Abstract
Mitochondria possess an outer membrane (OMM) and an inner membrane (IMM), which folds into invaginations called cristae. Lipid composition, membrane potential, and proteins in the IMM influence organization of cristae. Here we show an essential role of the OMM protein Sam50 in the maintenance of the structure of cristae. Sam50 is a part of the sorting and assembly machinery (SAM) necessary for the assembly of β-barrel proteins in the OMM. We provide evidence that the SAM components exist in a large protein complex together with the IMM proteins mitofilin and CHCHD3, which we term the mitochondrial intermembrane space bridging (MIB) complex. Interactions between OMM and IMM components of the MIB complex are crucial for the preservation of cristae. After destabilization of the MIB complex, we observed deficiency in the assembly of respiratory chain complexes. Long-term depletion of Sam50 influences the amounts of proteins from all large respiratory complexes that contain mitochondrially encoded subunits, pointing to a connection between the structural integrity of cristae, assembly of respiratory complexes, and/or the maintenance of mitochondrial DNA (mtDNA).
INTRODUCTION
Mitochondria are organelles of bacterial origin, with two membranes that divide them into four subcompartments: the outer membrane (OMM), the intermembrane space (IMS), the inner membrane (IMM), and the matrix. Cellular respiration, one of many important functions of mitochondria, takes place at the IMM. During electron transfer, which involves four protein complexes, protons are pumped across the IMM, forming a proton gradient used by the F1FO ATPase, also known as complex V, to produce ATP (38).
Respiratory chain complexes contain both nucleus- and mitochondrion-encoded subunits. In mammalian mitochondria, 13 subunits are encoded by the mitochondrial DNA (mtDNA), and they are a part of NADH dehydrogenase (ubiquinone) or complex I (7 subunits), cytochrome b-c1 complex or complex III (1 subunit), cytochrome c oxidase or complex IV (3 subunits), and of the F1FO ATPase (2 subunits) (25, 38). The nucleus-encoded subunits, which represent the majority, are produced in the cytosol and have to be imported into the mitochondria with the assistance of translocases in the OMM (the TOM complex) and the IMM (the TIM complexes) (3). The OMM of mitochondria also contains the sorting and assembly machinery (SAM) that is involved in the membrane assembly of mitochondrial β-barrel proteins (32, 46). In human mitochondria, this complex consists of metaxins 1 and 2 and the central component, Sam50, a pore-forming, β-barrel protein, conserved from bacteria to eukaryotes (18, 23, 24).
The IMM forms invaginations called cristae, connected to the rest of the IMM, the so-called inner boundary membrane, by crista junctions (5, 34). Cristae significantly increase the surface of the IMM. They are considered a distinct region of it because they differ in protein and lipid composition from the inner boundary membrane (12, 43, 48). Formation of cristae is known to be affected by the membrane potential (Δψ) and lipid and protein composition (10, 22, 37). Cardiolipin, a lipid produced and found exclusively in mitochondria, seems to be required for proper organization of cristae, as seen in patients suffering from cardiolipin deficiency-related Barth's syndrome (1). The importance of the F1FO ATPase in the maintenance of the shape of cristae has also been shown (33). Loss of the IMM protein optic atrophy 1 (OPA1) results in the widening of cristae and sensitization to apoptosis (9). Mitochondrial polynucleotide phosphorylase (PNPase), a protein involved in RNA import into mitochondria, influences the morphology of cristae, likely by controlling the amount of respiratory chain complexes (45). In addition, knockdown of mitofilin is also known to affect the structure of cristae (21).
Mitofilin is an abundant mitochondrial protein found mostly in the inner boundary membrane (21), where it exists in two isoforms of 88 and 90 kDa. It is anchored by its amino terminus in the IMM, with most of the protein exposed to the IMS (11, 30). Mitofilin has been found to interact with DISC1 (31) and PARP-1 (40), which affect mitochondrial function and mtDNA integrity, respectively. In two recent reports, mitofilin has been connected with several other proteins, such as CHCHD3, CHCHD6, Sam50, metaxins 1 and 2, and DnaJC11 (6, 49). The interaction of CHCHD3 with mitofilin and OPA1 has been proposed to be of major significance for the maintenance of the morphology of cristae (6).
Here we report the crucial role of the OMM protein Sam50 in the regulation of mitochondrial shape, the morphology of cristae, and the assembly of respiratory complexes. This function is performed together with mitofilin and CHCHD3. These three proteins are found in a >700-kDa-large complex, which we term the mitochondrial intermembrane space bridging (MIB) complex. The stability of the MIB complex depends strongly on Sam50. After Sam50 depletion, we observed severe defects in the mitochondrial ultrastructure and network formation. In addition, the steady-state levels and the assembly of large respiratory complexes that contain mitochondrion-encoded subunits were reduced. Taken together, our data establish a surprising connection between the OMM machinery for the assembly of β-barrel proteins, the organization of cristae, and the stability of respiratory chain complexes.
MATERIALS AND METHODS
Cell culture and isolation of mitochondria.
HeLa cells with an inducible short hairpin RNA (shRNA)-mediated knockdown were generated as described previously (24, 47). The sequence of sam50kd-3 shRNA is 5′-GCGGAATGTTGGTACCCATTG-3′, that of VDAC1kd-2 shRNA 5′-AAAGTGACGGGCAGTCTGGAA-3′, that of mflkd-2 shRNA 5′-GCATCCTCATCTTCTATAAGG-3′, that of NDUFS1kd-1 is 5′-GCATGCAGATCCCTCGATTCT-3′, that of coxVakd-2 shRNA is 5′-GCAGGACCTCATAAGGAAATC-3′, and that of CHCHD3kd-2 shRNA is 5′-TATCAGAAAGCTGCTGAAGAGGTGGAAGC-3′. shRNAs of sam50kd-2, tom40kd-2, and mtx2kd-2 were described previously (24). Single-cell clones were isolated for each shRNA, except NDUFS1kd-1. Cells were cultivated in RPMI 1640 or Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with 10% fetal calf serum (FCS) (Biochrom) and penicillin-streptomycin. Expression of shRNAs was induced by cultivating cells in medium additionally containing 1 μg/ml doxycycline (DOX) (BD Biosciences) for various time periods. Isolation of mitochondria was performed as described previously (24).
Microscopy. (i) TEM.
Cells were grown on coverslips and fixed for 1 h with 2.5% glutaraldehyde (50 mM cacodylate [pH 7.2], 50 mM KCl, and 2.5 mM MgCl2) at room temperature. Cells were then fixed for 2 h at 4°C with 2% OsO4 buffered with 50 mM cacodylate (pH 7.2), washed with H2O, and incubated overnight at 4°C with 0.5% uranyl acetate (in H2O). The cells were dehydrated and embedded in Epon 812. Sections stained with uranyl acetate and lead citrate were analyzed with a Zeiss EM10 or EM900 electron microscope (Zeiss, Oberkochen, Germany). Negatives were digitalized by scanning. For measuring of mitochondrial size, sam50kd-2 cells were embedded in Polybed epoxy resin (Fluka). Ultrathin sections were cut on an ultramicrotome (Leica), lead citrate contrasted in a transmission electron microscopy (TEM) stainer (Nanofilm), and analyzed in a Leo 906E TEM (Zeiss SMT) equipped with a side-mounted digital camera (Morada, Olympus SIS). Mitochondrial sizes were determined using the Image J software program by manually marking single mitochondria, measuring their area, and normalizing it against the scale bar.
(ii) Fluorescence microscopy.
Cells were grown on glass coverslips and stained by incubation with 150 nM MitoTracker stain (Molecular Probes) in cell culture medium for 30 min at 37°C. Samples were washed with phosphate-buffered saline (PBS), fixed in 3.7% paraformaldehyde (PFA), and analyzed with a Leica confocal microscope using Leica TCS software.
Protein import and electrophoresis.
Transcription and translation were performed in the presence of [35S]methionine/[35S]cysteine (PerkinElmer) using the TnT SP6 quick coupled system (Promega). Freshly isolated mitochondria from noninduced and induced knockdown cell lines were incubated with the radiolabeled proteins at 37°C in import buffer (250 mM sucrose, 20 mM HEPES [pH 7.4], 80 mM KCl, 5 mM MgCl2, 3% [wt/vol] bovine serum albumin [BSA], 2 mM KH2PO4, 5 mM methionine, 10 mM Na succinate, and 2 mM ATP) for appropriate time periods. For ferredoxin and F1β imports, potassium-acetate import buffer was used (250 mM sucrose, 5 mM Mg acetate, 80 mM K acetate, 20 mM HEPES [pH 7.4], 10 mM Na succinate, and 1 mM ATP). Samples were lysed in Laemmli sample buffer for SDS-PAGE or, for blue native PAGE (BN-PAGE), in 1% digitonin buffer (1% digitonin [Sigma] in 20 mM Tris–HCl, 0.1 mM EDTA, 50 mM NaCl, and 10% [vol/vol] glycerol, pH 7.4) and mixed with loading dye (5% [wt/vol] Coomassie brilliant blue G-250, 150 mM bis-Tris, and 500 mM ε-amino-n-caproic acid, pH 7.0). BN-PAGE samples were analyzed on 4 to 10% or 4 to 13% gels (41). 35S-labeled proteins were visualized by autoradiography using a Typhoon imaging system (GE Healthcare). Two-dimensional PAGE (2D-PAGE) was performed as described previously (24).
