Abstract
The mammalian TOB1 and TOB2 proteins have emerged as key players in repressing cell proliferation. Accumulating evidence indicates that TOBs regulate mRNA deadenylation. A recruitment model was proposed in which TOBs promote deadenylation by recruiting CAF1-CCR4 deadenylase complex to the 3′ end of mRNAs by simultaneously binding CAF1 and PABP. However, the exact molecular mechanism underlying TOB-promoted deadenylation remains unclear. It is also unclear whether TOBs' antiproliferative and deadenylation-promoting activities are connected. Here, we combine biochemical analyses with a functional assay directly monitoring deadenylation and mRNA decay to characterize the effects of tethering TOBs or their mutant derivatives to mRNAs. The results provide direct evidence supporting the recruitment model and reveal a link between TOBs' antiproliferative and deadenylation-promoting activities. We also find that TOBs' actions in deadenylation are independent of the phosphorylation state of three serines known to regulate antiproliferative actions, suggesting that TOBs arrest cell growth through at least two different mechanisms. TOB1 and TOB2 were interchangeable in the properties tested here, indicating considerable functional redundancy between the two proteins. We propose that their multiple modes of modulating mRNA turnover and arresting cell growth permit the TOB proteins to coordinate their diverse roles in controlling cell growth and differentiation.
INTRODUCTION
The two human TOB proteins, encoded by paralogous genes (TOB1 and TOB2) (20, 25), belong to a group of antiproliferative factors, the BTG/TOB family (26, 39). In addition to roles in cell proliferation, BTG/TOB proteins have also been implicated in embryonic development, cellular differentiation, cancer suppression, and apoptosis (21, 26, 27, 39). Levels of BTG/TOB proteins fluctuate during the cell cycle and can be induced by diverse stimuli, such as growth factors, tumor promoters, and genotoxic stress. Given the varied roles attributed to BTG/TOB proteins, the varied pathways controlling their expression, and the identification of varied associated proteins, the biological functions of BTG/TOB proteins are rather complicated.
In addition to the roles of BTG/TOB proteins in regulating mRNA production (reviewed in references 21, 26, 27, and 39), the detection of direct interaction between BTG/TOB proteins and the CAF1 deadenylase (13, 20, 28, 31) suggests a role of BTG/TOB proteins in deadenylation, a critical posttranscriptional step that regulates cytoplasmic mRNA levels (8, 16). CAF1 associates with CCR4 to form a deadenylase complex that plays a predominant role in shortening the mRNA poly(A) tail in eukaryotes (2, 9, 11, 35, 36, 38, 41). TOB proteins interact with CAF1 via their N-terminal domains (18, 27). Unlike other BTG/TOB family members, TOB proteins contain an extra-long C-terminal domain with two poly(A)-binding protein (PABP)-interacting motif 2 (PAM2) motifs (13, 22, 29; see also Fig. S1 in the supplemental material). Recently, we showed that TOB proteins can interact with CAF1 and PABP simultaneously (13). Overexpression of TOB promotes general deadenylation (13). We further demonstrated that the deadenylation-enhancing effect of TOB proteins is dependent upon their ability to bind PABP (13). Based on these results, we proposed that the interaction of TOB with CAF1 and PABP promotes deadenylation by recruiting the CAF1-CCR4 deadenylase complex to the 3′ end of mRNAs with a poly(A) tail (13). Nevertheless, the exact mechanism by which TOB promotes deadenylation remains unclear.
In mammals, TOB proteins are substrates for posttranslational modification, including phosphorylation by mitogen-activated protein kinases (MAPKs) (24, 34) and ubiquitination, which targets them for degradation (32). Upon serum or growth factor stimulation, TOB1 is rapidly phosphorylated by extracellular signal-regulated kinase 1 (ERK1) and ERK2 MAP kinases at serine residues 152, 154, and 164 (34; see also Fig. S1 in the supplemental material). When these serine residues were phosphorylated or mutated to glutamate, TOB's inhibitory effect on cell growth was blunted (24, 34). Our finding that TOB proteins promote mRNA deadenylation (13) raises important questions as to whether the antiproliferative action of TOB is linked to its ability to promote deadenylation and whether the deadenylation-enhancing ability of TOB is regulated through phosphorylation when cells reenter the cell cycle. Moreover, although ectopically overexpressed TOB accelerates deadenylation (13), it remains unknown whether TOB proteins promote the degradation of the mRNA body and reduce the cognate protein levels. Furthermore, as most studies thus far have focused on TOB1, it is unclear whether TOB2 differs mechanistically from TOB1.
In this study, we addressed these critical issues regarding human TOB1 and TOB2 using transcriptional pulse-chase and RNA-tethering approaches (7). The results from these functional assays provide crucial new insights into the mechanism by which TOB proteins downregulate gene expression. They also reveal a link between the TOB proteins' antiproliferative and mRNA deadenylation/decay-promoting actions.
MATERIALS AND METHODS
Plasmid constructs.
The plasmids for TOB1(FF) (F139A/F274A) and TOB2(FF) (F140A/F260A) mutants were created as described previously (13). To construct pcDNA6-MS2-TOB1 and pcDNA6-MS2-TOB2, a 360-bp fragment spanning the MS2 open reading frame (ORF) was amplified by PCR using pMBP-MS2 (23) (provided by J. Vilardell) as a template. The MS2 fragment was subcloned in-frame immediately upstream of the TOB1 or TOB2 ORF in the pTOB1-V5 or pTOB2-V5 plasmid (5) to create the final constructs. The various point mutations of TOB1 (mutants 3SA, S152A/S154A/S164A; 3SE, S152E/S154E/S164E; TM, E62A/K63A/D65A; and 3LG, L32G/L35G/L36G) or TOB2 (mutants 3SA, S153A/S155A/S163A; 3SE, S153E/S155E/S163E; TM, E62A/M63A/D65A; and 3LG, L32G/L35G/L36G) were introduced into the pcDNA6-MS2-TOB1 or pcDNA6-MS2-TOB2 plasmid using the QuikChange site-directed mutagenesis kit (Stratagene). The pTet-BBB+12bs plasmid containing 12 MS2-binding sites (bs) between the BglII and the SphI sites in the β-globin (BBB) 3′ untranslated region (UTR) was provided by J. Lykke-Andersen (10). To generate pTet-flag-EGFP+12bs, pTet-BBB+12bs was digested with BglII and SphI to cut out the fragment containing 12 MS2-binding sites. This fragment was treated with T4 DNA polymerase (New England BioLabs) to make it blunt ended, and it was then inserted into the EcoRV site in the 3′ UTR of the pTet-flag-GFP plasmid to create the final construct.
