 5H and I). As expected, no IRF7 signal was detected after ChIP-reChIP and amplification of the Gbp2 promoter in Irf7−/− MEFs. Furthermore, the background of IRF7 binding was similar between untreated WT MEFs and untreated Irf7−/− MEFs, suggesting that IRF7 is not constitutively associated with the Gbp2 promoter (Fig. 5J).
Taken together, our results indicate that IRF1 and IRF7 cooccupy the distal and proximal Gbp2 promoter, but binding of IRF1 precedes that of IRF7, since the IRF prototype binds already to the Gbp2 promoter 1 h after IFN-γ treatment (36) and reaches its maximum after 2 h. In contrast, maximal IRF7 binding was detected after 3 h of treatment. IRF7 binding was not reduced in Tbk1/Ikbke−/− MEFs, indicating that phosphorylation of IRF7 by either kinase is not required for nuclear translocation and DNA binding.
IRF7 is required for the sustained recruitment of RNA Pol II to the Gbp2 gene promoter.
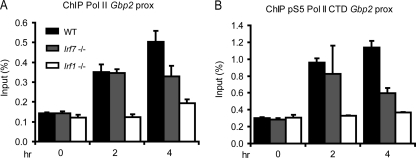
IRF1 was recently shown to contribute to Gbp2 gene induction by facilitating the recruitment of RNA Pol II (36). We therefore tested whether IRF7 similarly participates in the process of transcriptional initiation. ChIP assay analysis was performed to monitor the binding of both total RNA Pol II and the serine 5-phosphorylated enzyme to the Gbp2 promoter (Fig. 6). Serine 5 phosphorylation is an indication for a clearance-competent polymerase. It requires the action of the general transcription factor TFIIH and the associated kinase CDK7. The results demonstrate a clear reduction of both RNA Pol II recruitment and, as a consequence, its serine 5 phosphorylation 4 h after IFN-γ treatment of Irf7−/− MEFs. In line with the delayed impact of IRF7 on IFN-γ target gene expression, RNA Pol II recruitment was unaltered during the first 2 h after IFN-γ exposure. Our results indicate a direct role for IRF7 in delayed enhancement and maintenance of RNA Pol II binding for the sustained expression of IFN-γ target genes.
Fig 6.
Recruitment of RNA Pol II and of Pol II phosphorylated at serine 5 in its carboxy-terminal domain (pS5 RNA Pol II) to the proximal Gbp2 promoter after treatment with IFN-γ. WT, Irf7−/−, and Irf1−/− MEFs were treated with IFN-γ for the time periods indicated. The cells were processed for ChIP with the antibodies shown on top of the panels. The precipitates were amplified with primers flanking the proximal Gbp2 promoter and were analyzed by qPCR. Data are expressed as percentages of precipitate relative to input DNA. qPCR measurements were made in triplicate. All experiments were repeated at least three times.
IRF7 regulates antibacterial activity of activated macrophages.
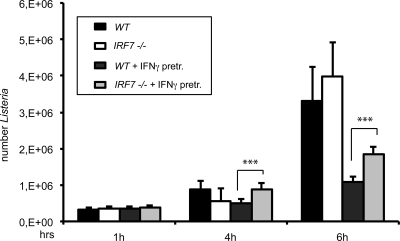
Macrophage activation by IFN-γ prevents the intracellular growth of phagocytosed bacteria. We examined whether IRF7 deficiency resulted in an increased cytoplasmic replication of the Gram-positive, facultative intracellular bacterium Listeria monocytogenes after infection of BMDMs. CFU assays were performed up to 6 h postinfection (51) (Fig. 7). Prior to infection, macrophages were pretreated with IFN-γ for 15 h to induce an antibacterial state or were left untreated. We observed equal bacterial uptake rates in WT and Irf7−/− macrophages, irrespective of pretreatment (see 1-h time point). Whereas bacterial replication was inhibited by IFN-γ in macrophages of both genotypes, Irf7−/− macrophages contained modestly but significantly increased amounts of viable bacteria after IFN-γ treatment compared to their WT counterparts. This result is in agreement with the notion that IRF7 enhances IFN-γ-induced antimicrobial gene expression but that it is not an absolute requirement. Deletion of the type I IFN receptor in a similar experimental setup did not increase or reduce bacterial replication. This rules out that the reduced antimicrobial activity in IRF7−/− macrophages resulted from a defect in Listeria-induced type I IFN synthesis (data not shown).