Measurement of mitochondrial Δψ.
Cells were seeded to equal densities on a 96-well black plate. Forty to fifty percent confluent cells were stained for 30 min with 30 nM tetramethyl rhodamine methyl ester (TMRM) in the absence or in the presence of 1 μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) to dissipate Δψ. Cells were washed once with PBS buffer and remained in PBS for fluorescence intensity measurements, which were performed using a Tecan plate reader. The difference between fluorescence intensities of samples without and with CCCP was taken as a measure of membrane potential (Δψ).
Antibodies and antibody shift experiment.
Tom40 and NDUFS1 (Fe-S protein 1 of complex I) antibodies were purchased from Santa Cruz, F1α from BD Biosciences and Invitrogen, OPA1, cytochrome c and Tom20 from BD Biosciences, mitofilin, mitofusin 1, mitofusin 2, CHCHD3, and VDAC from Abcam, CoreI, CoxI, CoxII, NDUFA9, and SDHA from Invitrogen, Bak from Upstate, Hsp60 from Stressgen, and isocitrate dehydrogenase (ICDH) from Biogenesis. Sam50, metaxin 1, and metaxin 2 antibodies were raised in rabbits against a full-length 10× His-tagged protein and affinity purified. The antibody shift experiment was performed after the import of radiolabeled protein by incubating mitochondria solubilized in 1% digitonin buffer on ice with the respective antibody as described previously (39).
Nano-LC/LTQ-Orbitrap mass spectrometry and data analysis.
For analysis, sam50kd-2 and sam50kd-3 cells were labeled by growing in 13C6 l-lysine/l-arginine-containing medium (Invitrogen) for 10 days in total. Knockdown was induced by addition of 1 μg/ml of doxycycline for a total of 7 days in unlabeled samples. Mitochondria were isolated, and the Coomassie G-250-stained single gel lanes of three biological replicates were excised for in-gel digestion with 0.1 μg of trypsin (Promega) in 20 μl 25 mM ammonium bicarbonate, pH 7.8, at 37°C for 16 h. The dried peptides were dissolved in 10 μl 1% formic acid in water, and 5 μl was injected into an Ultimate 3000 nano-LC system (Dionex) connected to a linear quadrupole ion trap-orbitrap (LTQ-Orbitrap XL) mass spectrometer (ThermoScientific) equipped with a nanoelectrospray ion source. An Acclaim PepMap 100 column (C18, 3 μm, 100 Å) (Dionex) with a capillary of 12-cm bed length was used for separation by liquid chromatography. A flow rate of 300 nl/min was employed with a solvent gradient of 7% B to 40% B in 87 min, followed by 40 to 80% B in 8 min, and subsequently from 40 to 80% B in 8 min. Solvent A was 0.1% formic acid, whereas aqueous 90% acetonitrile in 0.1% formic acid was used as solvent B. Other instrument parameters were previously described (41).
Instrument raw data were processed and quantified using the software program MaxQuant version 1.0.13.13 (4). For processing, the top 6 tandem mass spectrometry (MS/MS) peaks per 100 Da were used to generate msn files. These msn files were searched against the human fraction of the Swiss-Prot database (v.57.5; 20,329 sequences) containing both reversed sequences and contaminants using an in-house version of the Mascot search engine (v.2.2.1). A mass tolerance of 0.5 Da was used for MS/MS fragments. Trypsin was used as a protease, allowing up to one missed cleavage and peptide charges 2+ and 3+. As variable modifications, oxidation (met), pyro-Glu (N-terminal gln), and N-acetyl (protein) were allowed. At least two peptides, one of them being a unique peptide, were required for protein identification, as well as a false discovery rate (FDR) of 1% being applied. For protein quantification, at least two ratio counts were required, and razor peptides were included in the calculation of protein ratios. To assess the regulated proteins and gain control of the FDR, the MaxQuant significance B values were corrected for multiple hypotheses using the Benjamini-Hochberg algorithm. Proteins with a corrected significance of less than 0.05 were considered regulated.
Cardiolipin measurements. (i) Extraction.
Lipids were extracted according to the described method (2). One hundred fifty to three hundred milligrams of cells were mixed with 500 μl methanol and 250 μl chloroform, and 1 μg of 1′,3′-bis[1,2-dimyristoyl-sn-glycero-3-phospho]-sn-glycerol was added as an internal standard. The mixture was shaken for 3 min using a ball mill with three steel balls, followed by 3 min of sonication. After centrifugation, the tissue residue was reextracted with 250 μl chloroform, the organic phases were combined, and 250 μl of 0.88% potassium chloride was added. The mixture was shaken thoroughly. After centrifugation, the lower organic phase containing the lipids was removed and the solvent was removed in a stream of nitrogen. The residue was dissolved in 50 μl methanol and analyzed using liquid chromatography (LC)-MS/MS. LC-MS/MS analyses were performed using a Waters Acquity ultra-high-performance liquid chromatograph coupled to a Waters Micromass Quattro Premier triple-quadrupole mass spectrometer (Milford, MA) with an electrospray interface (ESI). All aspects of system operation and data acquisition were controlled using the MassLynx V 4.1 software program.
(ii) Chromatographic and mass spectrometric conditions.
Separation of cardiolipins was carried out by reversed-phase ion pair chromatography according to a published procedure (29), using an Acquity BEH C8 column (2.1 by 50 mm, 1.7-μm particle size, with a 2.1- by 5-mm guard column; Waters) with the following solvent systems: solvent A consisted of 450 ml acetonitrile, 50 ml water, 2.5 ml triethylamine, and 2.5 ml acetic acid; solvent B contained 450 ml 2-propanol, 50 ml water, 2.5 ml triethylamine and 2.5 ml acetic acid. A gradient elution was performed at a flow rate of 0.25 ml min−1 at a column temperature of 40°C from 25% to 100% B in 10 min, followed by 100% B for 2 min, and reconditioning at 25% B for 3 min. For the analysis of cardiolipins, the electrospray source was operated in the negative electrospray mode (ESI−) at 120°C and a capillary voltage of 3.25 kV. Nitrogen was used as the dissolvent and cone gas with flow rates of 800 liters h−1 at 450°C and 50 liters h−1, respectively; the cone voltage (CV) was adjusted to 35 V. Cardiolipin species were analyzed by multiple reaction monitoring (MRM) using Argon as a collision gas at a pressure of approximately 3 × 10−3 bar and a collision energy (CE) of 55 eV. The characteristic formation of fatty acid fragments ([M-H]− → [FA]−) for each individual molecular species during collision-induced dissociation (CID) was monitored for a scan time of 0.025 s per transition.
RESULTS
Depletion of Sam50 leads to changes in mitochondrial shape and the structure of cristae.
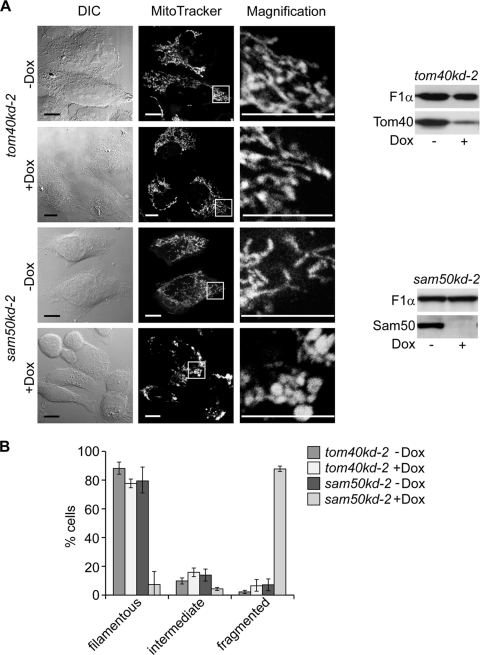
We studied the effect of the reduction of Sam50 and Tom40, the central components of the SAM and TOM complexes, respectively, on mitochondrial structure and composition. Proteins were depleted using cell lines containing doxycycline (Dox)-inducible shRNA constructs (sam50kd-2 and tom40kd-2) (24). After induction of knockdown (Fig. 1A), mitochondria were stained with MitoTracker, a Δψ-sensitive dye, and analyzed by confocal microscopy. Reduction of Tom40 led to only a slight decrease in the interconnectivity of mitochondria (Fig. 1A), indicating that a defect in protein import did not generally affect the mitochondrial network. In Sam50-depleted cells, however, mitochondria clumped while retaining the Δψ (Fig. 1A). A similar phenotype was observed in a cell line with an alternative shRNA against Sam50, sam50kd-3, excluding an off-target effect (data not shown). Control cells without Dox treatment displayed normal mitochondria. Quantification of cells with fragmented mitochondria confirmed these results (Fig. 1B). We conclude that depletion of Sam50 impairs mitochondrial network formation, leading to abnormal spherical mitochondria.