Cell culture and transfection.
NIH 3T3 cells or U2OS cells were transfected using Lipofectamine 2000 (Invitrogen) or FuGENE 6 (Roche), respectively, as described in the manufacturer's protocol. Briefly, NIH 3T3 B2A2 cells (40) maintained in the presence of 100 ng/ml tetracycline were split to a density of 3 × 106 per 10-cm dish 24 h before transfection. Each 10-cm dish of cells was transfected with 20 μg of total plasmid DNA, including carrier plasmid, reporter plasmid (1.3 μg pTet-BBB-12bs), internal control plasmid (1.3 μg pTet-BBB), and one of the TOB expression plasmids (13 μg of either wild-type or mutant derivatives of pcDNA6-MS2-TOB1-V5 or pcDNA6-MS2-TOB1-V5). In small interfering RNA (siRNA) knockdown experiments, 12 μg of nonspecific siRNA or 6 μg of CAF1a siRNA and 6 μg of CAF1b siRNA (Dharmacon) were mixed with 6.5 μg of either wild-type or mutant TOB expression plasmids, 1.3 μg of reporter plasmid pTet-BBB-12bs, and 1.3 μg of control plasmid pTet-BBB. The mixtures of DNA and siRNA were diluted into 1.5 ml of Opti-MEM, added to the Lipofectamine 2000 mixture, and incubated at room temperature for 25 min. The final mixtures were added to the cells and the dishes incubated at 37°C (5% CO2) for 18 h before the cells were split among 6-cm dishes (1.5 × 106 per 6-cm dish) and incubated at 37°C (8% CO2) for an additional 24 h. Procedures for time course experiments using the Tet-Off system for transcriptional pulsing were described previously (6). For cell growth assays, U2OS cells transfected with TOB plasmids or their mutant derivatives were enriched in the presence of blasticidin (15 μg/ml final concentration in culture medium) for 2 days. Equal cell numbers were then seeded in 10-cm plates and incubated for 5 days before the cells were harvested and counted.
Preparation and analysis of RNA samples.
At various time points after a transcription pulse driven by the Tet-Off promoter of the reporter plasmid in transfected cells (6), nuclei were spun out and cytoplasmic mRNA was isolated using the cytoplasmic lysis buffer RLN (50 mM Tris-HCl, pH 8, 140 mM NaCl, 1.5 mM MgCl2, and 0.5% NP-40) followed by RNeasy reagents (Sigma). For Northern blot analysis, RNA samples (8 to 10 μg for each time point) were separated using 1.4% formaldehyde agarose gel electrophoresis, transferred to GeneScreen hybridization transfer membrane (PerkinElmer), and probed with a 32P-labeled gene-specific DNA probe using ULTRAhyb hybridization buffer (Ambion). Gene-specific DNA probes were prepared with [α-32P]dCTP (>6,000 Ci/mmol; PerkinElmer) and the Rediprime II random prime labeling system (GE Healthcare). RNase H treatment of cytoplasmic mRNA after annealing to oligo(dT) (Invitrogen) was used to generate RNA lacking the poly(A) tail [poly(A)− RNA] as described previously (33). All time course experiments were performed at least two times to ensure the reproducibility of the results. To measure mRNA decay rates, data from autoradiograms were scanned and quantitated by using a Syngene bioimaging system. mRNA half-lives were determined by least-square analysis of semilogarithmic plots of normalized mRNA concentration as a function of time. Half-lives were estimated from the slope of each decay curve. Real-time quantitative reverse transcription (RT)-PCR analysis of RNA samples was performed as described previously (43).
Western blot analysis and immunoprecipitation.
Cytoplasmic lysates (20 to 30 μg protein) were resolved on a 12% SDS–polyacrylamide gel, transferred to polyvinylidene difluoride (PVDF) membranes, and probed with the relevant primary antibody, as follows: rabbit anti-CAF1-B peptide antibody (1/1,000) (41); rabbit anti-PABP-N antibody (1/4,000; R. Lloyd, Baylor College of Medicine); mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibody (1/10,000; Research Diagnostics); mouse anti-α-tubulin monoclonal antibody (1/10,000; Sigma); anti-eGFP (enhanced green fluorescent protein) antibody (1/2,000; Abcam); or rabbit anti-Flag (1/500; Sigma). Immunoreactive bands were visualized with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibodies (1/4,000; Bethyl) or goat anti-mouse antibodies (1/5,000; Bio-Rad) and peroxide-luminol chemiluminescence reagent (Pierce). For coimmunoprecipitation (Co-IP), a 10-cm dish of cells expressing wild-type TOB proteins or their mutant derivatives was lysed at 48 h after transfection by incubating for 10 min at 4°C in 600 μl of lysis buffer (20 mM Tris-HCl, pH 7.4, containing 150 mM NaCl, 1% NP-40, 1 mM Na orthovanadate, 1 mM Na pyrophosphate, and 1 mM NaF supplemented with protease inhibitor cocktail [Roche]). Fifty microliters of each cell lysate was preserved as input, and the remainder was incubated with a monoclonal anti-V5-agarose conjugate (Sigma-Aldrich) in the presence of 0.1 μg/ml RNase A at 4°C for 4 h. The agarose beads were then washed five times with lysis buffer, and the immunoprecipitated proteins were separated by electrophoresis and probed by Western blotting using the appropriate antibodies.
RESULTS
Tethering TOBs to mRNAs promotes deadenylation and decay of the transcripts.
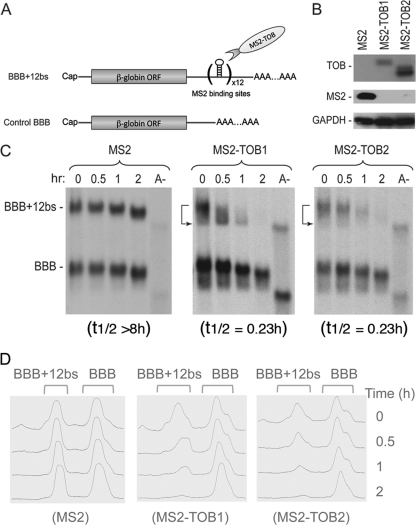
To directly monitor the effects of TOB proteins on mRNA deadenylation and decay and to identify the functionally relevant features of TOBs, we performed RNA-tethering assays (10), an approach more specific and sensitive than simply overexpressing TOB proteins. TOB proteins fused with the bacteriophage MS2 coat protein (MS2-TOB) were coexpressed in mouse NIH 3T3 cells with a stable β-globin mRNA reporter (BBB+12bs mRNA) carrying 12 specific hairpins that bind MS2 with high affinity and specificity (Fig. 1A). The expression of the reporter mRNAs was under the control of the Tet-Off-driven promoter. The kinetics of reporter deadenylation and decay were measured using Tet promoter-driven transcriptional pulse-chase and Northern blot analysis (6). This combination allows us to monitor nearly synchronous degradation of mRNA molecules that are similar in age, thus unequivocally dissecting deadenylation and decay kinetics, as well as revealing precursor-product relationships.