Fig 7.
Effect of Irf7−/− on intracellular growth of L. monocytogenes. Growth of L. monocytogenes LO28 was assessed by plating of serial dilutions of cellular lysates at the indicated time points. WT or Irf7−/− BMDMs were pretreated with IFN-γ for 15 h or left without pretreatment and infected with L. monocytogenes LO28 at a multiplicity of infection of 10. CFU counts represent mean values of six experimental values. The experiment was repeated three times. Data sets were analyzed by Student's t test (two tailed, equal variance). ***, P < 0.001.
DISCUSSION
Regulation of many IFN-γ-induced genes requires functional cooperation of STATs and IRFs. This subset of genes responds with delayed kinetics to IFN-γ because the participating IRF must be synthesized first as part of the immediate response. IRF1 is most often assigned to this task and has been shown in numerous studies to act in concert with STAT1 (5, 9, 18, 23, 32, 35). At the Gbp2 promoter, STAT1 is needed for the recruitment of the HAT CPB/p300 and subsequent histone hyperacetylation. Binding of Pol II and subsequent Gbp2 transcription requires the additional association of IRF1 with the promoter ISRE sequence (36). In search of IRF family members additionally involved in the regulation of Gbp2 expression, we uncovered a more general role of IRF7 in the regulation of IFN-γ-responsive genes. This finding is consistent with recent reports that IRF7 is expressed after IFN-γ administration to various cells of different organisms, ranging from a monocyte/macrophage cell line of rainbow trout to murine astrocytes and human fetal microglial cells, NIH 3T3 cells, or rat oligodendroglial progenitor cells (13, 15, 38).
The IRF7 promoter contains two IFN-γ response regions, a distal GAS and intronic ISRE sequences. The two response regions mediate, respectively, the association with STAT1 homodimers and with a STAT1/IRF9 complex which differs from ISGF3. Using mouse genetics and ChIP we provide the first evidence that cells employ complexes of STAT1 and IRF9 for the regulation of IFN-γ-induced genes.
IRF3-deficient cells also showed reduced expression of IFN-γ-induced genes. For the reasons given in the text describing the results shown in Fig. 2, the role of IRF3 is most likely indirect and results from reduced STAT1 expression and tyrosine phosphorylation. In further disagreement with a direct role of IRF3 in the regulation of IFN-γ-induced genes is our finding that in spite of constitutive IRF3 expression in fibroblasts, transfection of its activating kinase TBK1 did not stimulate the Gbp2 promoter (Fig. 4B). This result suggests that unlike IRF7, IRF3 has limited or no ability to regulate the expression of IFN-γ-induced genes.
Compared to IRF1, IRF7 deficiency had a less pronounced effect on IFN-γ-mediated induction of the Gbp2 gene (36). In part, this results from the participation of IRF1 in the regulation of Irf7 gene expression; hence, IRF1−/− cells contain reduced amounts of IRF7 besides lacking IRF1. This finding also explains the delay in IRF7 expression compared to IRF1. The data suggest that IRF1 and IRF7 are not simply redundant in the context of the transcriptional IFN-γ response but that they exert their predominant effects during overlapping but kinetically separated phases of transcription, with IRF1 preceding IRF7. Additionally, the results of our reporter gene analyses support the conclusion that IRF1 and IRF7 enhance each other's activity when simultaneously present at target promoters. IRF7 association with ISRE sequences to regulate IFN-γ-induced genes is consistent with a recent report showing that ISRE sites in the promoters of type I IFN-stimulated genes can be targeted by IRF7 to induce a type I IFN-like antiviral response in the absence of ISGF3 (42).