Fig 1.
The mitochondrial network structure is affected by Sam50 depletion. (A) tom40kd-2 and sam50kd-2 cells were cultivated in the presence or absence of doxycycline (Dox) for 7 days. Mitochondria in cells untreated (−Dox) or treated with Dox (+Dox) were stained with 150 nM MitoTracker Orange and analyzed by confocal microscopy. DIC, differential interference contrast. Scale bar, 10 μm. Western blots show the level of knockdown of the respective protein. Tom40, translocase of the outer mitochondrial membrane 40 protein; Sam50, sorting and assembly machinery 50 protein; F1α, subunit α of F1FO ATPase. (B) One hundred cells from 3 independent experiments were quantified according to the appearance of their mitochondria. The graph shows the mean values ± SD.
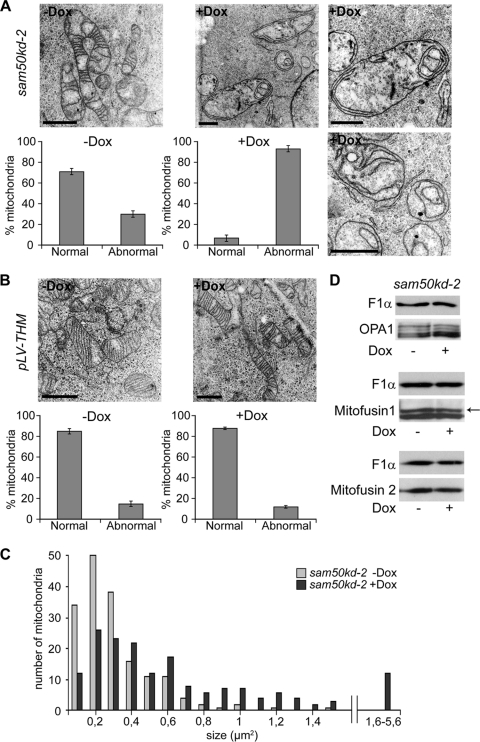
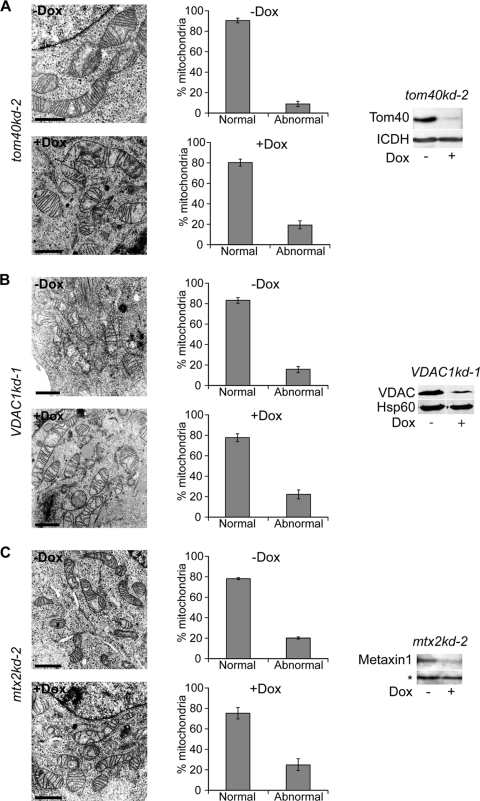
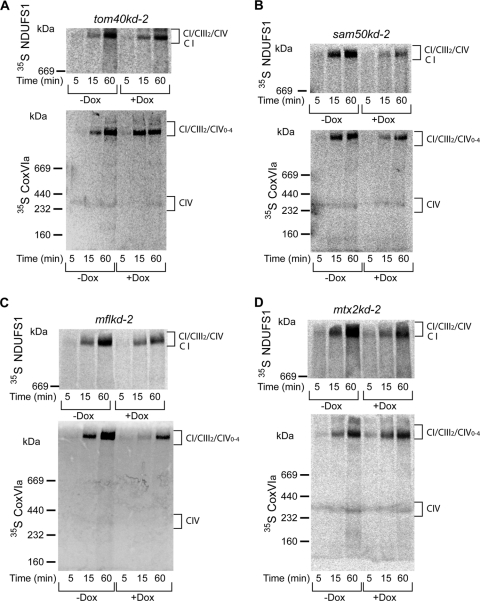
We then analyzed the mitochondrial ultrastructure in sam50kd-2 cells by transmission electron microscopy (TEM). Virtually all mitochondria in Dox-treated cells lacked the typical structure of cristae, although remnants of the membranes of cristae were preserved. The IMM often appeared in the form of one or more concentric circles (Fig. 2A). In contrast, mitochondria of the untreated sam50kd-2 cells revealed conventional cristae (Fig. 2A). In the sam50kd-3 cell line, we again observed a striking rearrangement of the structure of mitochondrial cristae after Sam50 depletion (data not shown). By using the empty vector cell line pLV-THM as a control, we could rule out that the Dox treatment induced the described alterations (Fig. 2B). Depletion of Sam50 results in a reduction in the amounts of its substrates, the β-barrel proteins Tom40 and voltage-dependent anion-selective channel (VDAC), and of other components of the SAM complex, metaxins 1 and 2 (24). We therefore investigated whether the reduction of any of these proteins had an impact on the structure of cristae, using cell lines containing inducible shRNAs (tom40kd-2, mtx2kd-2 [24], and VDAC1kd-1 [this study]). In all of these cell lines, the depletion of the respective protein was equal to or higher than the depletion we observed after Sam50 knockdown (Fig. 3A, B and C, 4B, and 5A). However, reduction of these proteins did not significantly affect the structure of cristae (Fig. 3A, B, and C).
Fig 2.
Mitochondria lacking Sam50 show an altered structure of cristae. (A and B) sam50kd-2 and pLV-THM cells were induced with doxycycline for 7 days and fixed with glutaraldehyde. Noninduced (−Dox) and induced (+Dox) cells were embedded in resin, and ultrathin sections were analyzed by TEM. Scale bar, 1 μm. For quantification, at least 50 mitochondria were counted from several sections of two independent samples. The graphs show the mean values ± SD. (C) The size of 170 mitochondria from sam50kd-2 cells without (−Dox) or after 7 days of (+Dox) Dox treatment was determined from TEM pictures using the ImageJ software program. (D) Levels of the mitochondrial fusion proteins mitofusins 1 and 2 and an organizer of cristae, OPA1, were assessed by SDS-PAGE and Western blotting of mitochondria from sam50kd-2 cells with or without Dox induction for 7 days. F1α was used as a loading control. The arrow marks the specific mitofusin 1 band as indicated by the antibody manufacturer.
Fig 3.
Reduction of Tom40, VDAC1, and metaxins does not affect the structure of cristae. (A, B, and C) tom40kd-2 cells were treated for 5 days and VDAC1kd-1 and mtx2kd-2 cells were treated for 7 days with doxycycline (Dox). Cells were then analyzed by TEM, and the effect on the morphology of cristae was quantified by counting at least 50 mitochondria from several sections of two different samples. Graphs represent mean values ± SD. Scale bar, 1 μm. The knockdown of the respective protein was assessed by SDS-PAGE and Western blotting. ICDH, isocitrate dehydrogenase; Hsp60, heat shock protein 60. The asterisk represents a cross-reactive band used as a loading control. The efficiency of the metaxin 2 knockdown was assessed by monitoring the reduction in the amount of metaxin 1.
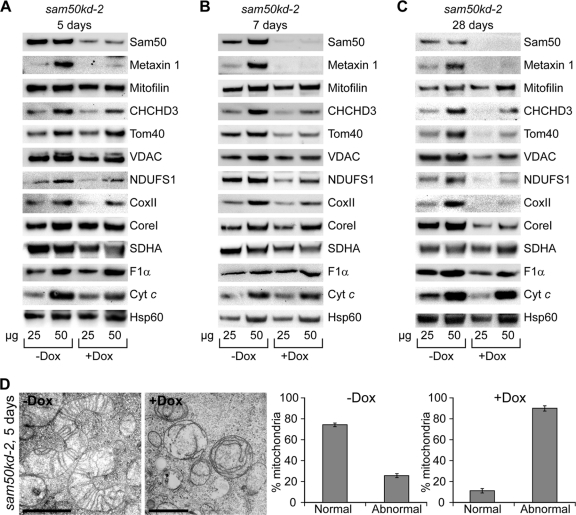
Fig 4.