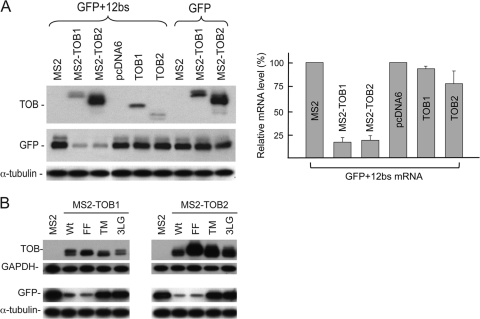
Fig 1.
Tethering TOB proteins to mRNAs promotes deadenylation and decay of the transcripts. NIH 3T3 B2A2 cells were cotransfected with Tet promoter-regulated plasmids encoding the BBB+12bs and the BBB mRNAs, along with plasmid coding for either MS2 or one of the MS2-TOB proteins as indicated. (A) Diagram showing structures of BBB and BBB+12bs mRNAs. TOB proteins with C terminus fused to RNA-binding domain derived from bacteriophage MS2 coat protein bind to the MS2-specific elements (bs [binding site]) inserted in the BBB+12bs mRNA. (B) Western blot analysis of ectopically expressed MS2, MS2-TOB1, and MS2-TOB2 proteins, with GAPDH as loading control. (C) Northern blot analyses showing that tethering MS2-TOBs (middle and right) but not MS2 alone (left) to the otherwise stable BBB+12bs mRNA triggers highly processive deadenylation and rapid decay. BBB mRNA lacking the MS2-binding sites served as control. Samples of mRNA were taken at the indicated times after tetracycline addition to terminate a short pulse of mRNA synthesis. Poly(A)− RNA (A−) was prepared in vitro by treating an RNA sample from an early time point with oligo(dT) and RNase H. mRNA half-life (t1/2) determination is described in Materials and Methods. (D) Deadenylation profiles illustrating how poly(A) shortening kinetics change during BBB+12bs mRNA turnover upon tethering of TOBs.
The results show that when coexpressed with MS2 alone, the BBB+12bs reporter mRNA remained stable (with a half-life over 8 h) and underwent slow deadenylation, similar to control β-globin (BBB) mRNA lacking MS2-binding sites (Fig. 1B and C, left). In contrast, when coexpressed with MS2-TOB1 or MS2-TOB2 proteins (Fig. 1C, middle and right), the BBB+12bs reporter mRNA underwent rapid decay (with a half-life of about 14 min) in addition to the highly accelerated deadenylation (Fig. 1D); at 2 h, the reporter mRNA was barely detectable. The enhancing effects of ectopically expressed MS2-TOB proteins on the BBB+12bs reporter (Fig. 1C, middle and right, top bands) are more dramatic than the effects on a BBB reporter lacking the MS2-binding sites (Fig. 1C, middle and right, bottom bands), indicating that the RNA-tethering strategy gives a larger effect than simply coexpressing TOB proteins and mRNAs. When tethered to the target mRNA, both of the TOB proteins not only promote deadenylation but also accelerate subsequent decay of the mRNA body (Fig. 1C, middle and right, top bands). The results also suggest that there are two distinct aspects to the effects of TOB proteins on mRNA expression. While TOBs are able to promote nonspecific size reduction of poly(A) tails in the general mRNA population (13), they can also specifically accelerate both deadenylation and decay (Fig. 1C and D) of particular transcripts through interactions with sequence-specific RNA-binding proteins that “tether” them to the transcripts (see Discussion).
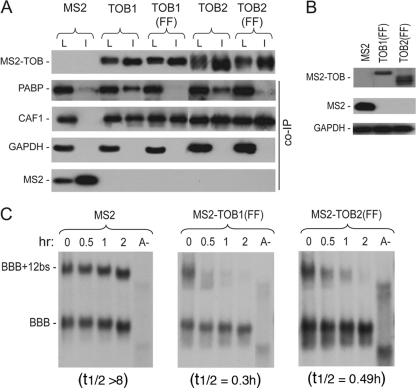
The ability of TOBs to enhance deadenylation and decay of their targets becomes independent of PABP binding when TOBs are tethered to the mRNAs.
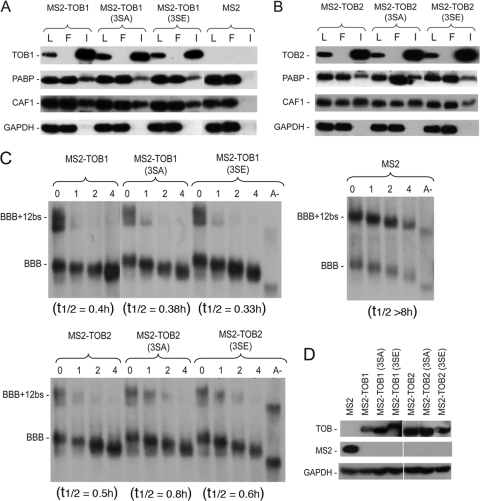
Previously, we showed that abolishing interaction between TOB and PABP by mutating the two PAM2 motifs in TOB (see Fig. S1 in the supplemental material) diminished TOB's ability to promote deadenylation (13). We hypothesized that interaction between TOB and PABP brings the TOB-CAF1 complex near the 3′-end poly(A) tail–PABP complex, increasing the accessibility of CAF1 deadenylase to the 3′-end poly(A) tail and promoting rapid deadenylation. If this model is correct, tethering TOB proteins with the PAM2 mutations [TOB(FF) mutants] to the BBB+12bs reporter mRNA should elicit rapid deadenylation and decay of the reporter because TOB(FF) proteins can still bind CAF1 (13). To test this hypothesis, we fused MS2 coat protein to the TOB1(FF) and TOB2(FF) mutants. Co-IP–Western blot analysis (Fig. 2A) confirmed that both MS2-TOB(FF) mutants bound to CAF1 but not to PABP. In time course experiments, ectopically expressed MS2-TOB(FF) mutants promoted rapid deadenylation and decay of BBB+12bs mRNA (Fig. 2B and C), as did the wild-type TOB proteins fused with MS2 (Fig. 1A, middle and right). Collectively, our data demonstrate that when TOB is tethered to a target mRNA, its ability to enhance deadenylation and decay of the target becomes independent of PABP binding. These results support the model that TOB proteins promote rapid mRNA deadenylation and decay by recruiting CAF1 to the 3′ poly(A) tail-PABP complex.