Using a ChIP assay to monitor RNA Pol II recruitment to the Gbp2 promoter, we found that IRF7 stimulated RNA Pol II binding during the delayed phase of the transcriptional response to IFN-γ. Hence, both IRF1 and IRF7 affect promoter binding of Pol II, but the maximal activity of IRF7 is delayed with respect to that of IRF1. Both IRF family members may additionally contribute to the recruitment of the Pol II kinases CDK7-TFIIH and/or CDK9-pTEFb, which are needed to efficiently engage RNA Pol II into active transcription.
Unlike IRF1, activation of the Gbp2 promoter by IRF7 occurred only in the presence of the kinases TBK1 and/or IKKε. Absence of both kinases led to a reduction of Gbp2 and Tap1 gene expression comparable to that observed in Irf7-deficient cells. This led us to conclude that phosphorylated IRF7 activates the corresponding promoters. Our notion was further strengthened by the finding that IRF7 was unable to stimulate the expression of the Gbp2 promoter-luciferase construct in Tbk1/Ikbke double-deficient MEFs, unless TBK1 was reexpressed. Treatment with IFN-γ did not enhance the TBK1/IKKε-dependent transcriptional activity of IRF7. Therefore, TBK1/IKKε may not be activated downstream of the IFNGR or, alternatively, its activation by IFN-γ is irrelevant for the genes examined in our study. We propose that basal activity of TBK1/IKKε suffices to bring about the necessary IRF7 phosphorylation. Support for this hypothesis was obtained by 2D PAGE analysis, suggesting that basal TBK1/IKKε activity generates at least three IRF7 phosphoisoforms in WT but not in Tbk1/Ikbke−/− MEFs. This is, to our knowledge, the first report of a constitutive activity of the TBK1/IKKε kinase model with a clear biological impact not on the virus-induced pattern recognition pathways through which the kinases are usually activated but on an independent pathway. Our data thus describe a novel mode of employing the same kinase module in distinct pathways of the immune system.
The IRF7 regulatory region contains eight serines as possible targets for TBK1/IKKε, and a series of IRF7 phosphoisoforms is generated upon viral infection and activation of the two kinases (6). Only three isoforms could be detected in our experiments, and the analysis of several different IRF7 mutants revealed that a combination of phosphorylated serines, especially S425-S426 and S437-S438, are required for IRF7 transcriptional activity in the context of IFN-γ signaling. The M15 mutant containing phosphomimetic aspartate residues of all regulatory domain serines except S425 and S426 was active in the absence of TBK/IKKε, but its activity could be enhanced by selective introduction of TBK1 to exceed that of WT IRF7. In contrast, transfection of the same mutant into MEFs expressing both TBK1 and IKKε resulted in GBP2 expression levels comparable to or lower than those of WT IRF7. This phenomenon can best be explained by a negative effect of IKKε. Further unpublished studies in our lab are consistent with this assumption. Negative regulation of TBK1 activity by IKKε could result from competition for common adapter proteins (33) or from a direct inhibitory phosphorylation of either TBK1 or IRF7 by IKKε.
Contrasting the findings for Gbp2 expression, Socs1 expression was reduced in Irf7−/− MEFs but still fully responsive to IFN-γ in Tbk1/Ikbke−/− MEFs. Hence, IRF7 presence is required for full expression of IFN-γ-induced genes containing IRF binding sites, but its phosphorylation is required only for a subset of such genes. We speculate that phosphorylation of IRF7 may be necessary at the promoters of Gbp2-like genes to recruit cofactors and, perhaps, to introduce activating chromatin marks whereas, in contrast, these prerequisites for transcriptional activation may be implemented at promoters of Socs1-like genes independently of IRF7 recruitment. In this context, it is interesting to note that the input of IRF7 to transcriptional initiation of the Gbp2 promoter, but not its nuclear translocation or the recruitment to target promoters, requires TBK1-mediated phosphorylation. This is demonstrated both by the TBK1-independent effect of IRF7 on SOCS1 expression and the binding of IRF7 to the Gbp2 promoter in Tbk1/Ikbke−/− cells.