Depletion of Sam50 affects levels of several other mitochondrial proteins. (A, B, and C) sam50kd-2 cells were grown in the absence (−Dox) or presence (+Dox) of doxycycline for 5, 7, and 28 days. Mitochondria were prepared from noninduced and induced samples, and 25 and 50 μg of mitochondrial protein was analyzed by SDS-PAGE and Western blotting. NDUFS1, Fe-S protein 1 of complex I; CoxII, subunit 2 of complex IV; CoreI, core protein 1 of complex III; SDHA, subunit A flavoprotein of complex II; Cyt c, cytochrome c. For other designations, see the text and the legends for Fig. 1 and 3. (D) Mitochondria from panel A were analyzed by TEM, and the number of morphologically changed mitochondria was quantified by counting at least 50 mitochondria from different sections of two independent samples. The graph shows the mean values ± SD.
Fig 5.
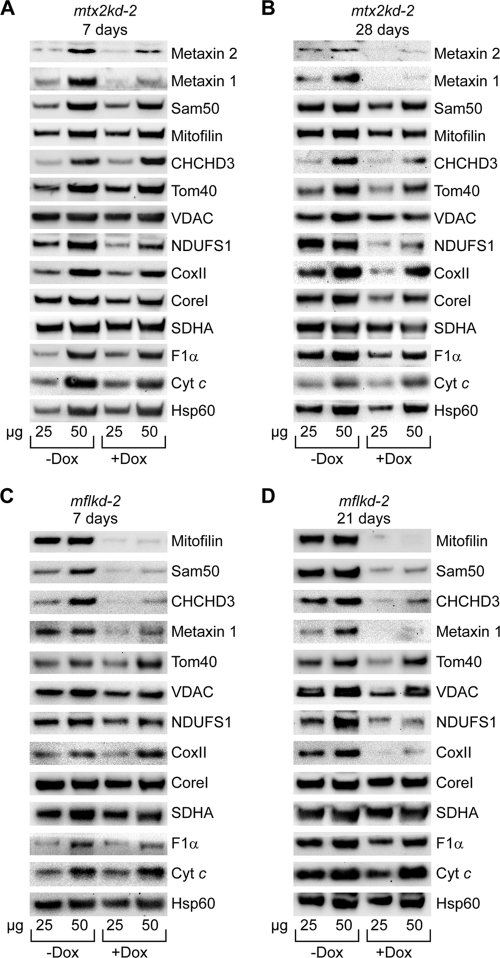
Depletion of mitofilin and metaxins affects mitochondrial proteins in a way similar to that with Sam50 reduction. (A and B) Mitochondria from mtx2kd-2 cells were isolated after doxycycline (Dox) treatment for 7 and 28 days. Protein levels of 25 and 50 μg of mitochondrial protein were analyzed by SDS-PAGE and Western blotting. (C and D) Knockdown of mitofilin was induced for 7 and 21 days with Dox, and mitochondria were isolated from noninduced and induced samples and analyzed by SDS-PAGE and Western blotting. For designations, see the text and the legends for Fig. 1, 3, and 4.
Interestingly, we observed that the Dox-treated sam50kd-2 cells contained visibly less mitochondria than the same number of untreated cells. Therefore, we determined the size of mitochondria. Whereas mitochondria in the noninduced cells had an average size of 0.3 (±0.2) μm2, we measured an average size of 0.7 (±0.7) μm2 of mitochondria in Dox-treated cells, displaying a variability ranging between 0.1 and 5.6 μm2 (Fig. 2C). In comparison, mitochondria from Tom40 knockdown cells revealed an average size of 0.4 (±0.3) μm2 in both Dox-treated and untreated cells (not shown). This suggests a fission/fusion abnormality in Sam50-depleted cells. Considering our previous observation that mitochondria appear fragmented after Sam50 knockdown, we determined the amounts of proteins involved in mitochondrial fusion (mitofusins 1 and 2) and of a known organizer of crista junction, OPA1. The levels of all three proteins were unchanged in mitochondria lacking Sam50 (Fig. 2D). Therefore, we conclude that Sam50 plays a role in maintenance of both the mitochondrial network and the structure of cristae, which is likely independent of its function in the assembly of β-barrel proteins.
The knockdown of Sam50 leads to changes in levels of other mitochondrial proteins.
It is possible that in addition to Tom40, VDAC, and metaxins, other as yet unidentified proteins are also diminished in the absence of Sam50, leading to the defect in cristae that we observed. Recent reports connected mitofilin and coiled-coil helix, coiled-coil helix domain-containing protein 3 (CHCHD3) to Sam50, and both proteins were also shown to influence the organization of cristae (6, 21, 49). Thus, we performed stable isotope labeling with amino acids in cell culture (SILAC) combined with mass spectrometry to determine which proteins were reduced after Sam50 depletion and to what extent. Three samples of sam50kd-2 mitochondria and one of sam50kd-3 mitochondria were analyzed after 7 days of knockdown induction. The list of reduced proteins included expected targets, such as Sam50 itself, metaxin 1, Tom40, and two isoforms of VDAC (Table 1). Metaxin 2 was not identified in a sufficient number of samples, possibly due to the low abundance of the protein. Interestingly, mitofilin and CHCHD3, although identified, were not significantly reduced in knockdown samples. Surprisingly, the list of proteins reduced after Sam50 knockdown included members of respiratory complexes I, III, and IV (Table 1; see also the supplemental material).
Table 1.
Proteins significantly downregulated after Sam50 depletiona
| UniProt no. | Name | Mass (kDa) | Ratio (H/L) |
|---|---|---|---|
| Q9Y512 | Sorting and assembly machinery component 50 homolog (Sam50) | 51.98 | 5.20 |
| Q13505 | Metaxin 1 | 35.78 | 4.53 |
| Q9UI09 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 12 | 17.11 | 2.40 |
| Q9P0J0 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 13 | 16.70 | 2.17 |
| O95168 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 4 | 15.21 | 2.09 |
| P49821 | NADH dehydrogenase (ubiquinone) flavoprotein 1 | 50.82 | 1.98 |
| P28331 | NADH dehydrogenase (ubiquinone) 75-kDa subunit | 79.47 | 1.94 |
| Q9Y6M9 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 9 | 21.83 | 1.89 |
| P45880 | Voltage-dependent anion-selective channel protein 2 | 31.57 | 1.89 |
| O00217 | NADH dehydrogenase (ubiquinone) iron-sulfur protein 8 | 23.71 | 1.87 |
| P19404 | NADH dehydrogenase (ubiquinone) flavoprotein 2 | 27.39 | 1.86 |
| P14927 | Cytochrome b-c1 complex subunit 7 | 13.53 | 1.84 |
| O96008 | Mitochondrial import receptor subunit TOM40 homolog | 37.89 | 1.83 |
| P00403 | Cytochrome c oxidase subunit 2 | 25.56 | 1.83 |
| O75251 | NADH dehydrogenase (ubiquinone) iron-sulfur protein 7 | 23.56 | 1.82 |
| P21796 | Voltage-dependent anion-selective channel protein 1 | 30.77 | 1.80 |
| O96000 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 10 | 20.78 | 1.74 |
| P13073 | Cytochrome c oxidase subunit 4 isoform 1 | 19.58 | 1.72 |
sam50kd-2 and sam50kd-3 cells were induced by doxycycline for 7 days. Noninduced samples were grown in 13C6 l-lysine/l-arginine-containing medium (H). Samples with induced knockdown of Sam50 remained unlabeled (L). The complete list can be found in the supplemental material.
To verify this, we analyzed protein levels by Western blotting in mitochondria from sam50kd-2 cells where knockdown was induced for 5, 7, and 28 days. After 5 days, levels of Sam50 and metaxin 1 were significantly diminished, and a mild reduction could be seen in the levels of CHCHD3 and of the respiratory complex I protein NDUFS1 (Fig. 4A). After 7 days, apart from metaxin 1, we could see a significant reduction in the amounts of Tom40 and VDAC, the IMM protein CHCHD3, and the respiratory chain proteins NDUFS1 (complex I) and CoxII (complex IV) (Fig. 4B). Long-term depletion of Sam50 for 28 days did not further significantly reduce the amount of CHCHD3, but Tom40, VDAC, NDUFS1, and CoxII levels continued to diminish. In addition, CoreI and F1α, belonging to complex III and F1FO ATPase, were also depleted. The level of mitofilin remained unaffected (Fig. 4C). Therefore, Western blot data were in agreement with the results of SILAC/mass spectrometry analysis, with the exception of the reduction in the CHCHD3 levels. However, the effect on the morphology of cristae observed after Sam50 depletion cannot be explained simply by a reduction in the amount of CHCHD3, considering that even after 5 days of Sam50 knockdown, when levels of CHCHD3 were not significantly altered, the structure of cristae was already severely disturbed (Fig. 4D). We conclude that the effect of Sam50 on the shape of cristae is not the consequence of CHCHD3 reduction. In the beginning phase of Sam50 depletion, mitochondrial function seems to be preserved, since practically all of the mitochondrial proteins we tested were not affected, but the structure of cristae is completely lost. The long-term absence of Sam50 negatively reflects on the amount of proteins from respiratory complexes I, III, IV, and F1FO ATPase—all large complexes that contain mitochondrially encoded subunits.