Fig 2.
The ability of TOBs to enhance deadenylation and decay of their targets is independent of PABP binding when TOBs are directly tethered to the mRNAs. (A) Co-IP–Western blot analysis showing interactions of MS2-TOB1 and MS2-TOB2 and their FF mutant derivatives with PABP and CAF1 deadenylase. Cells were transfected with plasmids encoding V5-tagged MS2-TOBs, MS2-TOB(FF) mutants, or MS2 alone. RNase A-treated total lysates were incubated with anti-V5 antibody conjugated to agarose beads, and the immunoprecipitates (I) were analyzed by SDS-PAGE and Western blotting. An aliquot (8%) of each lysate (L) was analyzed for comparison. GAPDH served as the loading and nonspecific control. (B) Western blot analysis of MS2 and MS2-TOB(FF) mutant protein levels in cells ectopically expressing BBB+12bs and BBB mRNAs. GAPDH served as loading control. (C) Northern blot analyses showing that tethering of MS2-TOB(FF) mutants (middle and right) but not of MS2 alone (left) to the otherwise stable BBB+12bs mRNA triggers highly processive deadenylation and rapid decay of the transcript. Cell transfection and poly(A)− mRNA (A−) preparation were as described in the Fig. 1 legend. mRNA samples were taken at the indicated times after tetracycline addition.
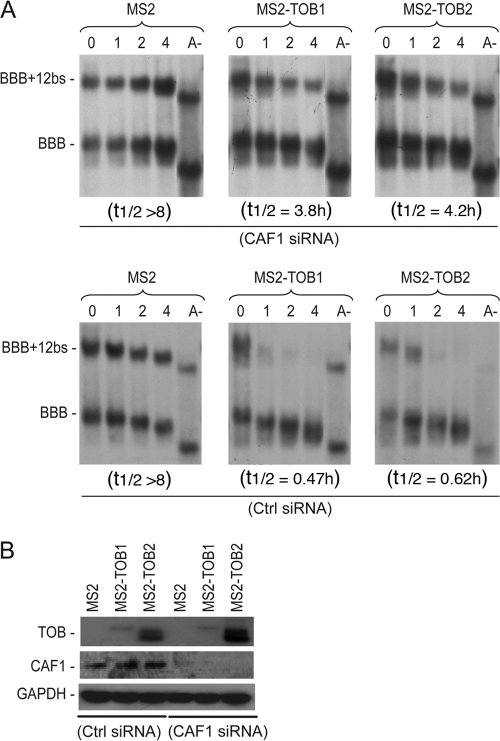
CAF1 binding is necessary for TOBs to promote rapid mRNA deadenylation and decay.
To further test the recruitment model, we combined siRNA-mediated gene knockdown and RNA tethering to see whether knocking down CAF1 would affect the ability of TOB to promote mRNA deadenylation and decay. The results (Fig. 3A, top) show that when endogenous CAF1 was knocked down, tethering TOB1 or TOB2 protein did not promote rapid mRNA deadenylation and decay. In contrast, the deadenylation and decay promoted by TOB was not affected in controls treated with nonspecific siRNA (Fig. 3A, bottom [Ctrl siRNA]). Western blot analysis confirmed that CAF1 expression was knocked down by specific but not control siRNA (Fig. 3B). These results indicate that the ability of TOB to enhance mRNA deadenylation and decay requires CAF1 poly(A) nuclease.
Fig 3.
CAF1 is required for TOB-promoted deadenylation and decay. (A) Northern blot analyses of the effects of Caf1 knockdown (top) or control knockdown (bottom) on deadenylation and decay of BBB+12bs mRNA promoted by tethering MS2-TOBs to the transcript. NIH 3T3 B2A2 cells were cotransfected with Tet promoter-regulated plasmids encoding the BBB+12bs and the BBB control mRNAs and either the MS2 plasmid or one of the MS2-TOB plasmids as indicated, as well as either the control siRNA (Ctrl siRNA, bottom) or CAF1 siRNA (top). Samples were taken at the indicated times after tetracycline addition. The preparation of poly(A)− RNA (A−) was as described in the Fig. 1 legend. (B) Western blot analysis showing expression levels of MS2, MS2-TOB1, and MS2-TOB2 proteins (top), CAF1 protein levels (middle), and GAPDH loading control (bottom) in cells transfected with either the control siRNA (Ctrl siRNA, left) or CAF1 siRNA (right).
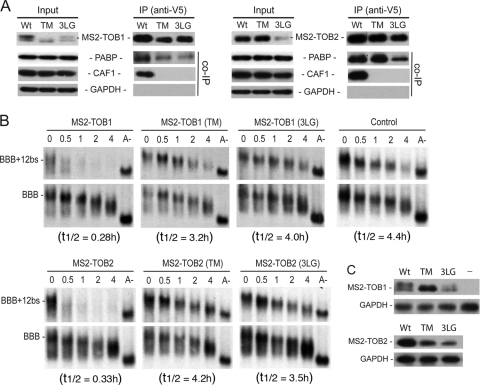
To determine if interaction between TOB and CAF1 is required for TOB to promote mRNA deadenylation and decay, we created TOB mutants that do not interact with CAF1 and tested whether tethering these mutants affected the deadenylation and decay of target mRNAs. According to the crystallographic structure of a complex between the highly conserved N-terminal domain of TOB1 (BTG domain) and CAF1 (18), we predicted that residues 61, 62, and 64 of TOB1 are critical for the interaction of TOB with CAF1. We changed residues 61, 62, and 64 of human TOB1 and TOB2 to alanine to create MS2-TOB1(TM) and MS2-TOB2(TM) (see Fig. S1 in the supplemental material). These residues are in a loop region, so the alanine substitutions are unlikely to have a negative impact on the N-terminal domain integrity (see Fig. S2 in the supplemental material). Co-IP and Western blot analysis (Fig. 4A) showed that the triple-alanine mutations (TM) in TOB1 and TOB2 indeed abolished interactions of the TOB proteins with CAF1 but not with PABP, whereas the wild-type (Wt) TOB1 and TOB2 proteins pulled down both CAF1 and PABP. When the MS2-TOB1(TM) or MS2-TOB2(TM) mutant was coexpressed with the BBB+12bs reporter mRNA, neither promoted rapid deadenylation and decay (Fig. 4B and C). This contrasts with the behavior of the wild-type MS2-TOB proteins (Fig. 4B and C). These results indicate that TOB must interact with CAF1 in order to accelerate mRNA deadenylation and decay.