In addition to these mechanistic statements, our data allow speculation about immunological consequences of IFN-γ-induced IRF7 expression. Most cells in an organism do not express IRF7 constitutively, consistent with its classification as a class II transcription factor. A notable exception from this rule is a subset of dendritic cells, plasmacytoid dendritic cells (pDCs), often referred to as interferon-producing cells (IPCs) due to their capability to produce large amounts of type I IFNs very soon after their exposure to a pathogen. A hallmark of pDCs is the constitutive expression of IRF7, the “master regulator” of type I IFN production. Since type I IFNs are mainly produced in response to intracellular pathogens, pDCs play a fundamental role in the defense against various viral infections (14, 27). Our results, together with recent work by others, indicate that IRF7 is expressed in a variety of cell types upon IFN-γ treatment and may participate in the induction of an antiviral state either alone or in synergy with type I IFNs (13, 15, 38, 42). By elevating the basal levels of IRF7 in non-pDCs, IFN-γ may play a prominent role in enhancing the innate immune response against invading intracellular pathogens.
Whereas the ability of interferons to stimulate IRF7 production explains their priming activity for transcription of type I IFN genes, it remains an open question why activation of the IFN-β or IFN-α promoters, but not stimulation of interferon-induced genes, requires full activation of TBK1/IKKε by pattern recognition receptor signaling prior to IRF7 phosphorylation. One potential explanation for this is that the basal TBK/IKKε activity which appears to suffice for the stimulation of IFN-induced genes does not produce the phosphoisoforms needed to activate the type I IFN gene promoters. In line with this assumption, several IRF7 isoforms not present in unstimulated or IFN-γ-stimulated cells are found in virus-infected cells (6). Alternatively, basal TBK1 activity may not suffice to produce transcriptionally active IRF3. These hypotheses will be tested in future work.
ACKNOWLEDGMENTS
We gratefully acknowledge Shizuo Akira (University of Osaka, Japan) for providing fibroblasts deficient for both Tbk1 and Ikbke.
Work in the lab of T.D. was funded by the Austrian Research Foundation through SFB-28.
Footnotes
Published ahead of print 17 January 2012
REFERENCES
- 1. Au WC, Pitha PM. 2001. Recruitment of multiple interferon regulatory factors and histone acetyltransferase to the transcriptionally active interferon A promoters. J. Biol. Chem. 276:41629–41637 [DOI] [PubMed] [Google Scholar]
- 2. Au WC, Yeow WS, Pitha PM. 2001. Analysis of functional domains of interferon regulatory factor 7 and its association with IRF-3. Virology 280:273–282 [DOI] [PubMed] [Google Scholar]
- 3. Baccarini M, Bistoni F, Lohmann-Matthes ML. 1985. In vitro natural cell-mediated cytotoxicity against Candida albicans: macrophage precursors as effector cells. J. Immunol. 134:2658–2665 [PubMed] [Google Scholar]
- 4. Bluyssen HA, et al. 1995. Combinatorial association and abundance of components of interferon-stimulated gene factor 3 dictate the selectivity of interferon responses. Proc. Natl. Acad. Sci. U. S. A. 92:5645–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Briken V, et al. 1995. Interferon regulatory factor 1 is required for mouse Gbp gene activation by gamma interferon. Mol. Cell. Biol. 15:975–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caillaud A, Hovanessian AG, Levy DE, Marié IJ. 2005. Regulatory serine residues mediate phosphorylation-dependent and phosphorylation-independent activation of interferon regulatory factor 7. J. Biol. Chem. 280:17671–17677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan LLY, Cheung BKW, Li JCB, Lau ASY. 2010. A role for STAT3 and cathepsin S in IL-10 down-regulation of IFN-gamma-induced MHC class II molecule on primary human blood macrophages. J. Leukoc. Biol. 88:303–311 [DOI] [PubMed] [Google Scholar]
- 8. Chau T-L, et al. 2008. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem. Sci. 33:171–180 [DOI] [PubMed] [Google Scholar]