Sam50, metaxins, mitofilin, and CHCHD3 are functionally connected.
We next analyzed protein levels in mitochondria after short- and long-term depletion of metaxin 2 and mitofilin using mtx2kd-2 (24) and mflkd-2 (this study) cell lines. Metaxin 2 depletion for 7 days led to a reduction in metaxin 1, as observed previously (24), and in NDUFS1 levels, but other proteins were unaffected (Fig. 5A). However, after 28 days of metaxin 2 knockdown, levels of the β-barrel proteins Tom40 and VDAC were reduced. The amounts of CHCHD3, mitofilin, and Sam50 were also affected. Also depleted were NDUFS1, CoxII, CoreI, and F1α, although to a lesser extent than after Sam50 knockdown (Fig. 5B). The observed effects were not a consequence of Dox treatment of cells, because after the knockdown of Tom40 in the tom40kd-2 cell line, only the TOM complex components Tom40 and Tom20 were diminished, whereas other proteins were not affected (data not shown). We conclude that metaxins together with Sam50 are important for the stability of respiratory complexes.
Mitofilin depletion strongly influenced the levels of Sam50, CHCHD3, and metaxins, but other proteins were not affected (Fig. 5C). Even after 21 days of mitofilin knockdown, levels of the β-barrel proteins Tom40 and VDAC remained stable, but we observed a reduction in proteins belonging to complexes I and IV (Fig. 5D). The depletion of CHCHD3, in turn, affects the amounts of mitofilin and Sam50 (see Fig. 7C) (6). Therefore, we conclude that a functional connection exists between the components of the SAM complex in the OMM and mitofilin and CHCHD3 in the IMM.
Fig 7.
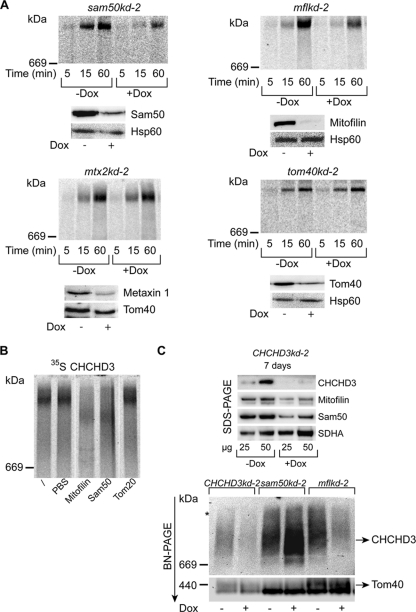
CHCHD3 exists in the same protein complex as Sam50 and mitofilin. (A) Mitochondria isolated from noninduced (−Dox) and doxycycline-induced (+Dox) tom40kd-2 cells (5 days of induction) and sam50kd-2, mflkd-2, and mtx2kd-2 cells (7 days of induction) were incubated with 35S-labeled CHCHD3 for the indicated time periods and analyzed by BN-PAGE and autoradiography. Western blot analyses show the efficiency of the respective protein knockdown. Metaxin 1 levels were used to estimate the efficiency of the metaxin 2 knockdown. Decoration with Hsp60 or Tom40 antibodies served as a loading control. (B) Mitochondria isolated from HeLa cells were incubated with 35S-labeled CHCHD3 for 1 h before solubilization in 1% digitonin buffer. One sample was left untreated (/), and others were treated with PBS buffer or with antibodies against mitofilin, Sam50, and Tom20. Samples were then analyzed by BN-PAGE and autoradiography. Irrelevant lanes were excised. (C) Mitochondria isolated from CHCHD3kd-2, sam50kd-2, and mflkd-2 cells after 7 days of Dox induction were analyzed by SDS-PAGE or BN-PAGE and Western blotting using antibodies against CHCHD3, mitofilin, Sam50, SDHA, and Tom40. The occasionally observed larger complex is marked with an asterisk.
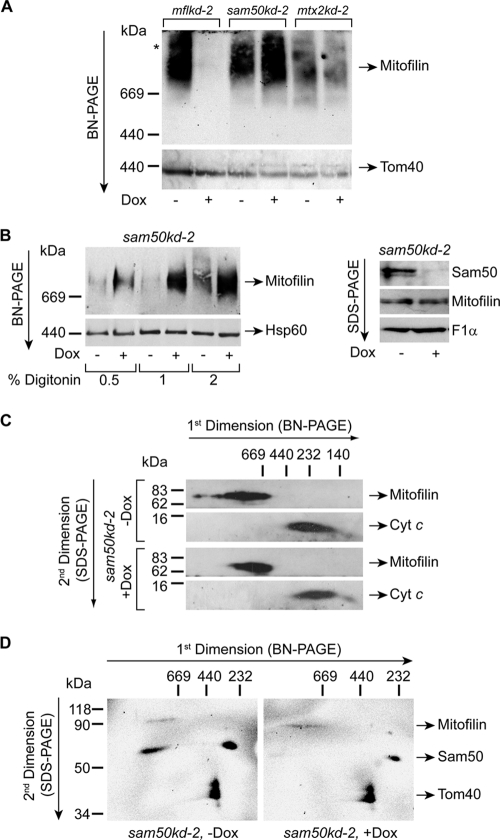
Previously, we were able to detect metaxins 1 and 2 in a high-molecular-mass protein complex (>700 kDa) (24). When we analyzed mitochondria from mflkd-2, sam50kd-2, and mtx2kd-2 cells by blue native polyacrylamide gel electrophoresis (BN-PAGE) and Western blotting, we observed that antibodies against mitofilin specifically recognized a protein complex larger than 700 kDa. This complex was not detected after knockdown of mitofilin but was more abundant after Sam50 depletion and unchanged after metaxin 2 knockdown. In comparison, levels of the TOM complex were equal in all samples but that of Sam50 plus Dox, where they were slightly reduced (Fig. 6A). The amount of mitofilin complex detected also increased with the concentration of detergent digitonin used, and much more of this protein complex was detected in Sam50-depleted samples, in spite of equal quantities of loaded material and mitofilin (Fig. 6B). When analyzed by BN-PAGE in the first dimension and sodium dodecyl sulfate (SDS)-PAGE in the second dimension, followed by Western blotting, we could see that the protein complex containing mitofilin was shifting slightly to the lower-molecular-mass range in the absence of Sam50, indicating that Sam50 might be a component of this complex (Fig. 6C).
Fig 6.
Sam50 and mitofilin are found together in a high-molecular-mass protein complex. (A) Mitochondria were isolated from mflkd-2, sam50kd-2, and mtx2kd-2 cells where the knockdown of the respective protein was induced with doxycycline (Dox) for 7 days. Fifty micrograms of mitochondrial protein was solubilized in 1% digitonin buffer and analyzed by BN-PAGE and Western blotting with antibodies against mitofilin and Tom40. An asterisk marks the larger mitofilin complex that is sometimes observed. (B) Fifty micrograms of mitochondria isolated from sam50kd-2 cells with (+Dox) or without (−Dox) 7 days of doxycycline induction were solubilized using increasing amounts of digitonin as indicated and analyzed by BN-PAGE and Western blotting using antibodies against mitofilin and Hsp60. The same mitochondria were also analyzed by SDS-PAGE and Western blotting using antibodies against mitofilin, Sam50, and F1α as a loading control. (C) Samples of mitochondria isolated from untreated (−Dox) or Dox-treated (+Dox) sam50kd-2 cells were solubilized in 2% digitonin buffer and separated by BN-PAGE in the first and SDS-PAGE in the second dimension. After transfer by Western blotting, the membranes were probed for mitofilin and cytochrome c as an internal control. (D) Fifty micrograms of sam50kd-2 −Dox and +Dox mitochondria described for panel A were solubilized in 1% digitonin buffer and analyzed by BN-PAGE in the first dimension and SDS-PAGE in the second dimension, followed by Western blotting using antibodies against mitofilin, Sam50, and Tom40. Individual images were overlaid to enable better comparison between complexes.
We and other groups previously located Sam50 in only one protein complex of ∼250 kDa (18, 24). To address this issue again, we raised another Sam50 antibody in rabbit against the full-length protein and affinity purified it. Using this antibody, we could detect Sam50 in two complexes after 2D BN/SDS-PAGE: one corresponding to the previously described ∼250-kDa complex, and the other of a much larger size of >700 kDa. Mitofilin could also be detected in the complex of similar size, whereas the control protein, Tom40, was found only in the TOM complex of ∼400 kDa (Fig. 6D).