Fig 4.
Interaction with CAF1 is necessary for TOBs to elicit rapid deadenylation and decay. (A) Co-IP–Western blot analysis of interactions between CAF1 deadenylase and MS2-TOB1, MS2-TOB2, and their mutant derivatives. Cells were transfected with plasmids encoding V5-tagged MS2, MS2-TOB1, MS2-TOB2, or their TM or 3LG mutants. RNase A-treated total lysates were incubated with anti-V5 antibody conjugated to agarose beads, and the immunoprecipitates (I) were analyzed by SDS-PAGE–Western blotting. An aliquot (8%) of each lysate (L) was analyzed for comparison, and GAPDH served as loading and nonspecific control. (B) Northern blot analyses of the effects of tethering MS2-TOBs, MS2-TOB mutants, or MS2 alone (control) on deadenylation and decay of BBB+12bs mRNA. Cell transfection and poly(A)− mRNA (A−) preparation were as described in the Fig. 1 legend. Samples were taken at the indicated times after tetracycline addition. (C) Western blot analysis of MS2-TOB(TM) or (3LG) mutant proteins in cells ectopically expressing BBB+12bs mRNA and BBB mRNA. GAPDH served as loading control. Wt, wild type.
To further corroborate the results described above, we changed three highly conserved leucine residues close to the N terminus of TOB1 and TOB2 (positions 32, 35, and 36) to glycine to create mutants MS2-TOB1(3LG) and MS2-TOB2(3LG) (see Fig. S1 in the supplemental material). These mutations were predicted to disrupt an α-helix that is critical for the interaction of TOB with CAF1 (18). Co-IP and Western blot analysis (Fig. 4A) showed that these mutations, like the TM mutations described above, abolished interaction of the TOB proteins with CAF1 but not with PABP. The results from the corresponding time course experiments (Fig. 4B and C) showed that neither MS2-TOB1(3LG) nor MS2-TOB2(3LG) promoted rapid mRNA deadenylation or decay. Taken together, our results show that CAF1 binding is necessary for TOB1 and TOB2 to promote rapid mRNA deadenylation and decay.
Interaction with CAF1 is required for TOBs to repress protein production of target mRNAs.
To determine the effect of tethering TOBs on the protein production from target mRNAs, we introduced 12 MS2 binding sites into the 3′ UTR of a green fluorescent protein (GFP) mRNA to create the GFP+12bs reporter mRNA. The results from Western blot analysis (Fig. 5A, MS2-TOB1 and MS2-TOB2) showed that tethering TOB proteins to the GFP mRNA greatly reduced the protein levels derived from the reporter. In contrast, no appreciable decrease in the cognate protein was observed when GFP mRNA lacking MS2-binding sites was coexpressed with either MS2-TOB1 or MS2-TOB2 or when GFP+12bs mRNA was coexpressed with pcDNA6 vector alone, MS2 alone, or TOB proteins lacking the MS2 domain. Real-time RT-PCR quantification of GFP+12bs mRNA levels (Fig. 5A, right) further showed that the changes in the levels of GFP+12bs mRNA directly parallel the changes in the corresponding protein levels. Taken together, these data demonstrate that tethering TOBs represses protein production from the target mRNAs through promoting decay of the transcripts.
Fig 5.
Effects of tethering TOB proteins to mRNAs on protein production from target transcripts. (A) Top left, levels of ectopically expressed TOBs, MS2, MS2-TOB1, and MS2-TOB2; middle left, levels of GFP protein; bottom left, levels of α-tubulin (loading control); right, bar graph showing changes in the relative levels of GFP+12bs mRNA upon tethering the corresponding TOB proteins and their mutant derivatives. mRNA levels were quantified by real-time RT-PCR and are presented as the normalized percentage ± standard deviation (n = 3). (B) Top, levels of ectopically expressed MS2-TOBs detected by anti-V5 antibody, with GAPDH as loading control; bottom, levels of GFP detected by anti-Flag antibody, with α-tubulin as loading control.
We then tested whether interaction of TOB protein with CAF1 is required for TOBs to repress protein production from the GFP+12bs mRNA. The results show that tethering TOB(TM) or TOB(3LG) mutant proteins, which do not bind CAF1, had little effect on the protein levels derived from GFP+12bs mRNA (Fig. 5B, TM and 3LG). This contrasts with the effect of tethering the wild-type TOB or TOB(FF) mutant proteins (Fig. 5B, Wt and FF). Collectively, these results indicate that interaction with CAF1 is required for TOB1 and TOB2 to downregulate gene expression, further supporting the recruitment model.
The ability of TOBs to promote mRNA deadenylation and decay is independent of the phosphorylation status of three regulatory serines for antiproliferative action.
Phosphorylation of serines 152, 154, and 164 of TOB1 (see Fig. S1 in the supplemental material) appears to play a role in regulating its antiproliferative function (24, 34). Inhibition of cell growth was decreased when these serines were replaced with glutamic acid residues to mimic constitutive phosphorylation. Furthermore, serum stimulation of quiescent NIH 3T3 cells is accompanied by phosphorylation of TOB at these serine residues and a decreased ability of TOB to exert growth arrest. To test whether the phosphorylation status of these three conserved serine residues in TOB1 and TOB2 proteins affects their ability to promote mRNA deadenylation and decay, we carried out RNA-tethering assays with two TOB phosphomimetics, MS2-TOB1(3SE) and MS2-TOB2(3SE), created by substituting glutamate at S152, S154, and S164 of TOB1 and S153, S155, and S163 of TOB2 (see Fig. S1 in the supplemental material). We also created nonphosphorylatable mutants, MS2-TOB1(3SA) and MS2-TOB2(3SA), by converting the target serines to alanine (see Fig. S1). The results from Co-IP and Western blot analysis (Fig. 6A and B) showed that all four mutant MS2-TOB proteins pulled down both CAF1 and PABP, and time course experiments showed that all four mutants were each able to elicit rapid deadenylation and decay of the BBB+12bs reporter mRNA, both in proliferating cells (Fig. 6C and D) and in serum-stimulated quiescent cells (see Fig. S3 in the supplemental material). These data indicate that the phosphorylation status of these three conserved serine residues does not affect the ability of TOB proteins to promote mRNA deadenylation and decay.