- 9. Decker T, Kovarik P. 1999. Transcription factor activity of STAT proteins: structural requirements and regulation by phosphorylation and interacting proteins. Cell. Mol. Life Sci. 55:1535–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Génin P, Vaccaro A, Civas A. 2009. The role of differential expression of human interferon-A genes in antiviral immunity. Cytokine Growth Factor Rev. 20:283–295 [DOI] [PubMed] [Google Scholar]
- 11. Grandvaux N, et al. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harada H, et al. 1996. Regulation of IFN-alpha/beta genes: evidence for a dual function of the transcription factor complex ISGF3 in the production and action of IFN-alpha/beta. Genes Cells 1:995–1005 [DOI] [PubMed] [Google Scholar]
- 13. Holland JW, et al. 2008. Molecular characterization of IRF3 and IRF7 in rainbow trout, Oncorhynchus mykiss: functional analysis and transcriptional modulation. Mol. Immunol. 46:269–285 [DOI] [PubMed] [Google Scholar]
- 14. Honda K, et al. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777 [DOI] [PubMed] [Google Scholar]
- 15. Horiuchi M, Itoh A, Pleasure D, Ozato K, Itoh T. 2011. Cooperative contributions of interferon regulatory factor 1 (IRF1) and IRF8 to interferon-γ-mediated cytotoxic effects on oligodendroglial progenitor cells. J. Neuroinflammation 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanno Y, Levi B-Z, Tamura T, Ozato K. 2005. Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J. Interferon Cytokine Res. 25:770–779 [DOI] [PubMed] [Google Scholar]
- 17. Kastner R, et al. 2011. LipA, a tyrosine and lipid phosphatase involved in the virulence of Listeria monocytogenes. Infect. Immun. 79:2489–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kimura T, et al. 1994. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science 264:1921–1924 [DOI] [PubMed] [Google Scholar]
- 19. Kovarik P, et al. 2001. Specificity of signaling by STAT1 depends on SH2 and C-terminal domains that regulate Ser727 phosphorylation, differentially affecting specific target gene expression. EMBO J. 20:91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kovarik P, Stoiber D, Novy M, Decker T. 1998. Stat1 combines signals derived from IFN-gamma and LPS receptors during macrophage activation. EMBO J. 17:3660–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laricchia-Robbio L, et al. 2005. Partner-regulated interaction of IFN regulatory factor 8 with chromatin visualized in live macrophages. Proc. Natl. Acad. Sci. U. S. A. 102:14368–14373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levy DE, Darnell JE. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3:651–662 [DOI] [PubMed] [Google Scholar]
- 23. Lew DJ, Decker T, Strehlow I, Darnell JE. 1991. Overlapping elements in the guanylate-binding protein gene promoter mediate transcriptional induction by alpha and gamma interferons. Mol. Cell. Biol. 11:182–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin R, Mamane Y, Hiscott J. 2000. Multiple regulatory domains control IRF-7 activity in response to virus infection. J. Biol. Chem. 275:34320–34327 [DOI] [PubMed] [Google Scholar]
- 25. Marié I, Durbin JE, Levy DE. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marié I, Smith E, Prakash A, Levy DE. 2000. Phosphorylation-induced dimerization of interferon regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol. Cell. Biol. 20:8803–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCullough KC, Ruggli N, Summerfield A. 2009. Dendritic cells—at the front-line of pathogen attack. Vet. Immunol. Immunopathol. 128:7–15 [DOI] [PubMed] [Google Scholar]