CHCHD3 was reported to interact and exist in the same complex with both mitofilin and Sam50, although the described complex was only ∼340 kDa in size (6, 49). We imported radiolabeled CHCHD3 into mitochondria isolated from Sam50, mitofilin, metaxin 2, and Tom40 knockdown cells. Radiolabeled CHCHD3 was found to assemble in a time-dependent manner into a large complex of >700 kDa. The assembly was minimally affected by the reduction of Tom40 and metaxins but was strongly diminished when either Sam50 or mitofilin was missing (Fig. 7A). The large CHCHD3 complex could be depleted by both the Sam50 and mitofilin antibodies but not by the Tom20 antibody, nor by PBS buffer alone (Fig. 7B). Next, we generated a cell line with inducible knockdown of CHCHD3, which we termed CHCHD3kd-2 cells. As reported previously (6), depletion of CHCHD3 leads to a reduction in Sam50 and mitofilin levels (Fig. 7C). Antibody against CHCHD3 detected a protein complex that strongly resembled the mitofilin complex (Fig. 6A). This complex migrated slightly above the 669-kDa marker and was diminished in CHCHD3 and mitofilin knockdown mitochondria but increased in amount after Sam50 knockdown (Fig. 7C). The CHCHD3 complex observed after Western blotting was significantly smaller than the one observed after the import of the radiolabeled protein. However, both CHCHD3 and mitofilin antibodies recognize small amounts of a larger protein complex, which could represent the one obtained after the import of radiolabeled CHCHD3 (Fig. 6A and 7C, asterisk). We conclude that the OMM proteins Sam50 and the metaxins exist together with the IMM proteins mitofilin and CHCHD3 in a large complex, possibly including other subunits as reported previously (49), and we name this complex the mitochondrial intermembrane space bridging (MIB) complex.
Absence of Sam50 affects assembly of respiratory complexes.
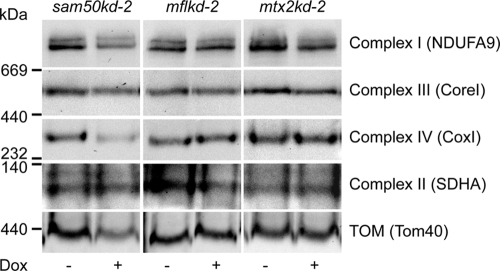
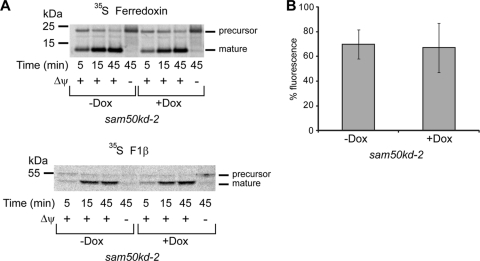
The amounts of respiratory complexes in BN-PAGE after knockdown of Sam50, mitofilin, or metaxin 2 (Fig. 8) corresponded to the levels of individual proteins in SDS-PAGE determined previously (Fig. 4 and 5). In sam50kd-2 knockdown mitochondria, in addition to the TOM complex, respiratory complexes I and IV were also reduced, and there was a mild reduction of complex III. Apart from that, we observed some reduction of complex I in mitochondria where mitofilin or metaxin 2 was depleted (Fig. 8). We then performed an in vitro import of radiolabeled components of complexes I and IV (NDUFS1 and CoxVIa) into mitochondria isolated from cell lines with inducible knockdown of Tom40, Sam50, metaxin 2, and mitofilin (Fig. 9). The assembly of imported radiolabeled subunits into mature respiratory complexes was only marginally affected by the reduction in Tom40 (Fig. 9A). This is explainable even though NDUFS1 and CoxVIa require the TOM complex to enter mitochondria: the depletion of Tom40 was not complete (Fig. 7A), and the remaining TOM complexes were most likely sufficient to efficiently import the precursors of these proteins. However, in the absence of Sam50, there was a strong defect in the assembly of newly imported respiratory chain subunits (Fig. 9B). Likewise, when mitofilin (and therefore also Sam50) was reduced, we again observed the effect on the assembly of subunits into complexes I and IV (Fig. 9C). Finally, after metaxin 2 depletion, only the assembly of complex I was slightly affected (Fig. 9D), corresponding to what we observe at the steady-state level (Fig. 8). The levels of Tom40 in Sam50-depleted mitochondria are reduced, but comparably to what we see in Tom40 knockdown cells (Fig. 4B). Considering that Tom40 depletion did not strongly influence the assembly of respiratory complexes (Fig. 9A), it is unlikely that this is the reason for the assembly defect we observe in Sam50 knockdown mitochondria. To exclude that mitochondria from cells with Sam50 knockdown are generally impaired in the ability to import proteins, we imported radiolabeled ferredoxin and F1β, a component of the F1FO ATPase whose amounts are not reduced after short-term Sam50 depletion. The import of ferredoxin was only slightly affected, whereas the F1β precursor was imported with same efficiency into control and Sam50-depleted mitochondria (Fig. 10A). The membrane potential of mitochondria after Sam50 knockdown was also only slightly reduced in comparison to that for the control (Fig. 10B). We conclude that Sam50-depleted mitochondria retain their ability to efficiently import proteins and that Sam50, mitofilin, and to some extent metaxins play specific roles in the proper assembly of respiratory complexes.
Fig 8.
Levels of Sam50 influence the amounts of respiratory complexes. Mitochondria were isolated after 7 days of knockdown induction with doxycycline (Dox) from sam50kd-2, mflkd-2, and mtx2kd-2 cells (see also Fig. 6A). Fifty micrograms of mitochondrial protein was solubilized in 1% digitonin buffer and analyzed by BN-PAGE and Western blotting. NDUFA9, subunit 9 of alpha subcomplex of complex I; CoxI, subunit 1 of complex IV. See the legends for Fig. 1 and 4 for other designations.
Fig 9.
Assembly of respiratory complexes I and IV is affected after depletion of Sam50, mitofilin, and metaxins. (A, B, C, and D) NDUFS1 (Fe-S protein 1 of NADH dehydrogenase [ubiquinone]) and CoxVIa (cytochrome c oxidase subunit VIa) were transcribed/translated in vitro in the presence of [35S]methionine/[35S]cysteine. Radiolabeled proteins were imported for indicated time periods into mitochondria isolated from tom40kd-2 cells induced for 5 days with doxycycline (Dox) (A) or sam50kd-2, mflkd-2, and mtx2kd-2 cells Dox-induced for 7 days (B, C, and D). Mitochondria were solubilized in 1% digitonin buffer and analyzed by BN-PAGE and autoradiography. CIV, respiratory complex IV; CI/CIII2/CIV0–4, supercomplexes of respiratory complexes I, III, and IV.
Fig 10.
Sam50-depleted mitochondria are not significantly impaired in protein import or membrane potential. (A) Mitochondria were isolated from sam50kd-2 cells grown in the absence (−Dox) or in the presence (+Dox) of doxycycline for 7 days. Mitochondria were incubated with precursors of radiolabeled ferredoxin and F1β for the indicated time periods. In the case of F1β, mitochondria were treated with protease K after import. Membrane potential (Δψ) was dissipated using 1 μM CCCP and 1 μM valinomycin. (B) Noninduced and sam50kd-2 cells induced with Dox for 7 days were seeded on a 96-well plate to the same density. Cells were stained with the Δψ-sensitive dye tetramethyl rhodamine methyl ester (TMRM). The graph represents the mean value ± SD of fluorescence signals from noninduced (−Dox) or induced (+Dox) samples normalized against the fluorescence intensity of the sample of −Dox cells with the strongest signal.
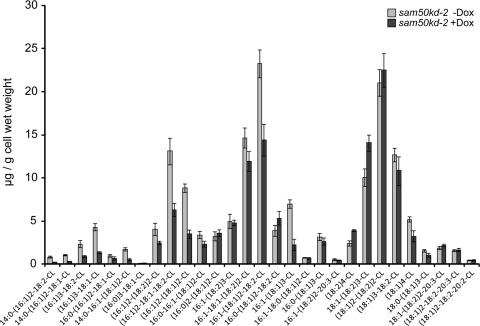
Numerous reports connect the morphology of mitochondrial cristae, stability of respiratory supercomplexes, and levels of cardiolipin in mitochondria (1, 13, 20, 22, 35). Mutations in the yeast homologue of metaxin 1, a component of the SAM complex, have been shown to diminish cardiolipin levels in yeast mitochondria (14). Therefore, we were curious whether depletion of Sam50 and the SAM complex leads to significant changes in cardiolipin levels in humans. We analyzed amounts of cardiolipin in sam50kd-2 cells after 7 days of knockdown induction with Dox. The overall abundance of cardiolipin was slightly reduced after Sam50 depletion, and the pattern of cardiolipin species was moderately changed—the amounts of cardiolipins with shorter acyl chains were reduced, whereas those cardiolipin species with prevalent longer acyl chains were enriched in knockdown samples (Fig. 11). However, none of these differences were striking enough to explain the complete loss of cristae we observe in the absence of Sam50 (Fig. 2).
Fig 11.
Cardiolipin levels and species are mildly affected after reduction of Sam50. Levels and distribution of cardiolipin species were measured in 5 independent samples of sam50kd-2 cells grown in the absence (−Dox) or presence (+Dox) of doxycycline for 7 days. The graph shows the mean values ± SD.