Fig 6.
The ability of TOBs to promote deadenylation and decay is not affected by the phosphorylation status of three conserved serines. (A and B) Co-IP–Western blot analysis of interactions of MS2-TOB1 and MS2-TOB2 and their mutant derivatives with PABP and CAF1 deadenylase. Cells were transfected with plasmids encoding V5-tagged MS2, MS2-TOB1, MS-TOB2, the 3SA mutants, or the 3SE mutants. RNase A-treated total lysates were incubated with anti-V5 antibody conjugated to agarose beads, and the immunoprecipitates (I) were analyzed by SDS-PAGE and Western blotting; the flowthrough (F) fractions were also examined. An aliquot (8%) of each lysate (L) was analyzed. GAPDH served as loading and nonspecific control. (C) Northern blot analyses of deadenylation and decay of reporter transcript (BBB+12bs mRNA) in cells expressing MS2, MS2-TOB1, MS2-TOB2, or the 3SA and 3SE mutants of the MS2-TOBs. Cell transfection and poly(A)− mRNA (A−) preparation were as described in the Fig. 1 legend. Samples were taken at the indicated times after tetracycline addition. (D) Western blot analysis of protein levels for MS2-TOBs and their mutant derivatives, with GAPDH served as loading control.
The antiproliferative and the mRNA deadenylation/decay-promoting effects of TOB proteins are linked.
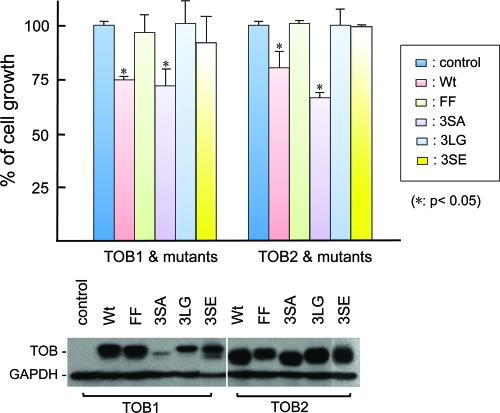
Earlier studies found that knocking down CAF1 or ectopically expressing a CAF1 mutant unable to bind TOB1 compromised the ability of TOB1 to arrest cell growth or to reduce foci formation in cancer cells (18), suggesting that interaction between TOB1 and CAF1 is involved in the antiproliferative effect of TOB1. However, as efficient cell proliferation requires CAF1 (4), it is unclear whether the observed loss of cell growth inhibition was simply due to a loss of proper CAF1 function or whether binding to CAF1 is necessary for TOB1 to achieve its antiproliferative effect. To address this issue, we performed cell growth assays using the U2OS cell line, widely used for growth arrest and cell cycle studies, to examine the cell growth arrest effects of TOB1 and TOB2 mutant proteins unable to bind PABP or CAF1. TOB1 and its 3SA and 3SE mutants were used as controls (34).
The results (Fig. 7) show that TOB2 and TOB2(3SA) significantly slowed cell growth, as did the positive controls, TOB1 and TOB1(3SA). On the other hand, TOB2(3SE) lost its ability to inhibit cell growth, as did the negative control, TOB1(3SE). Moreover, the TOB1(FF) and TOB2(FF) mutants, which lack PABP-binding ability, and the TOB1(3LG) and TOB2 (3LG) mutants, which lack CAF1-binding ability, did not inhibit cell growth. These results indicate that for both TOB1 and TOB2 proteins, there is a link between the ability to promote mRNA deadenylation and decay and the ability to suppress proliferation. It is worth noting that TOB(3SA) mutants exhibited a stronger growth-inhibitory effect than wild-type TOB. Although TOB1(3SA) is expressed at much lower levels than all others, it still exhibited a strong growth-inhibitory effect, comparable to that of wild-type TOB1. Given that phosphorylation of three conserved serines of TOB1 reduced its ability to exert growth arrest (34) but did not affect its ability to promote deadenylation and decay of mRNAs (Fig. 6; see also Fig. S3 in the supplemental material), our observations suggest that the TOB proteins exert their antiproliferative effects through more than one pathway.
Fig 7.
Growth arrest actions of TOB1, TOB2, and their FF, 3SA, 3SE, and 3LG mutants. Top, cell growth in U2OS cells transfected with the indicated wild-type (Wt) or mutant TOB plasmids. Data represent the average ± standard deviation (n = 3) for cell numbers at the end of the test period, normalized to the results for the controls. The asterisks denote a significant difference analyzed by a paired t test. Bottom, Western blot analysis of protein levels for ectopically expressed TOB1, TOB2, or their mutant derivatives from one of the cell growth experiments whose results are shown in the top panel. GAPDH served as loading control.
DISCUSSION
Recruitment model for TOB-promoted deadenylation.
Observations that TOB can interact simultaneously with CAF1 and PABP and that the PABP binding capacity of TOB1 and TOB2 proteins was required for accelerated mRNA deadenylation (13) prompted a recruitment model for TOB-enhanced deadenylation. The model posits that TOB proteins form a complex with CCR4-CAF1 deadenylases, with the CCR4-CAF1-TOB complex then recruited to the mRNA poly(A) tail through binding of TOB to PABP, thereby leading to accelerated deadenylation. In the present study, the combination of an RNA-tethering strategy with functional assays that directly monitor deadenylation kinetics and precursor-product relationships during mRNA decay has provided several new lines of evidence supporting the recruitment model.
A key element of the recruitment model is that PABP serves as the docking protein at the mRNA poly(A) tail for the TOB-CAF1-CCR4 complex; this predicts that the dependence on PABP binding of TOB-enhanced deadenylation/decay can be bypassed if TOB is directly tethered to the target mRNA. This prediction is confirmed by the data in Fig. 2 showing that TOB mutants unable to bind PABP promote rapid deadenylation and decay only if tethered to the target mRNA. The recruitment model also predicts that TOB proteins will lose the ability to promote deadenylation if their interaction with CAF1 deadenylase is removed. This prediction is confirmed by the results from tethering experiments (Fig. 4) with TOB mutants that cannot bind CAF1 but remain able to bind PABP. Moreover, when CAF1 was knocked down, tethering wild-type TOB proteins no longer promoted rapid deadenylation and decay of target mRNA (Fig. 3). Collectively, these separate lines of kinetic data provide critical mechanistic evidence to validate the recruitment model for the actions of TOB proteins.