- 28. Ni Z, et al. 2005. Apical role for BRG1 in cytokine-induced promoter assembly. Proc. Natl. Acad. Sci. U. S. A. 102:14611–14616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ning S, Pagano JS, Barber GN. 2011. IRF7: activation, regulation, modification and function. Genes Immun. 12:399–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nissen RM, Yamamoto KR. 2000. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 14:2314–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Panne D, Maniatis T, Harrison SC. 2007. An atomic model of the interferon-beta enhanceosome. Cell 129:1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pattenden SG, Klose R, Karaskov E, Bremner R. 2002. Interferon-gamma-induced chromatin remodeling at the CIITA locus is BRG1 dependent. EMBO J. 21:1978–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paz S, et al. 2009. Ubiquitin-regulated recruitment of IκB kinase ε to the MAVS interferon signaling adapter. Mol. Cell. Biol. 29:3401–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perry AK, Chow EK, Goodnough JB, Yeh W-C, Cheng G. 2004. Differential requirement for TANK-binding kinase-1 in type I interferon responses to Toll-like receptor activation and viral infection. J. Exp. Med. 199:1651–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Piskurich JF, Linhoff MW, Wang Y, Ting JP. 1999. Two distinct gamma interferon-inducible promoters of the major histocompatibility complex class II transactivator gene are differentially regulated by STAT1, interferon regulatory factor 1, and transforming growth factor β. Mol. Cell. Biol. 19:431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramsauer K, et al. 2007. Distinct modes of action applied by transcription factors STAT1 and IRF1 to initiate transcription of the IFN-gamma-inducible gbp2 gene. Proc. Natl. Acad. Sci. U. S. A. 104:2849–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reis LF, Ruffner H, Stark G, Aguet M, Weissmann C. 1994. Mice devoid of interferon regulatory factor 1 (IRF-1) show normal expression of type I interferon genes. EMBO J. 13:4798–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saha B, Jyothi Prasanna S, Chandrasekar B, Nandi D. 2010. Gene modulation and immunoregulatory roles of interferon gamma. Cytokine 50:1–14 [DOI] [PubMed] [Google Scholar]
- 39. Sato M, et al. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539–548 [DOI] [PubMed] [Google Scholar]
- 40. Savitsky D, Tamura T, Yanai H, Taniguchi T. 2010. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol. Immunother. 59:489–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schlüter G, Boinska D, Nieman-Seyde SC. 2000. Evidence for translational repression of the SOCS-1 major open reading frame by an upstream open reading frame. Biochem. Biophys. Res. Commun. 268:255–261 [DOI] [PubMed] [Google Scholar]
- 42. Schmid S, Mordstein M, Kochs G, GarcíA-Sastre A, Tenoever BR. 2010. Transcription factor redundancy ensures induction of the antiviral state. J. Biol. Chem. 285:42013–42022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sheehan KCF, et al. 2006. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 26:804–819 [DOI] [PubMed] [Google Scholar]
- 44. Stockinger S, et al. 2004. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J. Immunol. 173:7416–7425 [DOI] [PubMed] [Google Scholar]
- 45. Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623–655 [DOI] [PubMed] [Google Scholar]
- 46. Taniguchi T, Takaoka A. 2001. A weak signal for strong responses: interferon-alpha/beta revisited. Nat. Rev. Mol. Cell Biol. 2:378–386 [DOI] [PubMed] [Google Scholar]
- 47. TenOever BR, et al. 2004. Activation of TBK1 and IKKvarε kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J. Virol. 78:10636–10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Varinou L, et al. 2003. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-gamma-dependent innate immunity. Immunity 19:793–802 [DOI] [PubMed] [Google Scholar]
- 49. Xiong H, et al. 2003. Complex formation of the interferon (IFN) consensus sequence-binding protein with IRF-1 is essential for murine macrophage IFN-gamma-induced iNOS gene expression. J. Biol. Chem. 278:2271–2277 [DOI] [PubMed] [Google Scholar]
- 50. Zhang L, Pagano JS. 1997. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17:5748–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zwaferink H, Stockinger S, Reipert S, Decker T. 2008. Stimulation of inducible nitric oxide synthase expression by beta interferon increases necrotic death of macrophages upon Listeria monocytogenes infection. Infect. Immun. 76:1649–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]