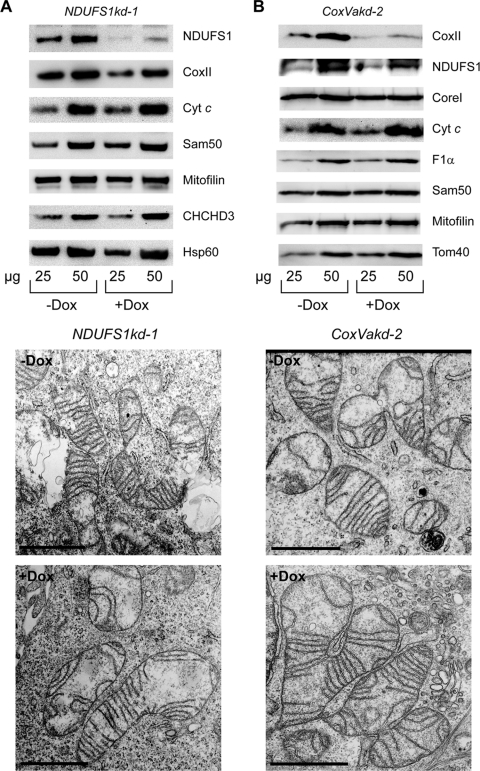
Finally, to clarify the cause and consequence regarding loss of the structure of cristae and reduction in respiratory complexes, we generated cell lines with an inducible knockdown of the complex I component NDUFS1 and complex IV component CoxVa (NDUFS1kd-1 and CoxVakd-2 cells) (Fig. 12). The knockdown of CoxVa was assessed through the reduction of CoxII, another component of the same complex (Fig. 12B). Despite the reduction in the respective respiratory proteins, the morphology of mitochondrial cristae was not severely affected when analyzed by TEM (Fig. 12A and B). We observed only some shortening of the cristae when NDUFS1 was depleted and an increase in autophagic vesicles after the knockdown of CoxVa. We conclude that the loss of cristae after Sam50 depletion seems rather to be the cause than the consequence of changes in cardiolipin species and of defects in the assembly of respiratory complexes.
Fig 12.
Depletion of respiratory proteins from complexes I and IV does not significantly affect mitochondrial crista morphology. (A and B) NDUFS1kd-1 (pool) and CoxVakd-2 (single clone) cells were grown for 7 days in the absence (−Dox) or presence (+Dox) of doxycycline. Cells were then analyzed by TEM (lower panels), and mitochondria were isolated in parallel and analyzed by SDS-PAGE and Western blotting. Scale bar, 1 μm. For designations, see the text and the legends for Fig. 1, 3, and 4.
DISCUSSION
In this report, we describe a novel role of Sam50 in maintaining the mitochondrial shape and the morphology of cristae. We provide evidence for the existence of a large complex that connects OMM and IMM and for the role this connection plays in structuring of the IMM and stability of mitochondrial respiratory complexes.
Previously, mitochondria in yeast mutants of Sam50, Sam37 (yeast metaxin homolog), and Mdm10 (a component of the yeast SAM complex not found in humans) were found to clump and failed to be transferred to daughter cells. Defects in Tom40 led to the same effects in yeast (28). In humans, however, the absence of Sam50 leads to enlarged, spherical mitochondria (Fig. 1), but cells are able to survive for a relatively long time period and the transfer of mitochondria during cell division does not seem to be significantly affected. In addition, the reduction in Tom40 amounts had no effect on mitochondrial morphology (Fig. 3). The SAM complex components therefore seem to be important for the maintenance of the mitochondrial network both in yeast and in humans. However, in human mitochondria the evidence speaks in favor of a direct role of the SAM complex in this process.
Strikingly, even a moderate Sam50 reduction leads to the complete loss of cristae (Fig. 2 and 4D). In addition, Sam50 absence leads to the depletion of respiratory complexes (Table 1). We do not believe that mild changes in cardiolipin levels and species that occur after Sam50 knockdown are responsible for this (Fig. 11). It has in fact been shown that the biogenesis of cardiolipin depends on the intact Δψ and functional respiratory complexes (13). What is then causing such drastic reorganization of cristae and defects in components of the respiratory chain?
Two recent reports indicate a connection between the SAM complex in the OMM and several proteins in the IMM, such as CHCHD3 and mitofilin (6, 49). It has been reported that depletion of CHCHD3 affects levels of Sam50 and mitofilin (6). In support of these findings, we observe that when mitofilin levels are reduced, both Sam50 and CHCHD3 amounts are strongly diminished (Fig. 5C). We are able to detect a protein complex of high molecular mass that contains mitofilin, Sam50, and CHCHD3 (Fig. 6 and 7), which we term the MIB complex. Importantly, radiolabeled CHCHD3 assembles in a complex containing Sam50 and mitofilin, which is of much larger size than the complex detected by mitofilin (Fig. 6A) or CHCHD3 (Fig. 7C) antibodies. The amount of the smaller complex we detect by Western blotting increases significantly when Sam50 is depleted (Fig. 6A and 7C) or when we use more-stringent solubilization conditions (Fig. 6B). We therefore presume that the larger complex represents the holo-MIB complex, consisting of both outer and inner membrane components. This complex corresponds in size to the previously described mitofilin complex of an estimated molecular weight slightly less than 1.2 MDa (21) and not to the ∼350-kDa large mitofilin complex described by Xie and colleagues (49). The smaller complex detected by antibodies could represent the MIB complex lacking Sam50 (and other SAM components). We propose that the MIB complex described here plays the role of a scaffolding construction that maintains the structure of IMM cristae. The work of Darshi and colleagues assigns the central role in the regulation of the architecture of cristae to CHCHD3 interacting with mitofilin and OPA1 in the IMM (6). We, on the other hand, show that the outer membrane protein Sam50 is in fact the central player in this process. This establishes a dual role for Sam50: first in the assembly of the β-barrel proteins in the OMM and second in the maintenance of the structure of cristae.
In the initial stages of Sam50 depletion, levels of both CHCHD3 and mitofilin are nearly unaffected, but mitochondrial cristae are completely lost (Fig. 4A and D). The amount of OPA1 is also unchanged at the similar stage (Fig. 2D). This excludes reduction in the mitofilin/CHCHD3/OPA1 complex as the main cause for the observed defect in cristae as proposed by Darshi et al. (6) and points to the importance of the communication of the OMM and IMM components of the MIB complex in keeping cristae properly structured. Metaxins 1 and 2, found by Xie and colleagues in the same complex together with Sam50, mitofilin, and CHCHD3 (49), appear to be peripheral OMM subunits of the MIB complex, because only a long-term depletion leads to a moderate reduction in the levels of other MIB components (Fig. 5B). Mitofilin could be important primarily for stabilizing CHCHD3 and in this way influences the amount of Sam50. This could partially explain why the observed phenotype of cristae after mitofilin depletion, which consists of multiple layers of IMM forming an onion-like structure (21; data not shown), does not entirely resemble the defect in cristae detected after the knockdown of CHCHD3 (6) or Sam50 (Fig. 2A).
Communication between two mitochondrial membranes has been connected with maintenance of cristae and mtDNA in yeast. For example, the yeast OMM proteins Mmm2, Mdm10, and Mdm12 cooperate with two IMM proteins, Mdm31 and Mdm32, in maintenance of mitochondrial morphology and mtDNA (7, 50). Defects in preservation of mtDNA eventually lead to defects in respiration, considering that mtDNA encodes a number of crucial subunits of respiratory chain complexes. First affected are probably respiratory complexes that contain the largest number of mitochondrially encoded subunits. In human mitochondria, these are complexes I and IV. This is what we observe after Sam50 reduction (Fig. 4B and 8). In addition, long-term Sam50 knockdown influences other respiratory complexes that contain mitochondrially encoded subunits (Fig. 4C), pointing to a potential role of the MIB complex in the upkeep of mtDNA. Also worth considering is the fact that all of the affected respiratory complexes undergo complicated assembly pathways (8), and this process could be impaired by changes in the morphology of cristae.
Analysis of frozen-hydrated rat liver mitochondria shows an existence of numerous 10- to 15-nm large particles in the IMS that keep the constant separation of two membranes (26, 27) and could represent the MIB complex. Such bridging particles are detected not only in mitochondria but in other organelles as well (42), indicating a possible universal mechanism in keeping membrane structures in a certain shape.