A previous study reported that ectopic expression of BTG2, another member of the BTG/TOB family, also resulted in rapid deadenylation of mRNA (28). In contrast with the TOB proteins, which bind to both CAF1 and PABP, BTG2 can bind to CAF1 but not to PABP (27). This suggests that BTG2 has a different mechanism for accelerating deadenylation than that used by TOB1 and TOB2.
Linkage between deadenylation-promoting and antiproliferative actions of TOB proteins.
The antiproliferative effects of TOB1 have been suggested to involve the CAF1-CCR4 deadenylase complex (18), but the supporting evidence was open to other interpretations. This ambiguity has now been removed by results from our experiments with TOB1 and TOB2 mutants that are selectively unable to bind either PABP or CAF1 (Fig. 1, 2, and 4 to 7). The new data provide direct evidence establishing a link between the ability of TOB proteins to promote mRNA deadenylation/decay and their ability to arrest cell growth.
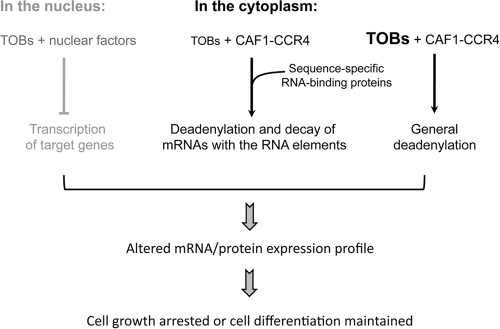
Given the linkage between the deadenylation-promoting and antiproliferative actions of the TOB proteins, the finding (Fig. 6) that the ability of TOB1 and TOB2 to promote mRNA deadenylation and decay is independent of the phosphorylation status of three regulatory serines critical for antiproliferative action (34) suggests that the TOB proteins exert their growth inhibition effect through at least two distinct mechanisms; one is modulated by the phosphorylation status of the three conserved serine residues and the other is not. However, it remains possible that the TOB(3SE) mutants may not be true phosphomimetics and they may be impaired in a step in deadenylation/decay that is rendered nonlimiting by directly tethering them to the reporter mRNA. In any case, while TOB was reported to exert cell growth arrest through altering transcription of target genes (reviewed in reference 21), the linkage between the deadenylation-promoting and antiproliferative actions of TOB1 and TOB2 (Fig. 7) also suggests that TOB proteins can exert their antiproliferative function by modulating mRNA turnover (Fig. 8).
Fig 8.
Proposed modes of TOB-mediated effects on mRNA decay and growth arrest.
Proximity to mRNA intensifies actions of TOBs.
The present data show that tethering TOBs to the reporter mRNA not only promotes rapid deadenylation but also enhances subsequent mRNA decay (Fig. 1) and significantly reduces protein production (Fig. 5) from the target mRNA. They further demonstrate that TOBs exert their gene silencing effect through promoting mRNA destabilization. On the other hand, when TOB proteins are ectopically expressed without engineered targeting to a reporter mRNA, the effect is a modest increase in deadenylation but unaltered decay of the RNA body (Fig. 1C, control BBB mRNA) (13). These observations suggest that there are two distinct aspects to the effects of TOB proteins on mRNA expression in mammalian cells (Fig. 8). On one hand, TOB expression can result in a nonspecific size reduction of poly(A) tails in the general mRNA population, which may lead to an overall change in mRNA translation efficiency and protein expression. On the other hand, interactions with sequence-specific RNA-binding proteins may localize or “tether” TOB proteins to a subset of transcripts and specifically intensify the rapid deadenylation and decay of these particular transcripts, thus attenuating production of the cognate proteins. Supporting the notion of an endogenous system for TOB tethering, a recent study (19) showed that TOB1 interacts with a sequence-specific RNA-binding protein, cytoplasmic polyadenylation element-binding protein 3 (CPEB3), recruiting the TOB1-CAF1-CCR4 complex to an mRNA bearing the CPE, resulting in low expression of that transcript. It is possible that a transient increase in TOB protein levels could shut off gene expression by slowing translation of mRNAs through shortening their poly(A) without degrading the transcripts during G1 to G0 transition. On the other hand, some TOB proteins could be tethered to target mRNAs through interacting partners that can bind mRNA in a sequence-specific manner and degrade the transcripts without affecting the general mRNA population. The latter mode of TOB's action may be operating in a cell type- or tissue-specific manner, such as is seen in naïve T cells (37).
Functional redundancy of TOB1 and TOB2.
The TOB1 and TOB2 genes are paralogs of one another. TOB1 and TOB2 single-knockout mice are viable and do not exhibit any pathologically significant phenotypes (1, 42). This suggests that either of the TOB proteins can provide the critical functionality needed for viability. Our current and previous studies have made several observations indicating functional similarity for the TOB1 and TOB2 proteins. We found that both proteins (i) bind simultaneously with PABP and CAF1 (13); (ii) promote mRNA deadenylation and decay via a PABP-dependent mechanism (13) that is probably independent of the phosphorylation status of three regulatory serines for antiproliferative action (Fig. 6); and (iii) suppress cell growth via a pathway connected with their deadenylation-enhancing function (Fig. 7). Despite these functional similarities, the maintenance of TOB1 and TOB2 as separate proteins suggests that each protein has some distinct function that remains to be discovered. In this regard, a recent report showed that miR-378 participates in oncogenic transformation by inhibition of TOB2 and not of TOB1, leading to transcriptional activation of cyclin D1 (14). In contrast, a study reported that TOB1 is constitutively expressed in liver and its downregulation is required for normal liver regeneration (17). Furthermore, TOB2 and not TOB1 was found to colocalize with mRNA-processing bodies (13) that contain nontranslating mRNAs, as well as proteins involved in translational inhibition and mRNA degradation (3, 12, 15, 30, 44). It will be interesting to identify the different features of the TOB proteins that are responsible for these specific functions and to decipher the mechanisms controlling and/or coordinating the multiple actions of these two functionally important but redundant proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Kulmacz for critical reading of the manuscript and for plotting deadenylation profiles, J. Lykke-Andersen (University of California, San Diego) and J. Vilardell (Molecular Biology Institute, Barcelona, Spain) for providing plasmids, R. Lloyd (Baylor College of Medicine) for rabbit anti-PABP-N antibody, K. Rogers and Z. Xia for technical assistance, and A. Chadee for helping analyze the X-ray crystal structure of the CAF1-BTG domain complex.
This work was supported by grants from the National Institutes of Health (RO1 GM046454 to A.-B. Shyu) and from the Houston Endowment, Inc. (to A.-B. Shyu).
Footnotes
Published ahead of print 17 January 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Ajima R, et al. 2008. Osteoporotic bone formation in mice lacking tob2; involvement of Tob2 in RANK ligand expression and osteoclasts differentiation. FEBS Lett. 582:1313–1318 [DOI] [PubMed] [Google Scholar]
- 2. Albert TK, et al. 2000. Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res. 28:809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson P, Kedersha N. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10:430–436 [DOI] [PubMed] [Google Scholar]
- 4. Aslam A, Mittal S, Koch F, Andrau J-C, Winkler GS. 2009. The Ccr4-NOT deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol. Biol. Cell 20:3840–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen AC-Y, Ezzeddine N, Shyu AB. 2008. Messenger RNA half-life measurements in mammalian cells. Methods Enzymol. 448:335–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C-YA, et al. 2007. Versatile applications of transcriptional pulsing to study mRNA turnover in mammalian cells. RNA 13:1775–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C-YA, Zheng D, Xia Z, Shyu A-B. 2009. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 16:1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen CYA, Shyu AB. 2011. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2:167–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen J, Chiang Y-C, Denis CL. 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21:1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clement SL, Lykke-Andersen J. 2008. A tethering approach to study proteins that activate mRNA turnover in human cells. Methods Mol. Biol. 419:121–133 [DOI] [PubMed] [Google Scholar]
- 11. Dupressoir A, et al. 2001. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eulalio A, Behm-Ansmant I, Izaurralde E. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8:9–22 [DOI] [PubMed] [Google Scholar]
- 13. Ezzeddine N, et al. 2007. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol. Cell. Biol. 27:7791–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng M, et al. 2011. Myc/miR-378/TOB2/cyclin D1 functional module regulates oncogenic transformation. Oncogene 30:2242–2251 [DOI] [PubMed] [Google Scholar]
- 15. Franks TM, Lykke-Andersen J. 2008. The control of mRNA decapping and P-body formation. Mol. Cell 32:605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goldstrohm AC, Wickens M. 2008. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 9:337–344 [DOI] [PubMed] [Google Scholar]
- 17. Ho KJ, et al. 2010. Tob1 is a constitutively expressed repressor of liver regeneration. J. Exp. Med. 207:1197–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Horiuchi M, et al. 2009. Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J. Biol. Chem. 284:13244–13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hosoda N, et al. 2011. Anti-proliferative protein Tob negatively regulates CPEB3 target by recruiting Caf1 deadenylase. EMBO J. 30:1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ikematsu N, et al. 1999. Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene 18:7432–7441 [DOI] [PubMed] [Google Scholar]
- 21. Jia S, Meng A. 2007. Tob genes in development and homeostasis. Dev. Dyn. 236:913–921 [DOI] [PubMed] [Google Scholar]
- 22. Lim NS, et al. 2006. Comparative peptide binding studies of the PABC domains from the ubiquitin-protein isopeptide ligase HYD and poly(A)-binding protein: implications for HYD function. J. Biol. Chem. 281:14376–14382 [DOI] [PubMed] [Google Scholar]
- 23. Macias S, Bragulat M, Tardiff DF, Vilardell J. 2008. L30 binds the nascent RPL30 transcript to repress U2 snRNP recruitment. Mol. Cell 30:732–742 [DOI] [PubMed] [Google Scholar]
- 24. Maekawa M, Nishida E, Tanoue T. 2002. Identification of the anti-proliferative protein Tob as a MAPK substrate. J. Biol. Chem. 277:37783–37787 [DOI] [PubMed] [Google Scholar]
- 25. Matsuda S, et al. 1996. Tob, a novel protein that interacts with p185erbB2, is associated with anti-proliferative activity. Oncogene 12:705–713 [PubMed] [Google Scholar]
- 26. Matsuda S, Rouault J-P, Magaud J-P, Berthet C. 2001. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 497:67–72 [DOI] [PubMed] [Google Scholar]
- 27. Mauxion F, Chen C-YA, Sèraphin B, Shyu A- B. 2009. BTG/TOB factors impact deadenylases. Trends Biochem. Sci. 34:640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mauxion F, Faux C, Seraphin B. 2008. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 27:1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okochi K, Suzuki T, Inoue J, Matsuda S, Yamamoto T. 2005. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes Cells 10:151–163 [DOI] [PubMed] [Google Scholar]
- 30. Parker R, Sheth U. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25:635–646 [DOI] [PubMed] [Google Scholar]
- 31. Rouault J-P, et al. 1998. Interaction of BTG1 and p53-regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J. Biol. Chem. 273:22563–22569 [DOI] [PubMed] [Google Scholar]
- 32. Sasajima H, Nakagawa K, Yokosawa H. 2002. Antiproliferative proteins of the BTG/Tob family are degraded by the ubiquitin-proteasome system. Eur. J. Biochem. 269:3596–3604 [DOI] [PubMed] [Google Scholar]
- 33. Shyu A-B, Belasco JG, Greenberg MG. 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5:221–232 [DOI] [PubMed] [Google Scholar]
- 34. Suzuki T, JK-Tsuzuku Ajima R, Nakamura T, Yoshida Y, Yamamoto T. 2002. Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation. Genes Dev. 16:1356–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. 2004. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 23:2862–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tucker M, et al. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377–386 [DOI] [PubMed] [Google Scholar]
- 37. Tzachanis D, et al. 2001. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat. Immunol. 2:1174–1182 [DOI] [PubMed] [Google Scholar]
- 38. Viswanathan P, Ohn T, Chiang Y-C, Chen J, Denis CL. 2004. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 279:23988–23995 [DOI] [PubMed] [Google Scholar]
- 39. Winkler GS. 2010. The mammalian anti-proliferative BTG/Tob protein family. J. Cell. Physiol. 222:66–72 [DOI] [PubMed] [Google Scholar]
- 40. Xu N, Loflin P, Chen C-YA, Shyu A-B. 1998. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 26:558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yamashita A, et al. 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12:1054–1063 [DOI] [PubMed] [Google Scholar]
- 42. Yoshida Y, et al. 2003. Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Genes Dev. 17:1201–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhai Y, et al. 2008. Coordinated changes in mRNA turnover, translation, and RNA processing bodies in bronchial epithelial cells following inflammatory stimulation. Mol. Cell. Biol. 28:7414–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng D, Chen C-YA, Shyu A-B. 2011. Unraveling regulation and new components of human P-bodies through a protein interaction framework and experimental validation. RNA 17:1619–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.