A novel protein, termed MOMA-1, was recently found to interact with the CHCHD3 and mitofilin homologues in Caenorhabditis elegans. The morphology of mitochondria and cristae was strongly affected after MOMA-1 reduction, similar to what we observe after Sam50 knockdown. MOMA-1 is related to two human proteins that show a weak homology to apolipoproteins. Homologues of MOMA-1 are also present in yeast (16). In addition, yeast also contains a mitofilin homolog, called Fcj1 (36). Its depletion produces a phenotype similar to what is observed after mitofilin knockdown in mammalian cells (21). During the review process of our article, three exciting reports appeared identifying a large protein complex in the IMM of yeast, which contains Fcj1/mitofilin and a set of newly identified IMM proteins, some of which are similar to CHCHD3 and homologous to MOMA-1 (15, 17, 44). This large complex has been termed MINOS, for mitochondrial inner membrane organizing system (44), MITOS, for mitochondrial organizing structure, or MICOS, for mitochondrial contact site (15). The work from von der Malsburg and colleagues (44) establishes a connection of the mitofilin complex with the TOM complex in the OMM and with Mia40, a part of the mitochondrial intermembrane space assembly machinery (MIA). However, the OMM connection of the MINOS complex is proposed to be important only for the transport of proteins and not for the organization of cristae (44). Hoppins et al. also establish an OMM/MITOS connection, but with the OMM proteins OM45 and porin, and propose that the MITOS complex functions in the IMM organization through its interaction with ATP synthase dimers (17). The work of Harner and colleagues is the only one to show that the components of the SAM complex are found in a common structure with the proteins from the mitofilin-containing MICOS complex (15), indicating that OMM/IMM bridging in mitochondria is likely conserved throughout evolution.
Finally, a large number of proteins involved in regulating mitochondrial morphology have been identified in a screen in C. elegans, showing that homeostasis of protein import, mitochondrial respiration, and metabolism all have an effect on mitochondrial structure (19). Investigations into the importance of and connection between various proteins regulating mitochondrial shape represent an exciting perspective for further research.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by DFG grants KO3882/1-1 to V.K.-P. and RU 631/7-1 to T.R.
Footnotes
Published ahead of print 17 January 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Acehan D, Xu Y, Stokes DL, Schlame M. 2007. Comparison of lymphoblast mitochondria from normal subjects and patients with Barth syndrome using electron microscopic tomography. Lab. Invest. 87:40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 3. Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. 2008. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 9:42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 [DOI] [PubMed] [Google Scholar]
- 5. Daems WT, Wisse E. 1966. Shape and attachment of the cristae mitochondriales in mouse hepatic cell mitochondria. J. Ultrastruct. Res. 16:123–140 [DOI] [PubMed] [Google Scholar]
- 6. Darshi M, et al. 2011. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem. 286:2918–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dimmer KS, Jakobs S, Vogel F, Altmann K, Westermann B. 2005. Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast. J. Cell Biol. 168:103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fernández-Vizarra E, Tiranti V, Zeviani M. 2009. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta 1793:200–211 [DOI] [PubMed] [Google Scholar]
- 9. Frezza C, et al. 2006. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126:177–189 [DOI] [PubMed] [Google Scholar]
- 10. Ghochani M, et al. 2010. Tensile forces and shape entropy explain observed crista structure in mitochondria. Biophys. J. 99:3244–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gieffers C, Korioth F, Heimann P, Ungermann C, Frey J. 1997. Mitofilin is a transmembrane protein of the inner mitochondrial membrane expressed as two isoforms. Exp. Cell Res. 232:395–399 [DOI] [PubMed] [Google Scholar]
- 12. Gilkerson RW, Selker JM, Capaldi RA. 2003. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 546:355–358 [DOI] [PubMed] [Google Scholar]
- 13. Gohil VM, et al. 2004. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. J. Biol. Chem. 279:42612–42618 [DOI] [PubMed] [Google Scholar]
- 14. Gratzer S, et al. 1995. Mas37p, a novel receptor subunit for protein import into mitochondria. J. Cell Biol. 129:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harner M, et al. 2011. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30:4356–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Head BP, Zulaika M, Ryazantsev S, van der Bliek AM. 2011. A novel mitochondrial outer membrane protein, MOMA-1, that affects cristae morphology in Caenorhabditis elegans. Mol. Biol. Cell 22:831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoppins S, et al. 2011. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195:323–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Humphries AD, et al. 2005. Dissection of the mitochondrial import and assembly pathway for human Tom40. J. Biol. Chem. 280:11535–11543 [DOI] [PubMed] [Google Scholar]
- 19. Ichishita R, et al. 2008. An RNAi screen for mitochondrial proteins required to maintain the morphology of the organelle in Caenorhabditis elegans. J. Biochem. 143:449–454 [DOI] [PubMed] [Google Scholar]
- 20. Jiang F, et al. 2000. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275:22387–22394 [DOI] [PubMed] [Google Scholar]
- 21. John GB, et al. 2005. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell 16:1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khalifat N, Puff N, Bonneau S, Fournier JB, Angelova MI. 2008. Membrane deformation under local pH gradient: mimicking mitochondrial cristae dynamics. Biophys. J. 95:4924–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kozjak V, et al. 2003. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 278:48520–48523 [DOI] [PubMed] [Google Scholar]
- 24. Kozjak-Pavlovic V, et al. 2007. Conserved roles of Sam50 and metaxins in VDAC biogenesis. EMBO Rep. 8:576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lazarou M, Thorburn DR, Ryan MT, McKenzie M. 2009. Assembly of mitochondrial complex I and defects in disease. Biochim. Biophys. Acta 1793:78–88 [DOI] [PubMed] [Google Scholar]
- 26. Mannella CA. 2006. The relevance of mitochondrial membrane topology to mitochondrial function. Biochim. Biophys. Acta 1762:140–147 [DOI] [PubMed] [Google Scholar]
- 27. Mannella CA, et al. 2001. Topology of the mitochondrial inner membrane: dynamics and bioenergetic implications. IUBMB Life 52:93–100 [DOI] [PubMed] [Google Scholar]
- 28. Meisinger C, et al. 2004. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell 7:61–71 [DOI] [PubMed] [Google Scholar]
- 29. Minkler PE, Hoppel CL. 2010. Separation and characterization of cardiolipin molecular species by reverse-phase ion pair high-performance liquid chromatography-mass spectrometry. J. Lipid Res. 51:856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Odgren P, Toukatly G, Bangs P, Gilmore R, Fey E. 1996. Molecular characterization of mitofilin (HMP), a mitochondria-associated protein with predicted coiled coil and intermembrane space targeting domains. J. Cell Sci. 109:2253–2264 [DOI] [PubMed] [Google Scholar]
- 31. Park Y-U, et al. 2010. Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with mitofilin. Proc. Natl. Acad. Sci. U. S. A. 107:17785–17790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paschen SA, et al. 2003. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature 426:862–866 [DOI] [PubMed] [Google Scholar]
- 33. Paumard P, et al. 2002. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21:221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perkins G, et al. 1997. Electron tomography of neuronal mitochondria: three-dimensional structure and organization of cristae and membrane contacts. J. Struct. Biol. 119:260–272 [DOI] [PubMed] [Google Scholar]
- 35. Pfeiffer K, et al. 2003. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278:52873–52880 [DOI] [PubMed] [Google Scholar]
- 36. Rabl R, et al. 2009. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 185:1047–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Renken C, et al. 2002. A thermodynamic model describing the nature of the crista junction: a structural motif in the mitochondrion. J. Struct. Biol. 138:137–144 [DOI] [PubMed] [Google Scholar]
- 38. Rich PR, Marechal A. 2010. The mitochondrial respiratory chain. Essays Biochem. 47:1–23 [DOI] [PubMed] [Google Scholar]
- 39. Ross K, Rudel T, Kozjak-Pavlovic V. 2009. TOM-independent complex formation of Bax and Bak in mammalian mitochondria during TNFα-induced apoptosis. Cell Death Differ. 16:697–707 [DOI] [PubMed] [Google Scholar]
- 40. Rossi MN, et al. 2009. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J. Biol. Chem. 284:31616–31624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schägger H, von Jagow G. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223–231 [DOI] [PubMed] [Google Scholar]
- 42. Senda T, Yoshinaga-Hirabayashi T. 1998. Intermembrane bridges within membrane organelles revealed by quick-freeze deep-etch electron microscopy. Anat. Rec. 251:339–345 [DOI] [PubMed] [Google Scholar]
- 43. Vogel F, Bornhovd C, Neupert W, Reichert AS. 2006. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 175:237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. von der Malsburg K, et al. 2011. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell 21:694–707 [DOI] [PubMed] [Google Scholar]
- 45. Wang G, et al. 2010. PNPASE regulates RNA import into mitochondria. Cell 142:456–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wiedemann N, et al. 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424:565–571 [DOI] [PubMed] [Google Scholar]
- 47. Wiznerowicz M, Trono D. 2003. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77:8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wurm CA, Jakobs S. 2006. Differential protein distributions define two sub-compartments of the mitochondrial inner membrane in yeast. FEBS Lett. 580:5628–5634 [DOI] [PubMed] [Google Scholar]
- 49. Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. 2007. The mitochondrial inner membrane protein Mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 581:3545–3549 [DOI] [PubMed] [Google Scholar]
- 50. Youngman MJ, Hobbs AE, Burgess SM, Srinivasan M, Jensen RE. 2004. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 164:677–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.